The Responses of the Ovary and Eyestalk in Exopalaemon carinicauda under Low Salinity Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Sample Collection
2.3. Biochemical Assays
2.4. Ovary Histology
2.5. RNA Isolation, Library Construction and Illumine Sequencing
2.6. Basic Analysis of Sequencing Data
2.7. Differential Expression Genes (DEGs) Analysis and Enrichment Analysis
2.8. RNA-Seq Data Validation by Real-Time Quantitative PCR and Statistical Analysis
3. Results
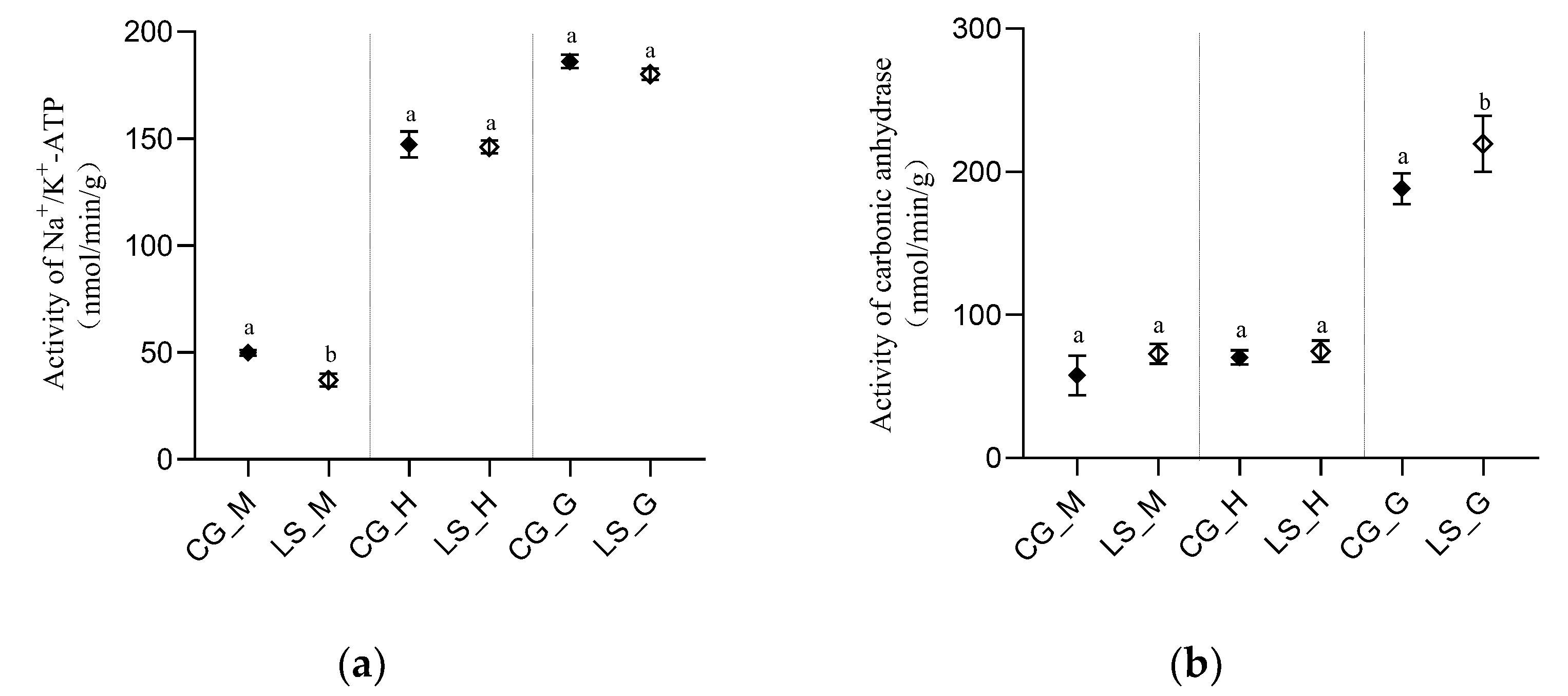
3.1. Variation of CARBONIC anhydrase and Na+/K+-ATPase Activity in the Tissue
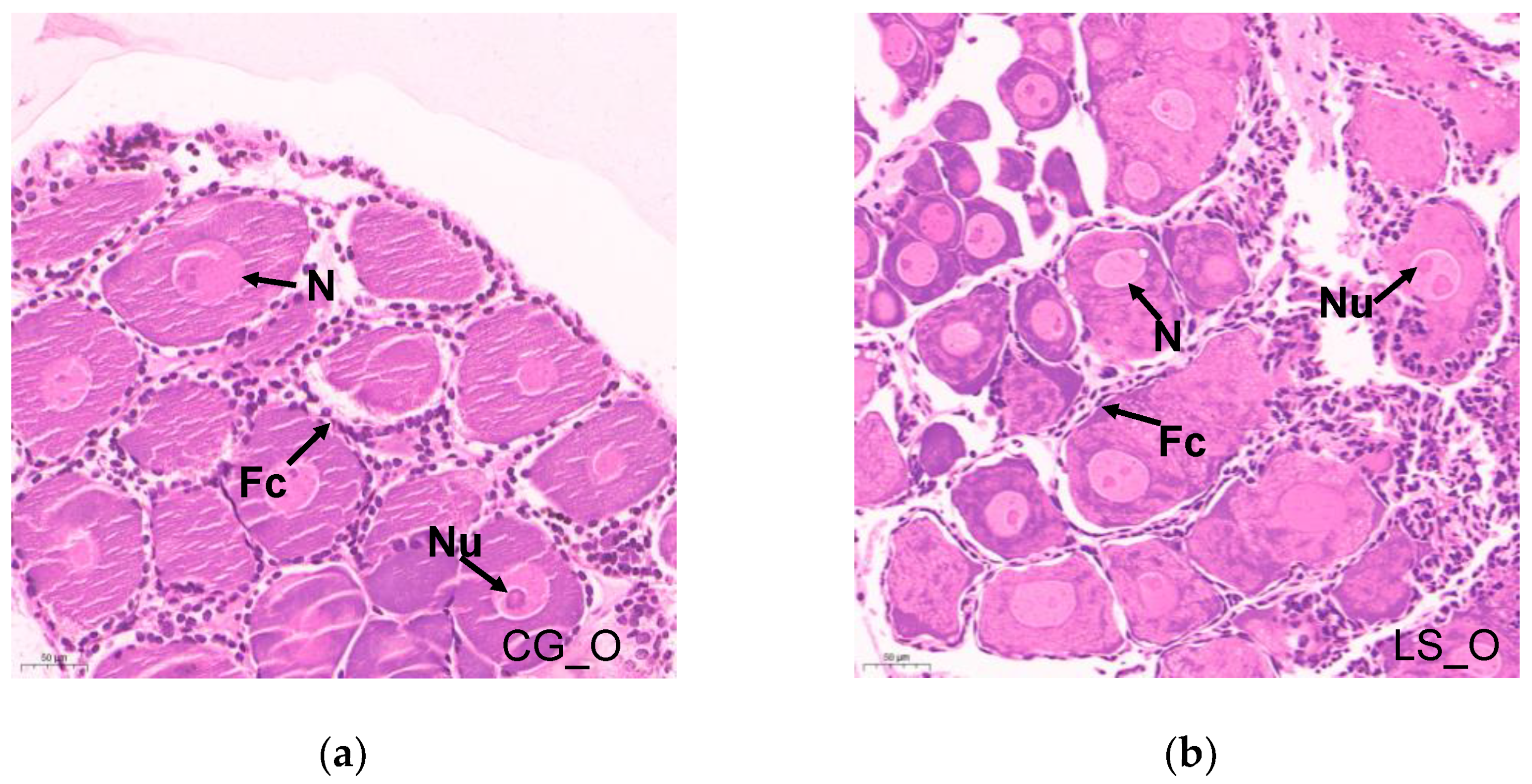
3.2. Histopathology of Ovary
3.3. Summary of the RNA-seq Data
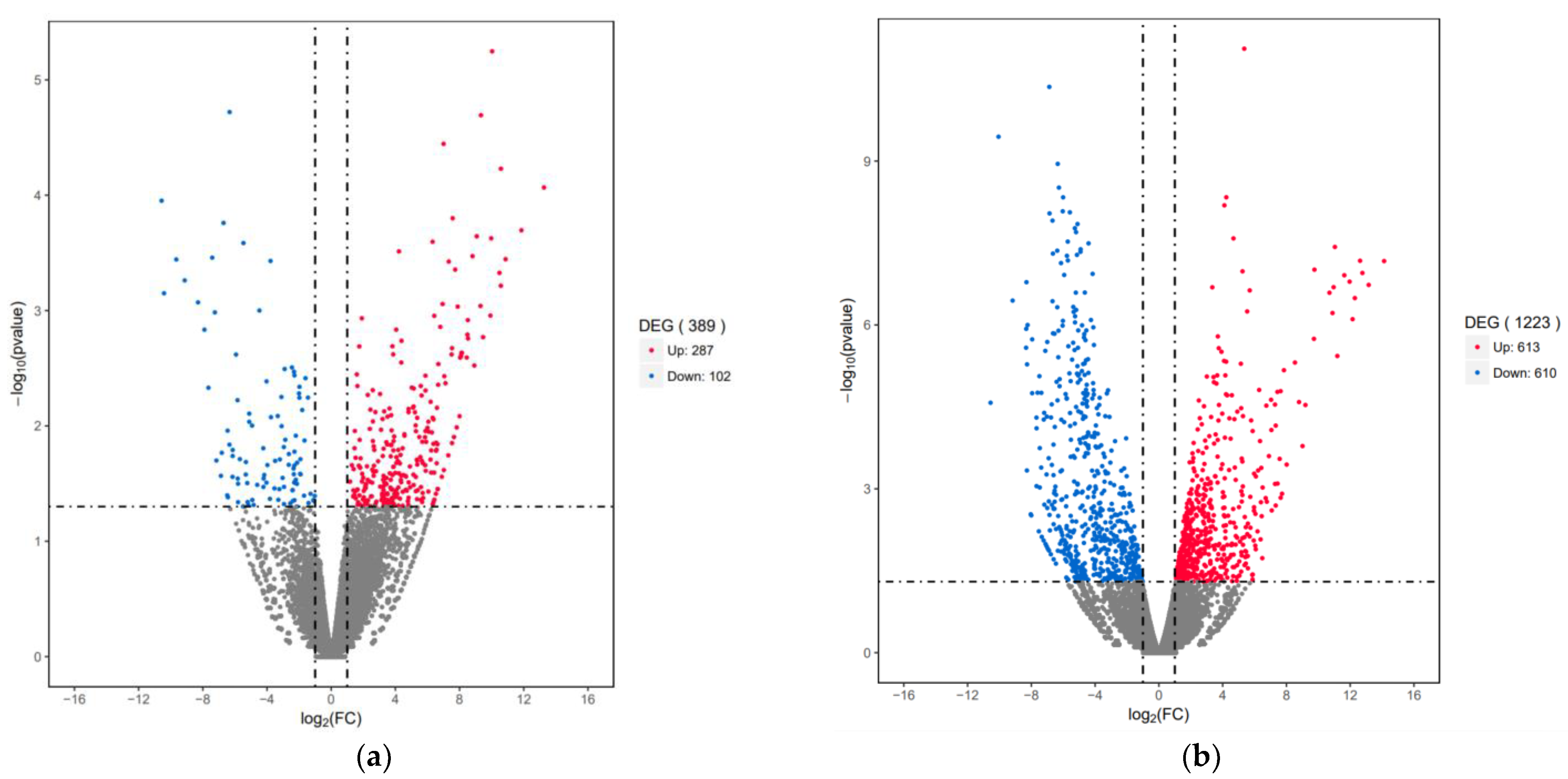
3.4. Differential Expression Genes (DEGs) Analysis
3.5. Gene Ontology (GO) Analysis of Significant DEGs
3.6. Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis
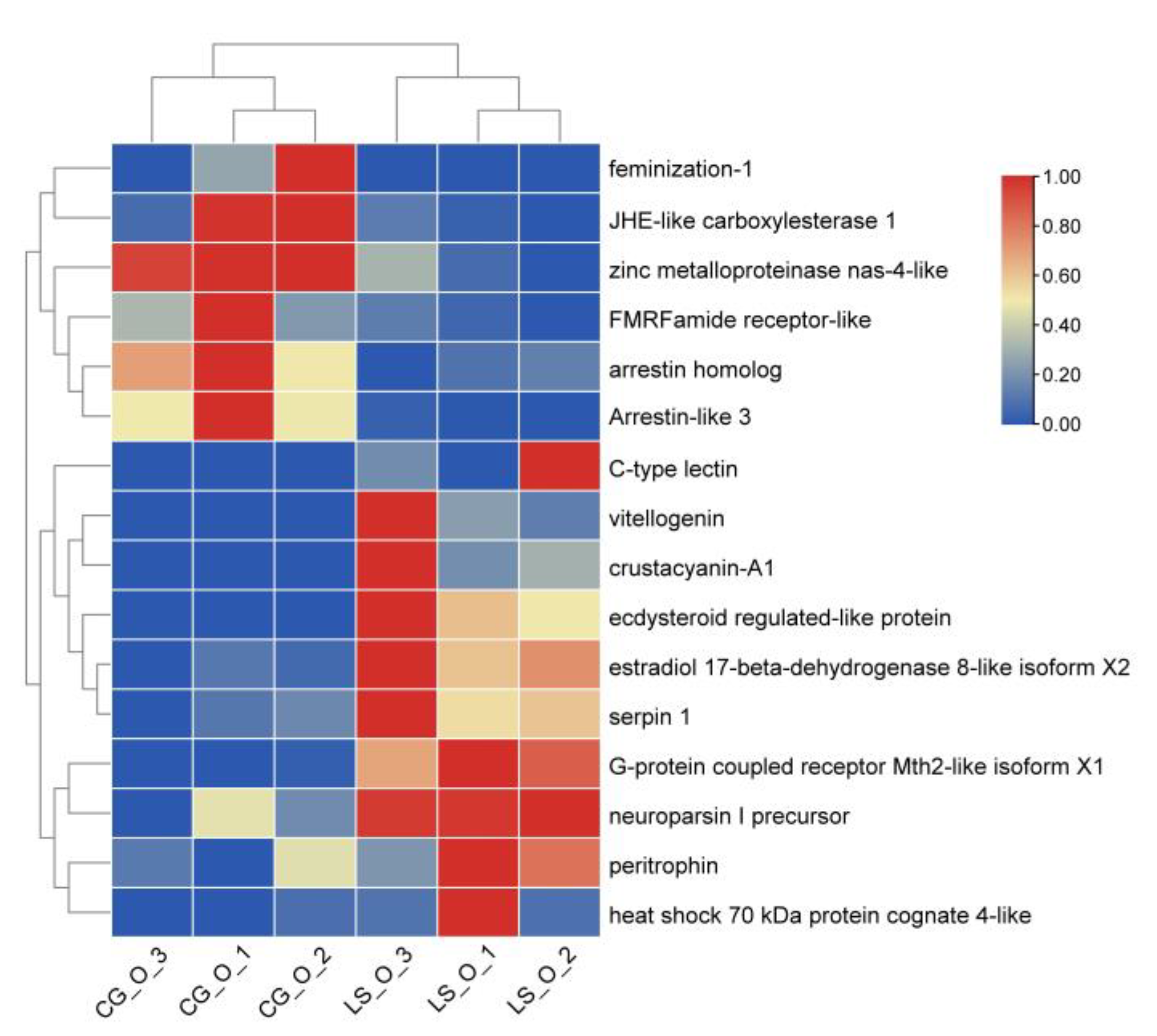
3.7. DEGs Involved in Ovarian Development
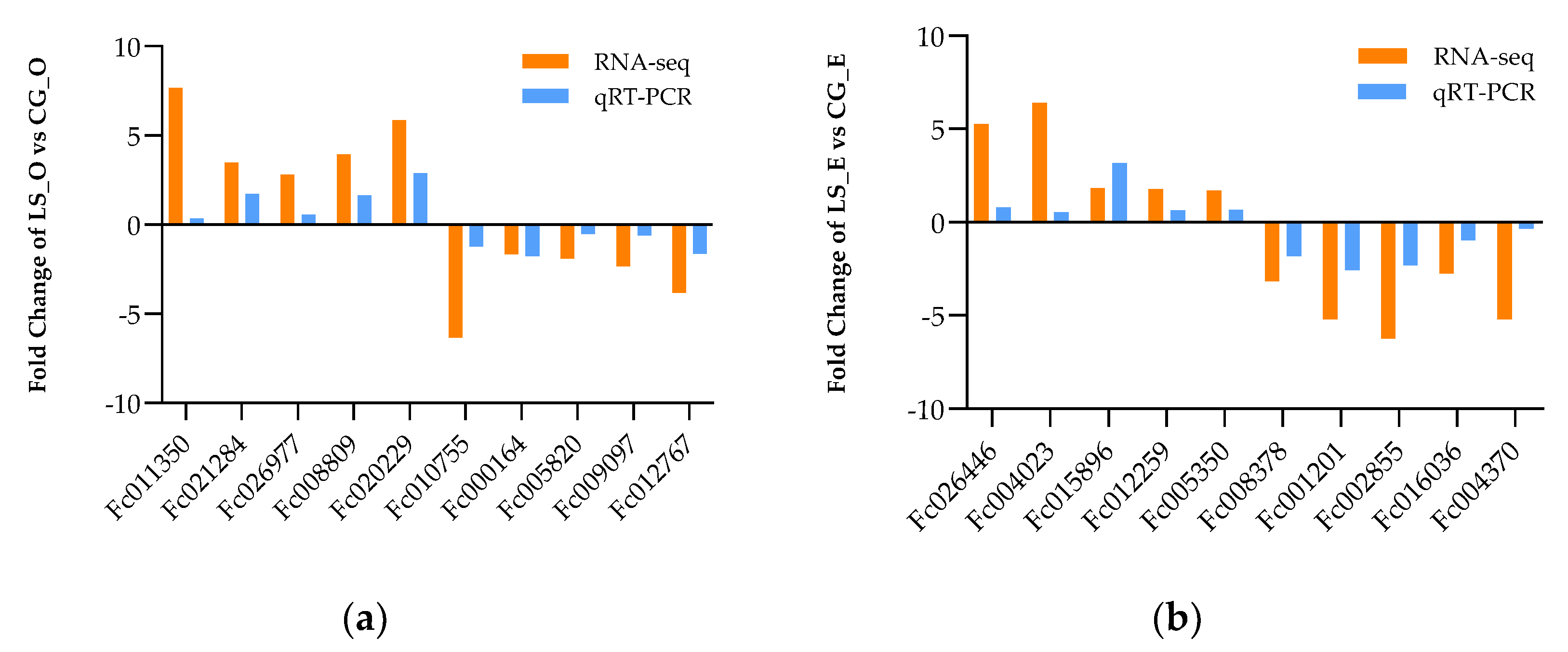
3.8. The Validation of Differently Expressional Genes by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- McFarland, K.; Donaghy, L.; Volety, A. Effect of acute salinity changes on hemolymph osmolality and clearance rate of the non-native mussel, Perna viridis, and the native oyster, Crassostrea virginica, in Southwest Florida. Aquat. Invasions 2013, 8, 299–310. [Google Scholar]
- Aguilar, C.; Raina, J.B.; Fôret, S.; Hayward, D.C.; Lapeyre, B.; Bourne, D.G.; Miller, D.J. Transcriptomic analysis reveals protein homeostasis breakdown in the coral Acropora millepora during hypo-saline stress. BMC Genom. 2019, 20, 148. [Google Scholar]
- Yang, W.K.; Chung, C.H.; Cheng, H.C.; Tang, C.H.; Lee, T.H. Different expression patterns of renal Na+/K+-ATPase α-isoform-like proteins between tilapia and milkfish following salinity challenges. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 2016, 202, 23–30. [Google Scholar]
- Wang, H.; Tang, L.; Wei, H.L.; Lu, J.K.; Mu, C.K.; Wang, C.L. Transcriptomic analysis of adaptive mechanisms in response to sudden salinity drop in the mud crab, Scylla paramamosain. BMC Genom. 2018, 19, 421. [Google Scholar]
- Huang, C.; Wu, Q.; Jiang, C.Y.; Xing, L.S.; Shi, G.L.; Zhang, B.; Qian, W.Q.; Li, Y.Z.; Xi, Y.; Yang, N.W.; et al. Identification and developmental expression of putative gene encoding juvenile hormone esterase (CpJHE-like) in codling moth, Cydia pomonella (L.). J. Integr. Agric. 2019, 18, 1624–1633. [Google Scholar]
- Mohanty, B.; Gupta, K.; Babu, G.; Pal, H.; Verma, P. Exposure to salinity stress cause ovarian disruption in a stenohaline freshwater teleost, Heteropneustes fossilis (Bloch, 1794). Aquac. Res. 2020, 51, 1964–1972. [Google Scholar]
- Mandal, B.; Sawant, P.B.; Dasgupta, S.; Chadha, N.K.; Sundaray, J.K.; Sawant, B.T.; Bera, A. Deviation of habitat salinity during seasonal gonad recrudescence affects plasma sex steroid levels and suppresses gonadal maturation in an euryhaline fish Etroplus suratensis. Aquac. Res. 2017, 48, 5973–5983. [Google Scholar]
- Kaeodee, M.; Pongsomboon, S.; Tassanakajon, A. Expression analysis and response of Penaeus monodon 14-3-3 genes to salinity stress. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2011, 159, 244–251. [Google Scholar]
- Romano, N.; Wu, X.G.; Zeng, C.S.; Genodepa, J.; Elliman, J. Growth, osmoregulatory responses and changes to the lipid and fatty acid composition of organs from the mud crab, Scylla serrata, over a broad salinity range. Mar. Biol. Res. 2014, 10, 460–471. [Google Scholar]
- Lu, S.K.; Li, R.H.; Zheng, W.B.; Chen, L.; Ren, Z.M.; Mu, C.K.; Song, W.W.; Wang, C.L. Long-term low-salinity stress affects growth, survival and osmotic related gamma-aminobutyric acid regulation in the swimming crab Portunus trituberculatus. Aquac. Res. 2019, 50, 2888–2895. [Google Scholar]
- Hu, D.X.; Pan, L.Q.; Zhao, Q.; Ren, Q. Transcriptomic response to low salinity stress in gills of the Pacific white shrimp, Litopenaeus vannamei. Mar. Genom. 2015, 24, 297–304. [Google Scholar]
- Long, X.W.; Wu, X.G.; Lei, Z.; Ye, H.H.; Cheng, Y.X.; Zeng, C.S. Physiological responses and ovarian development of female Chinese mitten crab Eriocheir sinensis subjected to different salinity conditions. Front. Physiol. 2017, 8, 1072. [Google Scholar]
- Feng, Y.Y.; Zhai, Q.Q.; Wang, J.J.; Li, J.T.; Li, J. Comparison of florfenicol pharmacokinetics in Exopalaemon carinicauda at different temperatures and administration routes. J. Vet. Pharmacol. Ther. 2019, 42, 230–238. [Google Scholar]
- Ge, Q.Q.; Li, J.; Wang, J.J.; Li, Z.D.; Li, J.T. Characterization, functional analysis, and expression levels of three carbonic anhydrases in response to pH and saline-alkaline stresses in the ridgetail white prawn Exopalaemon carinicauda. Cell Stress Chaperon. 2019, 24, 503–515. [Google Scholar]
- Qin, Z.; Ge, Q.Q.; Wang, J.J.; Li, M.D.; Liu, P.; Li, J.; Li, J.T. Comparative transcriptomic and proteomic analysis of Exopalaemon carinicauda in response to alkalinity stress. Front. Mar. Sci. 2021, 8, 1523. [Google Scholar]
- Xu, W.J.; Xie, J.J.; Shi, H.; Li, C.W. Hematodinium infections in cultured ridgetail white prawns, Exopalaemon carinicauda, in eastern China. Aquaculture 2010, 300, 25–31. [Google Scholar]
- Li, J.T.; Ma, P.; Liu, P.; Chen, P.; Li, J. The roles of Na+/K+-ATPase alpha-subunit gene from the ridgetail white prawn Exopalaemon carinicauda in response to salinity stresses. Fish Shellfish Immunol. 2015, 42, 264–271. [Google Scholar]
- Zhang, C.S.; Su, F.; Li, S.H.; Yu, Y.; Xiang, J.H.; Liu, J.G.; Li, F.H. Isolation and identification of the main carotenoid pigment from a new variety of the ridgetail white prawn Exopalaemon carinicauda. Food Chem. 2018, 269, 450–454. [Google Scholar]
- Ren, H.; Li, J.; Li, J.T.; Ying, Y.; Ge, H.X.; Li, D.L.; Yu, T.J. Cloning of catalase and expression patterns of catalase and selenium-dependent glutathione peroxidase from Exopalaemon carinicauda in response to low salinity stress. Acta Oceanol. Sin. 2015, 34, 52–61. [Google Scholar]
- Chang, Z.Q.; Neori, A.; He, Y.Y.; Li, J.T.; Qiao, L.; Preston, S.I.; Liu, P.; Li, J. Development and current state of seawater shrimp farming, with an emphasis on integrated multi-trophic pond aquaculture farms, in China-a review. Rev. Aquac. 2020, 12, 2544–2558. [Google Scholar]
- Liang, J.P.; Li, J.; Li, J.T.; Liu, P.; Zhao, F.Z.; Nie, G.X.; Li, X.J.; Kong, X.H. Effects of salinity on spawning and larval development of Exopalaemon carinicauda. Chin. J. Appl. Ecol. 2014, 25, 2105–2113. (In Chinese) [Google Scholar]
- Teets, N.M.; Peyton, J.T.; Colinet, H.; Renault, D.; Denlinger, D.L. Gene expression changes governing extreme dehydration tolerance in an Antarctic insect. Proc. Natl. Acad. Sci. USA 2013, 109, 20744–20749. [Google Scholar]
- Chen, K.; Li, E.C.; Li, T.Y.; Xu, C.; Wang, X.D.; Lin, H.Z.; Qin, J.G.; Chen, L.Q. Transcriptome and molecular pathway analysis of the hepatopancreas in the pacific white shrimp Litopenaeus vannamei under chronic low-salinity stress. PLoS ONE 2015, 10, e131505. [Google Scholar]
- Li, E.; Wang, S.; Li, C.; Wang, X.; Chen, K.; Chen, L. Transcriptome sequencing revealed the genes and pathways involved in salinity stress of Chinese mitten crab, Eriocheir sinensis. Physiol. Genom. 2014, 46, 177–190. [Google Scholar]
- Brady, P.; Elizur, A.; Williams, R.; Cummins, S.F.; Knibb, W. Gene expression profiling of the cephalothorax and eyestalk in Penaeus Monodon during ovarian maturation. Int. J. Biol. Sci. 2012, 8, 328–343. [Google Scholar]
- Qiao, H.; Xiong, Y.W.; Zhang, W.Y.; Fu, H.T.; Jiang, S.F.; Sun, S.M.; Bai, H.K.; Jin, S.B.; Gong, Y.S. Characterization, expression, and function analysis of gonad-inhibiting hormone in oriental river prawn, Macrobrachium nipponense and its induced expression by temperature. Comp. Biochem. Physiol. A-Mol. Integr. Physiol. 2015, 185, 1–8. [Google Scholar]
- Koshio, S.; Teshima, S.I.; Kanazawa, A. Effects of Unilateral Eyestalk Ablation and Feeding Frequencies on Growth, Survival, and Body Compositions of Juvenile Freshwater Prawn Macrobrachium rosenbergii. Nippon Suisan Gakkaishi 1992, 58, 1419–1425. [Google Scholar]
- Uawisetwathana, U.; Leelatanawit, R.; Klanchui, A.; Prommoon, J.; Klinbunga, S.; Karoonuthaisiri, N. Insights into eyestalk ablation mechanism to induce ovarian maturation in the black tiger shrimp. PLoS ONE 2011, 6, e24427. [Google Scholar]
- Robinson, M.D.; Mccarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Biogeosciences 2010, 26, 139–140. [Google Scholar]
- Bu, D.H.; Luo, H.T.; Huo, P.P.; Wang, Z.H.; Zhang, S.; He, Z.H.; Wu, Y.; Zhao, L.H.; Liu, J.J.; Guo, J.C.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, 317–325. [Google Scholar]
- Duan, Y.F.; Liu, P.; Li, J.T.; Li, J.; Gao, B.Q.; Chen, P. cDNA cloning, characterization and expression analysis of peroxiredoxin 5 gene in the ridgetail white prawn Exopalaemon carinicauda. Mol. Biol. Rep. 2013, 40, 6569–6577. [Google Scholar]
- Geers, C.; Gros, G. Carbon dioxide transport and carbonic anhydrase in blood and muscle. Physiol. Rev. 2000, 80, 681–715. [Google Scholar]
- Lucu, C.; Towle, D.W. Na++K+-ATPase in gills of aquatic crustacea. Comp. Biochem. Physiol. A-Mol. Integr. Physiol. 2003, 135, 195–214. [Google Scholar]
- Henry, R.P.; Gehnrich, S.; Weihrauch, D.; Towle, D.W. Salinity-mediated carbonic anhydrase induction in the gills of the euryhaline green crab, Carcinus maenas. Comp. Biochem. Physiol. A-Mol. Integr. Physiol. 2003, 136, 243–258. [Google Scholar]
- Ma, Q.; Kuang, J.; Liu, X.; Li, A.; Feng, W.; Zhuang, Z. Effects of osmotic stress on Na+/K+-ATPase, caspase 3/7 activity, and the expression profiling of sirt1, hsf1, and hsp70 in the roughskin sculpin (Trachidermus fasciatus). Fish Physiol. Biochem. 2020, 46, 135–144. [Google Scholar]
- Li, Z.G.; Zhang, C.S.; Li, F.H.; Xiang, J.H. Histological study on the gonadal development of Exopalaemon carinicauda (Holthuis, 1950). J. Fish. China 2014, 38, 362–370. (In Chinese) [Google Scholar]
- Kang, P.F.; Mao, B.; Fan, C.; Wang, Y.F. Transcriptomic information from the ovaries of red swamp crayfish (Procambarus clarkii) provides new insights into development of ovaries and embryos. Aquaculture 2019, 505, 333–343. [Google Scholar]
- Carnevali, O.; Cionna, C.; Tosti, L.; Lubzens, E.; Maradonna, F. Role of cathepsins in ovarian follicle growth and maturation. Gen. Comp. Endocrinol. 2006, 146, 195–203. [Google Scholar]
- Harwood, G.; Amdam, G. Vitellogenin in the honey bee midgut. Apidologie 2021, 52, 837–847. [Google Scholar]
- Rodriguez, E.M.; Medesani, D.A.; Greco, L.; Fingerman, M. Effects of some steroids and other compounds on ovarian growth of the red swamp crayfish, Procambarus clarkii, during early vitellogenesis. J. Exp. Zool. 2002, 292, 82–87. [Google Scholar]
- Quackenbush, L.S. Yolk synthesis in the marine shrimp, Penaeus vannamei. Comp. Biochem. Phys. A 1992, 103, 711–714. [Google Scholar]
- Yano, I.; Hoshino, R. Effects of 17 β-estradiol on the vitellogenin synthesis and oocyte development in the ovary of kuruma prawn (Marsupenaeus japonicus). Comp. Biochem. Phys. A 2006, 144, 18–23. [Google Scholar]
- Mayer, M.P.; Bukau, B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005, 62, 670–684. [Google Scholar]
- Romani, W.A.; Russ, D.W. Acute effects of sex-specific sex hormones on heat shock proteins in fast muscle of male and female rats. Eur. J. Appl. Physiol. 2013, 113, 2503–2510. [Google Scholar]
- Chan, S.F.; He, J.G.; Chu, K.H.; Sun, C.B. The shrimp heat shock cognate 70 functions as a negative regulator in vitellogenin gene expression. Biol. Reprod. 2014, 91, 1–11. [Google Scholar]
- Dhamad, A.E.; Zhou, Z.Q.; Zhou, J.H.; Du, Y.C. Systematic proteomic identification of the heat shock proteins (Hsp) that interact with estrogen receptor alpha (ER alpha) and biochemical characterization of the ER alpha-Hsp70 Interaction. PLoS ONE 2016, 11, e160312. [Google Scholar]
- Liu, M.; Pan, J.; Dong, Z.; Cheng, Y.; Gong, X. Comparative transcriptome reveals the potential modulation mechanisms of estradiol affecting ovarian development of female Portunus trituberculatus. PLoS ONE 2019, 14, e226698. [Google Scholar]
- Mak, A.S.C.; Choi, C.L.; Tiu, S.H.K.; Hui, J.H.L.; He, J.G.; Tobe, S.S.; Chan, S.M. Vitellogenesis in the red crab Charybdis feriatus: Hepatopancreas-specific expression and farnesoic acid stimulation of vitellogenin gene expression. Mol. Reprod. Dev. 2005, 70, 288–300. [Google Scholar]
- Yang, Y.; Ye, H.H.; Huang, H.Y.; Jin, Z.X.; Li, S.J. Cloning, expression and functional analysis of farnesoic acid O-methyltransferase (FAMeT) in the mud crab, Scylla paramamosain. Mar. Freshw. Behav. Phy. 2012, 45, 209–222. [Google Scholar]
- Miyakawa, H.; Toyota, K.; Sumiya, E.; Iguchi, T. Comparison of JH signaling in insects and crustaceans. Curr. Opin. Insect Sci. 2014, 1, 81–87. [Google Scholar]
- Borst, D.W.; Ogan, J.; Tsukimura, B.; Claerhout, T.; Holford, K.C. Regulation of the crustacean mandibular organ. Am. Zool. 2001, 41, 430–441. [Google Scholar]
- Liu, S.H.; Yangb, B.J.; Go, J.H.; Yao, X.M.; Zhang, Y.X.; Song, F.; Liu, Z.W. Molecular cloning and characterization of a juvenile hormone esterase gene from brown planthopper, Nilaparvata lugens. J. Insect Physiol. 2008, 54, 1495–1502. [Google Scholar]
- Tobe, S.S.; Young, D.A.; Khoo, H.W.; Baker, F.C. Farnesoic acid as a major product of release from crustacean mandibular organs In vitro. J. Exp. Zool. Part A 1989, 249, 165–171. [Google Scholar]
- Nagaraju, G. Reproductive regulators in decapod crustaceans: An overview. J. Exp. Biol. 2011, 214, 3–16. [Google Scholar]
- Jung, H.T.; Lyons, R.E.; Hurwood, D.A.; Mather, P. Genes and growth performance in crustacean species: A review of relevant genomic studies in crustaceans and other taxa. Rev. Aquacult. 2013, 5, 77–110. [Google Scholar]
- Rotllant, G.; Nguyen, T.V.; Aizen, J.; Suwansa-ard, S.; Ventura, T. Toward the identification of female gonad-stimulating factors in crustaceans. Hydrobiologia 2018, 825, 91–119. [Google Scholar]
- Huang, X.; Ye, H.; Huang, H.; Yu, K.; Huang, Y. Two beta-pigment-dispersing hormone (β-PDH) isoforms in the mud crab, Scylla paramamosain: Implication for regulation of ovarian maturation and a photoperiod-related daily rhythmicity. Anim. Reprod. Sci. 2014, 150, 139–147. [Google Scholar]
- Bao, C.; Yang, Y.; Huang, H.; Ye, H. Neuropeptides in the cerebral ganglia of the mud crab, Scylla paramamosain: Transcriptomic analysis and expression profiles during vitellogenesis. Sci. Rep. 2015, 5, 17055. [Google Scholar]
- Wei, L.L.; Chen, T.T.; Luo, B.Y.; Qiu, G.F. Evidences for red pigment concentrating hormone (RPCH) and beta-pigment dispersing hormone (beta-PDH) inducing oocyte meiotic maturation in the Chinese mitten crab, Eriocheir sinensis. Front. Endocrinol. 2021, 12, 802768. [Google Scholar]
- Jiang, Y.W.; Li, C.J.; Chen, L.; Wang, F.G.; Zhou, X. Potential role of retinoids in ovarian physiology and pathogenesis of polycystic ovary syndrome. Clin. Chim. Acta 2017, 469, 87–93. [Google Scholar]
- Kawai, T.; Yanaka, N.; Richards, J.S.; Shimada, M. De novo-synthesized retinoic acid in ovarian antral follicles enhances FSH-mediated ovarian follicular cell differentiation and female fertility. Endocrinology 2016, 157, 2160–2172. [Google Scholar]
- Damdimopoulou, P.; Chiang, C.; Flaws, J.A. Retinoic acid signaling in ovarian folliculogenesis and steroidogenesis. Reprod. Toxicol. 2019, 87, 32–41. [Google Scholar]
- Tahaei, L.S.; Eimani, H.; Yazdi, P.E.; Ebrahimi, B.; Fathi, R. Effects of retinoic acid on maturation of immature mouse oocytes in the presence and absence of a granulosa cell co-culture system. J. Assist. Reprod. Genet. 2011, 28, 553–558. [Google Scholar]
- Levi, L.; Ziv, T.; Admon, A.; Levavi-Sivan, B.; Lubzens, E. Insight into molecular pathways of retinal metabolism, associated with vitellogenesis in zebrafish. Am. J. Physiol.-Endocrinol. Metab. 2012, 302, 626–644. [Google Scholar]
- Wang, J.J.; Li, J.T.; Ge, Q.Q.; Li, W.Y.; Li, J. Full-Length transcriptome sequencing and comparative transcriptomic analysis provide insights into the ovarian maturation of Exopalaemon carinicauda. Front. Mar. Sci. 2022, 9, 906730. [Google Scholar]
- Lamming, D.W.; Bar-Peled, L. Lysosome: The metabolic signaling hub. Traffic 2019, 20, 27–38. [Google Scholar]
- Li, Y.; Wang, Z.; Andersen, C.L.; Ye, X. Functions of lysosomes in mammalian female reproductive system. Reprod. Dev. Med. 2020, 4, 109–122. [Google Scholar]
- Zhang, Y.N.; Jiang, S.F.; Qiao, H.; Xiong, Y.W.; Fu, H.T.; Zhang, W.Y.; Gong, Y.S.; Jin, S.B.; Wu, Y. Transcriptome analysis of five ovarian stages reveals gonad maturation in female updates Macrobrachium nipponense. BMC Genom. 2021, 22, 100868. [Google Scholar]
- Wu, P.; Qi, D.; Chen, L.Q.; Zhang, H.; Zhang, X.W.; Qin, J.G.; Hu, S.N. Gene discovery from an ovary cDNA library of oriental river prawn Macrobrachium nipponense by ESTs annotation. Comp. Biochem. Phys. D 2009, 4, 111–120. [Google Scholar]
- Luo, B.Y.; Qian, H.L.; Jiang, H.C.; Xiong, X.Y.; Ye, B.Q.; Liu, X.; Guo, Z.Q.; Ma, K.Y. Transcriptional changes revealed water acidification leads to the immune response and ovary maturation delay in the Chinese mitten crab Eriocheir sinensis. Comp. Biochem. Phys. D 2021, 39, 100863. [Google Scholar]
- Rettori, V.; McCann, S.M. Role of nitric oxide and alcohol on gonadotropin release in vitro and in vivo. Ann. N. Y. Acad. Sci. 1998, 840, 185–193. [Google Scholar]
- Lopez, F.J.; Moretto, M.; Merchenthaler, I.; Negro-Vilar, A. Nitric oxide is involved in the genesis of pulsatile LHRH secretion from immortalized LHRH neurons. J. Neuroendocrinol. 1997, 9, 647–654. [Google Scholar]
- Kim, H.W.; Batista, L.A.; Hoppes, J.L.; Lee, K.J.; Mykles, D.L. A crustacean nitric oxide synthase expressed in nerve ganglia, Y-organ, gill and gonad of the tropical land crab, Gecarcinus lateralis. J. Exp. Biol. 2004, 207, 2845–2857. [Google Scholar]
- Subramoniam, T. Mechanisms and control of vitellogenesis in crustaceans. Fish. Sci. 2011, 77, 1–21. [Google Scholar]



| Term | Significant/Annotated | Up/Down | ID | p-Value |
|---|---|---|---|---|
| Larval chitin-based cuticle development | 4/23 | up | 0008363 | 0.00039 |
| Ecdysteroid biosynthetic process | 4/27 | up | 0045456 | 0.00073 |
| Ecdysteroid metabolic process | 4/37 | up | 0045455 | 0.00244 |
| Sterol transport | 6/103 | up | 0015918 | 0.00517 |
| Molting cycle | 8/216 | up | 0042303 | 0.01884 |
| Steroid biosynthetic process | 6/150 | up | 0006694 | 0.02876 |
| Regulation of gastrulation | 3/50 | up | 0010470 | 0.04185 |
| Sterol transporter activity | 5/21 | up | 0015248 | 2.30×10−5 |
| Sterol binding | 6/48 | up | 0032934 | 0.00016 |
| Negative regulation of neurological system process | 3/52 | down | 0031645 | 0.00439 |
| Desensitization of G protein coupled receptor Signaling pathway | 2/18 | down | 0002029 | 0.00574 |
| Negative regulation of G protein coupled receptor Signaling pathway | 2/26 | down | 0045744 | 0.01181 |
| Positive regulation of reproductive process | 4/160 | down | 2000243 | 0.01891 |
| DNA/RNA helicase activity | 1/6 | down | 0033677 | 0.04114 |
| Term | Significant/Annotated | Up/Down | ID | p-Value |
|---|---|---|---|---|
| Regulation of ovulation | 3/12 | up | 0060278 | 0.00537 |
| Embryonic liver development | 3/12 | up | 1990402 | 0.00537 |
| Luteinization | 3/22 | up | 0001553 | 0.02989 |
| Positive regulation of ovulation | 2/10 | up | 0060279 | 0.03703 |
| Intracellular sterol transport | 4/42 | up | 0032366 | 0.04113 |
| Sterol metabolic process | 8/126 | up | 0016125 | 0.04343 |
| Molting cycle | 20/216 | down | 0042303 | 5.20 × 10−5 |
| Response to estrogen | 10/82 | down | 0043627 | 0.00047 |
| Molting cycle process | 13/131 | down | 0022404 | 0.00055 |
| Sterol homeostasis | 7/65 | down | 0055092 | 0.00663 |
| Estrogen secretion | 2/4 | down | 0035937 | 0.00672 |
| Positive regulation of estrogen secretion | 2/4 | down | 2000863 | 0.00672 |
| Sterol transport | 8/103 | down | 0015918 | 0.025 |
| Placenta development | 7/93 | down | 0001890 | 0.04033 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Wang, J.; Wang, C.; Li, W.; Ge, Q.; Qin, Z.; Li, J.; Li, J. The Responses of the Ovary and Eyestalk in Exopalaemon carinicauda under Low Salinity Stress. Fishes 2022, 7, 365. https://doi.org/10.3390/fishes7060365
Zhang X, Wang J, Wang C, Li W, Ge Q, Qin Z, Li J, Li J. The Responses of the Ovary and Eyestalk in Exopalaemon carinicauda under Low Salinity Stress. Fishes. 2022; 7(6):365. https://doi.org/10.3390/fishes7060365
Chicago/Turabian StyleZhang, Xiuhong, Jiajia Wang, Chengwei Wang, Wenyang Li, Qianqian Ge, Zhen Qin, Jian Li, and Jitao Li. 2022. "The Responses of the Ovary and Eyestalk in Exopalaemon carinicauda under Low Salinity Stress" Fishes 7, no. 6: 365. https://doi.org/10.3390/fishes7060365
APA StyleZhang, X., Wang, J., Wang, C., Li, W., Ge, Q., Qin, Z., Li, J., & Li, J. (2022). The Responses of the Ovary and Eyestalk in Exopalaemon carinicauda under Low Salinity Stress. Fishes, 7(6), 365. https://doi.org/10.3390/fishes7060365






