The Riddle of How Fisheries Influence Genetic Diversity
Abstract
:1. Introduction
2. Genetic Markers and Metrics
2.1. AFLPs
2.2. Mitochondrial DNA
2.3. Microsatellites
2.4. SNPs
3. Habitat Type
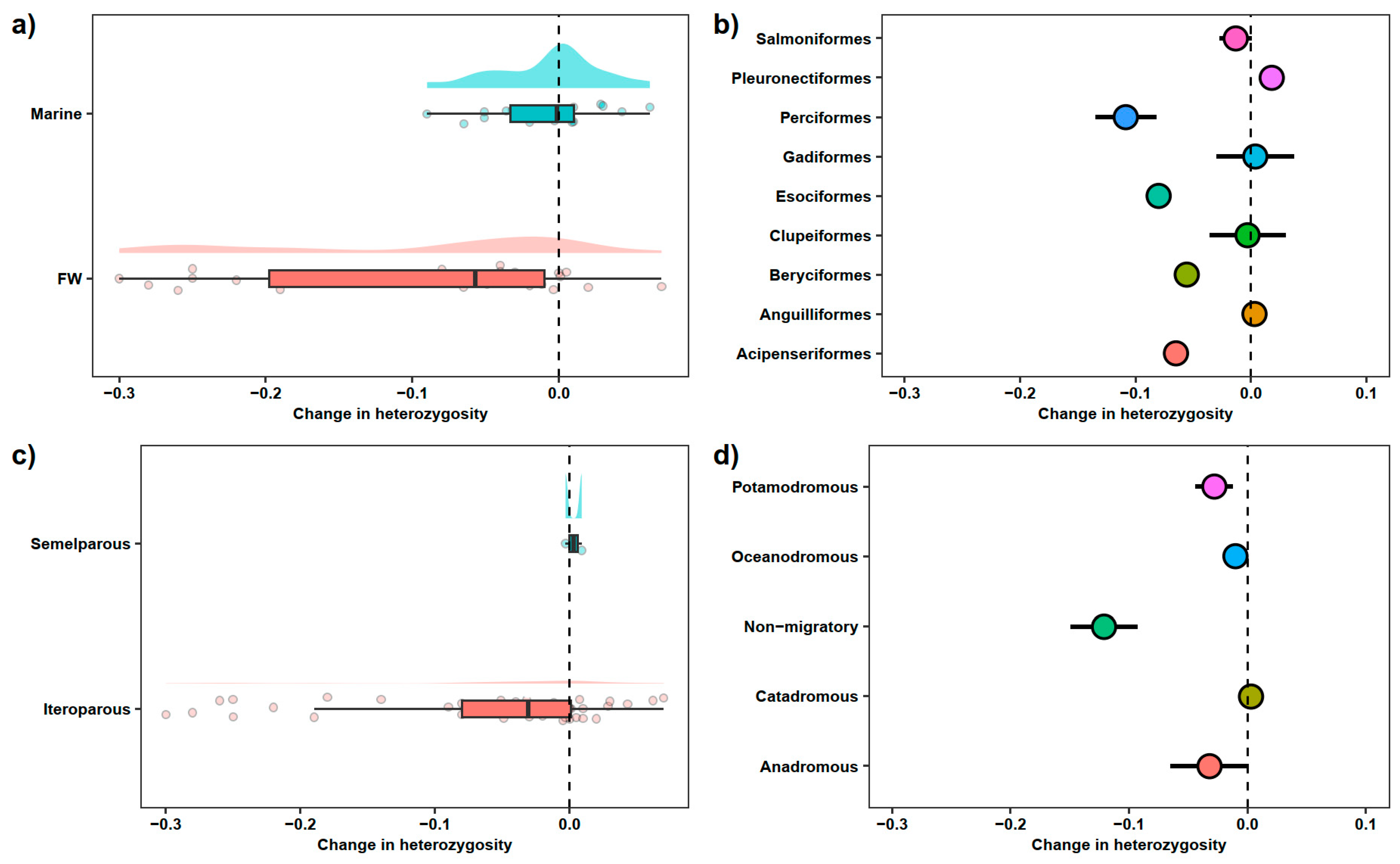
3.1. Freshwater vs. Saltwater
3.2. Latitude
3.3. Habitat Complexity
4. Population Demography
4.1. Population Size and Range
4.2. Sex Ratio
5. Life History and Behaviour
5.1. Life History
5.2. Migration
5.3. Behaviour
6. Coinciding Effects
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- FAO. The State of World Fisheries and Aquaculture 2020; Sustainability in Action; FAO: Rome, Italy, 2020. [Google Scholar]
- Halpern, B.S.; Silliman, B.R.; Olden, J.D.; Bruno, J.P.; Bertness, M.D. Incorporating positive interactions in aquatic restoration and conservation. Front. Ecol. Environ. 2007, 5, 153–160. [Google Scholar] [CrossRef]
- Jackson, J.B.C.; Kirby, M.X.; Berger, W.H.; Bjorndal, K.A.; Botsford, L.W.; Bourque, B.J.; Bradbury, R.H.; Cooke, R.; Erlandson, J.; Estes, J.A.; et al. Historical overfishing and the recent collapse of coastal ecosystems. Science 2001, 293, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Malmqvist, B.; Rundle, S. Threats to the running water ecosystems of the world. Environ. Conserv. 2002, 29, 134–153. [Google Scholar] [CrossRef]
- Therkildsen, N.O.; Nielsen, E.E.; Swain, D.P.; Pedersen, J.S. Large effective population size and temporal genetic stability in Atlantic cod (Gadus morhua) in the southern Gulf of St. Lawrence. Can. J. Fish. Aquat. Sci. 2010, 67, 1585–1595. [Google Scholar] [CrossRef]
- Worm, B. Averting a global fisheries disaster. Proc. Natl. Acad. Sci. USA 2016, 113, 4895–4897. [Google Scholar] [CrossRef]
- Worm, B.; Barbier, E.B.; Beaumont, N.; Duffy, J.E.; Folke, C.; Halpern, B.S.; Jackson, J.B.C.; Lotze, H.K.; Micheli, F.; Palumbi, S.R.; et al. Impacts of biodiversity loss on ocean ecosystem services. Science 2006, 314, 787–790. [Google Scholar] [CrossRef]
- Pinsky, M.L.; Palumbi, S.R. Meta-analysis reveals lower genetic diversity in overfished populations. Mol. Ecol. 2014, 23, 29–39. [Google Scholar] [CrossRef]
- Daskalov, G.M. Overfishing drives a trophic cascade in the Black Sea. Mar. Ecol. Prog. Ser. 2002, 225, 53–63. [Google Scholar] [CrossRef]
- Uusi-Heikkilä, S.; Whiteley, A.R.; Kuparinen, A.; Matsumura, S.; Venturelli, P.A.; Wolter, C.; Slate, J.; Primmer, C.R.; Meinelt, T.; Killen, S.S.; et al. The evolutionary legacy of size-selective harvesting extends from genes to populations. Evol. Appl. 2015, 8, 597–620. [Google Scholar] [CrossRef]
- Allendorf, F.W.; England, P.R.; Luikart, G.; Ritchie, P.A.; Ryman, N. Genetic effects of harvest on wild animal populations. Trends Ecol. Evol. 2008, 23, 327–337. [Google Scholar] [CrossRef]
- Fisher, R.A.; Bennett, H.; Fisher, R.A.; Bennett, H. (Eds.) The Genetical Theory of Natural Selection: A Complete Variorum Edition; Oxford University Press: Oxford, NY, USA, 1999. [Google Scholar]
- Holderegger, R.; Kamm, U.; Gugerli, F. Adaptive vs. neutral genetic diversity: Implications for landscape genetics. Landsc. Ecol. 2006, 21, 797–807. [Google Scholar] [CrossRef]
- Allendorf, F.W.; Berry, O.; Ryman, N. So long to genetic diversity, and thanks for all the fish. Mol. Ecol. 2014, 23, 23–25. [Google Scholar] [CrossRef] [PubMed]
- Hauser, L.; Adcock, G.J.; Smith, P.J.; Bernal Ramírez, J.H.; Carvalho, G.R. Loss of microsatellite diversity and low effective population size in an overexploited population of New Zealand snapper (Pagrus auratus). Proc. Natl. Acad. Sci. USA 2002, 99, 11742–11747. [Google Scholar] [CrossRef] [PubMed]
- Hoarau, G.; Boon, E.; Jongma, D.N.; Ferber, S.; Palsson, J.; Van der Veer, H.W.; Rijnsdorp, A.D.; Stam, W.T.; Olsen, J.L. Low effective population size and evidence for inbreeding in an overexploited flatfish, plaice (Pleuronectes platessa L.). Proc. R. Soc. B Biol. Sci. 2005, 272, 497–503. [Google Scholar] [CrossRef]
- Jones, A.T.; Lavery, S.D.; Le Port, A.; Wang, Y.-G.; Blower, D.; Ovenden, J. Sweepstakes reproductive success is absent in a New Zealand snapper (Chrysophrus auratus) population protected from fishing despite “tiny” Ne/N ratios elsewhere. Mol. Ecol. 2019, 28, 2986–2995. [Google Scholar] [CrossRef]
- Hutchinson, W.F.; van Oosterhout, C.; Rogers, S.I.; Carvalho, G.R. Temporal analysis of archived samples indicates marked genetic changes in declining North Sea cod (Gadus morhua). Proc. R. Soc. B Biol. Sci. 2003, 270, 2125–2132. [Google Scholar] [CrossRef]
- Poulsen, N.A.; Nielsen, E.E.; Schierup, M.H.; Loeschcke, V.; Grønkjær, P. Long-term stability and effective population size in North Sea and Baltic Sea cod (Gadus morhua). Mol. Ecol. 2006, 15, 321–331. [Google Scholar] [CrossRef]
- Ruzzante, D.E.; Taggart, C.T.; Doyle, R.W.; Cook, D. Stability in the historical pattern of genetic structure of Newfoundland cod (Gadus morhua) despite the catastrophic decline in population size from 1964 to 1994. Conserv. Genet. 2001, 2, 257–269. [Google Scholar] [CrossRef]
- Borovski, T.; Tadmor-Levi, R.; Shapiro, J.; Rubinstein, G.; Agyakwah, S.K.; Hulata, G.; David, L. Historical and recent reductions in genetic variation of the Sarotherodon galilaeus population in the Sea of Galilee. Conserv. Genet. 2018, 19, 1323–1333. [Google Scholar] [CrossRef]
- Euclide, P.T.; Kilpatrick, C.W.; Marsden, J.E. Genetic diversity and structure of lake whitefish (Coregonus clupeaformis) 100 years after closure of the commercial fishery. J. Great Lakes Res. 2019, 45, 1310–1319. [Google Scholar] [CrossRef]
- Laconcha, U.; Iriondo, M.; Arrizabalaga, H.; Manzano, C.; Markaide, P.; Montes, I.; Zarraonaindia, I.; Velado, I.; Bilbao, E.; Goñi, N.; et al. New nuclear SNP markers unravel the genetic structure and effective population size of Albacore tuna (Thunnus alalunga). PLoS ONE 2015, 10, e0128247. [Google Scholar] [CrossRef] [PubMed]
- D’Aloia, C.C.; Andrés, J.A.; Bogdanowicz, S.M.; McCune, A.R.; Harrison, R.G.; Buston, P.M. Unraveling hierarchical genetic structure in a marine metapopulation: A comparison of three high-throughput genotyping approaches. Mol. Ecol. 2020, 29, 2189–2203. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.E.; Welch, D.J. Marine fisheries management in a changing climate: A review of vulnerability and future options. Rev. Fish. Sci. 2009, 18, 106–124. [Google Scholar] [CrossRef]
- Rogers, A.; Blanchard, J.L.; Mumby, P.J. Vulnerability of coral reef fisheries to a loss of structural complexity. Curr. Biol. 2014, 24, 1000–1005. [Google Scholar] [CrossRef]
- Johnston, F.D.; Arlinghaus, R.; Dieckmann, U. Fish life history, angler behaviour and optimal management of recreational fisheries. Fish Fish. 2013, 14, 554–579. [Google Scholar] [CrossRef]
- Patrick, W.S.; Spencer, P.; Link, J.; Cope, J.; Field, J.; Kobayashi, D.; Lawson, P.; Gedamke, T.; Cortés, E.; Ormseth, O.; et al. Using productivity and susceptibility indices to assess the vulnerability of United States fish stocks to overfishing. Fish. Bull. 2010, 108, 305–322. [Google Scholar]
- Timm, L.E. A fair fight between molecular marker types in a seascape genetics setting. Mol. Ecol. 2020, 29, 2133–2136. [Google Scholar] [CrossRef] [PubMed]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van de Lee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef]
- Meudt, H.M.; Clarke, A.C. Almost forgotten or latest practice? AFLP applications, analyses and advances. Trends Plant Sci. 2007, 12, 106–117. [Google Scholar] [CrossRef]
- Mueller, U.G.; Wolfenbarger, L.L. AFLP genotyping and fingerprinting. Trends Ecol. Evol. 1999, 14, 389–394. [Google Scholar] [CrossRef]
- Wan, Q.-H.; Fang, S.-G.; Li, Y.-N. The loss of genetic diversity in Dabry’s sturgeon (Acipenser dabryanus, Dumeril) as revealed by DNA fingerprinting. Aquat. Conserv. Mar. Freshw. Ecosyst. 2003, 13, 225–231. [Google Scholar] [CrossRef]
- Schlötterer, C. The evolution of molecular markers—Just a matter of fashion? Nat. Rev. Genet. 2004, 5, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Ali, F.S.; Mamoon, A. Population genetic studies of genus Dicentrarchus reveal loss of genetic diversity in Egyptian waters. Reg. Stud. Mar. Sci. 2019, 31, 100783. [Google Scholar] [CrossRef]
- Diedericks, G.; Henriques, R.; von der Heyden, S.; Weyl, O.L.F.; Hui, C. The ghost of introduction past: Spatial and temporal variability in the genetic diversity of invasive smallmouth bass. Evol. Appl. 2018, 11, 1609–1629. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.M.; Kemp, B.M.; Thorgaard, G.H. Increased mitochondrial DNA diversity in ancient Columbia River basin Chinook salmon Oncorhynchus tshawytscha. PLoS ONE 2018, 13, e0190059. [Google Scholar] [CrossRef]
- Righi, T.; Splendiani, A.; Fioravanti, T.; Casoni, E.; Gioacchini, G.; Carnevali, O.; Caputo Barucchi, V. Loss of mitochondrial genetic diversity in overexploited Mediterranean swordfish (Xiphias gladius, 1759) population. Diversity 2020, 12, 170. [Google Scholar] [CrossRef]
- Zhai, D.-D.; Li, W.-J.; Liu, H.-Z.; Cao, W.-X.; Gao, X. Genetic diversity and temporal changes of an endemic cyprinid fish species, Ancherythroculter nigrocauda, from the upper reaches of Yangtze River. Zool. Res. 2019, 40, 427–438. [Google Scholar] [CrossRef]
- Liu, D.; Li, X.; Song, Z. No decline of genetic diversity in elongate loach (Leptobotia elongata) with a tendency to form population structure in the upper Yangtze River. Glob. Ecol. Conserv. 2020, 23, e01072. [Google Scholar] [CrossRef]
- Ruelas-Inzunza, J.; Rojas-Ruiz, E.; Spanopoulos-Hernández, M.; Barba-Quintero, G. Mercury in the blue tilapia Oreochromis aureus from a dam located in a mining region of NW Mexico: Seasonal variation and percentage weekly intake (PWI). Environ. Monit. Assess. 2015, 187, 233. [Google Scholar] [CrossRef]
- Wang, I.J. Choosing appropriate genetic markers and analytical methods for testing landscape genetic hypotheses. Mol. Ecol. 2011, 20, 2480–2482. [Google Scholar] [CrossRef]
- DeFaveri, J.; Viitaniemi, H.; Leder, E.; Merilä, J. Characterizing genic and nongenic molecular markers: Comparison of microsatellites and SNPs. Mol. Ecol. Res. 2013, 13, 377–392. [Google Scholar] [CrossRef]
- Hauser, S.S.; Athrey, G.; Leberg, P.L. Waste not, want not: Microsatellites remain an economical and informative technology for conservation genetics. Ecol. Evol. 2021, 11, 15800–15814. [Google Scholar] [CrossRef] [PubMed]
- Morin, P.A.; Luikart, G.; Wayne, R.K. The SNP Workshop Group. SNPs in ecology, evolution and conservation. Trends Ecol. Evol. 2004, 19, 208–216. [Google Scholar] [CrossRef]
- Payseur, B.A.; Cutter, A.D. Integrating patterns of polymorphism at SNPs and STRs. Trends Genet. 2006, 22, 424–429. [Google Scholar] [CrossRef]
- Miller, L.M.; Kapuscinski, A.R. Historical analysis of genetic variation reveals low effective population size in a Northern pike (Esox lucius) population. Genetics 1997, 147, 1249–1258. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.J.; Francis, R.I.C.C.; McVeagh, M. Loss of genetic diversity due to fishing pressure. Fish. Res. 1991, 10, 309–316. [Google Scholar] [CrossRef]
- Antao, T.; Pérez-Figueroa, A.; Luikart, G. Early detection of population declines: High power of genetic monitoring using effective population size estimators. Evol. Appl. 2011, 4, 144–154. [Google Scholar] [CrossRef]
- Cornuet, J.M.; Luikart, G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 1996, 144, 2001–2014. [Google Scholar] [CrossRef]
- Peery, M.Z.; Kirby, R.; Reid, B.N.; Stoelting, R.; Doucet-Bëer, E.; Robinson, S.; Vásquez-Carrillo, C.; Pauli, J.N.; Palsbøll, P.J. Reliability of genetic bottleneck tests for detecting recent population declines. Mol. Ecol. 2012, 21, 3403–3418. [Google Scholar] [CrossRef]
- Baillie, S.M.; Muir, A.M.; Scribner, K.; Bentzen, P.; Krueger, C.C. Loss of genetic diversity and reduction of genetic distance among lake trout Salvelinus namaycush ecomorphs, Lake Superior 1959 to 2013. J. Great Lakes Res. 2016, 42, 204–216. [Google Scholar] [CrossRef]
- Mirimin, L.; Macey, B.; Kerwath, S.; Lamberth, S.; der Merwe, A.B.-v.; Cowley, P.; Bloomer, P.; Roodt-Wilding, R. Genetic analyses reveal declining trends and low effective population size in an overfished South African sciaenid species, the dusky kob (Argyrosomus japonicus). Mar. Freshw. Res. 2015, 67, 266–276. [Google Scholar] [CrossRef]
- Pujolar, J.M.; Bevacqua, D.; Capoccioni, F.; Ciccotti, E.; De Leo, G.A.; Zane, L. No apparent genetic bottleneck in the demographically declining European eel using molecular genetics and forward-time simulations. Conserv. Genet. 2011, 12, 813–825. [Google Scholar] [CrossRef]
- Allendorf, F.W. Genetics and the conservation of natural populations: Allozymes to genomes. Mol. Ecol. 2017, 26, 420–430. [Google Scholar] [CrossRef]
- Allen, B.E.; Bowles, E.; Morris, M.R.J.; Rogers, S.M. Loss of SNP genetic diversity following population collapse in a recreational walleye (Sander vitreus) fishery. Can. J. Fish. Aquat. Sci. 2018, 75, 1644–1651. [Google Scholar] [CrossRef]
- Bowles, E.; Marin, K.; Mogensen, S.; MacLeod, P.; Fraser, D.J. Size reductions and genomic changes within two generations in wild walleye populations: Associated with harvest? Evol. Appl. 2020, 13, 1128–1144. [Google Scholar] [CrossRef]
- Therkildsen, N.O.; Wilder, A.P.; Conover, D.O.; Munch, S.B.; Baumann, H.; Palumbi, S.R. Contrasting genomic shifts underlie parallel phenotypic evolution in response to fishing. Science 2019, 365, 487–490. [Google Scholar] [CrossRef]
- Putman, A.I.; Carbone, I. Challenges in analysis and interpretation of microsatellite data for population genetic studies. Ecol. Evol. 2014, 4, 4399–4428. [Google Scholar] [CrossRef]
- Lemopoulos, A.; Prokkola, J.M.; Uusi-Heikkilä, S.; Vasemägi, A.; Huusko, A.; Hyvärinen, P.; Koljonen, M.-L.; Koskiniemi, J.; Vainikka, A. Comparing RADseq and microsatellites for estimating genetic diversity and relatedness—Implications for brown trout conservation. Ecol. Evol. 2019, 9, 2106–2120. [Google Scholar] [CrossRef]
- Uusi-Heikkilä, S.; Sävilammi, T.; Leder, E.; Arlinghaus, R.; Primmer, C.R. Rapid, broad-scale gene expression evolution in experimentally harvested fish populations. Mol. Ecol. 2017, 26, 3954–3967. [Google Scholar] [CrossRef]
- Barson, N.J.; Aykanat, T.; Hindar, K.; Baranski, M.; Bolstad, G.H.; Fiske, P.; Jacq, C.; Jensen, A.J.; Johnston, S.E.; Karlsson, S.; et al. Sex-dependent dominance at a single locus maintains variation in age at maturity in salmon. Nature 2015, 528, 405–408. [Google Scholar] [CrossRef]
- Weir, B.S.; Basten, C.J. Sampling strategies for distances between DNA sequences. Biometrics 1990, 46, 551–582. [Google Scholar] [CrossRef] [PubMed]
- Bowersox, B.J.; Corsi, M.P.; McCormick, J.L.; Copeland, T.; Campbell, M.R. Examining life history shifts and genetic composition in a hatchery steelhead population, with implications for fishery and ocean selection. Trans. Am. Fish. Soc. 2019, 148, 1056–1068. [Google Scholar] [CrossRef]
- Pinsky, M.L.; Eikeset, A.M.; Helmerson, C.; Star, B. Genomic stability through time despite decades of exploitation in cod on both sides of the Atlantic. Proc. Natl. Acad. Sci. USA 2021, 118, e2025453118. [Google Scholar] [CrossRef] [PubMed]
- Allendorf, F.W. Genetic drift and the loss of alleles versus heterozygosity. Zoo Biol. 1986, 5, 181–190. [Google Scholar] [CrossRef]
- Dawson, M.N. Species richness, habitable volume, and species densities in freshwater, the sea, and on land. Front. Biogeogr. 2012, 4, 105–116. [Google Scholar] [CrossRef]
- Arthington, A.H.; Dulvy, N.K.; Gladstone, W.; Winfield, I.J. Fish conservation in freshwater and marine realms: Status, threats and management. Aquat. Conserv. Mar. Freshw. Ecosyst. 2016, 26, 838–857. [Google Scholar] [CrossRef]
- Carrete Vega, G.; Wiens, J.J. Why are there so few fish in the sea? Proc. R. Soc. B Biol. Sci. 2012, 279, 2323–2329. [Google Scholar] [CrossRef]
- DeWoody, J.A.; Avise, J.C. Microsatellite variation in marine, freshwater and anadromous fishes compared with other animals. J. Fish Biol. 2000, 56, 461–473. [Google Scholar] [CrossRef]
- Lewin, W.-C.; Arlinghaus, R.; Mehner, T. Documented and potential biological impacts of recreational fishing: Insights for management and conservation. Rev. Fish. Sci. 2006, 14, 305–367. [Google Scholar] [CrossRef]
- Kaiser, M.J.; Spence, F.E.; Hart, P.J.B. Fishing-gear restrictions and conservation of benthic habitat complexity. Conserv. Biol. 2000, 14, 1512–1525. [Google Scholar] [CrossRef]
- Hasselman, D.J.; Ricard, D.; Bentzen, P. Genetic diversity and differentiation in a wide ranging anadromous fish, American shad (Alosa sapidissima), is correlated with latitude. Mol. Ecol. 2013, 22, 1558–1573. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, G. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Hillebrand, H. On the generality of the latitudinal diversity gradient. Am. Nat. 2004, 163, 192–211. [Google Scholar] [CrossRef] [PubMed]
- Stuart-Smith, R.D.; Bates, A.E.; Lefcheck, J.S.; Duffy, J.E.; Baker, S.C.; Thomson, R.J.; Stuart-Smith, J.F.; Hill, N.A.; Kininmonth, S.J.; Airoldi, L.; et al. Integrating abundance and functional traits reveals new global hotspots of fish diversity. Nature 2013, 501, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Watson, R.; Kitchingman, A.; Gelchu, A.; Pauly, D. Mapping global fisheries: Sharpening our focus. Fish Fish. 2004, 5, 168–177. [Google Scholar] [CrossRef]
- Papworth, S.K.; Rist, J.; Coad, L.; Milner-Gulland, E.J. Evidence for shifting baseline syndrome in conservation. Conserv. Lett. 2009, 2, 93–100. [Google Scholar] [CrossRef]
- Pinnegar, J.K.; Engelhard, G.H. The ‘shifting baseline’ phenomenon: A global perspective. Rev. Fish Biol. Fish. 2008, 18, 1–16. [Google Scholar] [CrossRef]
- Hanazaki, N.; Herbst, D.F.; Marques, M.S.; Vandebroek, I. Evidence of the shifting baseline syndrome in ethnobotanical research. J. Ethnobiol. Ethnomed. 2013, 9, 75. [Google Scholar] [CrossRef]
- Frankham, R. Stress and adaptation in conservation genetics. J. Evol. Biol. 2005, 18, 750–755. [Google Scholar] [CrossRef]
- Vitorino, C.A.; Nogueira, F.; Souza, I.L.; Araripe, J.; Venere, P.C. Low genetic diversity and structuring of the Arapaima (Osteoglossiformes, Arapaimidae) population of the Araguaia-Tocantins Basin. Front. Genet. 2017, 8, 159. [Google Scholar] [CrossRef]
- Beverton, R.J.H. Small marine pelagic fish and the threat of fishing; are they endangered? J. Fish Biol. 1990, 37, 5–16. [Google Scholar] [CrossRef]
- Frankham, R. Conservation genetics. Ann. Rev. Genet. 1995, 29, 305–327. [Google Scholar] [CrossRef] [PubMed]
- Ryman, N.; Utter, F.; Laikre, L. Protection of intraspecific biodiversity of exploited fishes. Rev. Fish Biol. Fish. 1995, 5, 417–446. [Google Scholar] [CrossRef]
- Bouzat, J.L. Conservation genetics of population bottlenecks: The role of chance, selection, and history. Conserv. Genet. 2010, 11, 463–478. [Google Scholar] [CrossRef]
- Weltz, K.; Lyle, J.M.; Semmens, J.M.; Ovenden, J.R. Population genetics of the endangered Maugean skate (Zearaja maugeana) in Macquarie Harbour, Tasmania. Conserv. Genet. 2018, 19, 1505–1512. [Google Scholar] [CrossRef]
- Otieno, O.N.; Kitaka, N.; Njiru, J.M. Length-weight relationship, condition factor, length at first maturity and sex ratio of Nile tilapia, Oreochromis niloticus in Lake Naivasha, Kenya. Int. J. Fish. Aquat. Stud. 2014, 2, 67–72. [Google Scholar]
- Uusi-Heikkilä, S. Implications of size-selective fisheries on sexual selection. Evol. Appl. 2020, 12, 487–1500. [Google Scholar] [CrossRef]
- Smith, G.H.; Murie, D.J.; Parkyn, D.C. Effects of sex-specific fishing mortality on sex ratio and population dynamics of Gulf of Mexico greater amberjack. Fish. Res. 2018, 208, 219–228. [Google Scholar] [CrossRef]
- Rosemond, R.C.; Nemeth, R.S.; Heppell, S.A. Demographic recovery of a reef fish population over 30 years of spawning aggregation site protection. Front. Mar. Sci. 2022, 9, 931409. [Google Scholar] [CrossRef]
- Madduppa, H.H.; Timm, J.; Kochzius, M. Reduced genetic diversity in the clown anemonefish Amphiprion ocellaris in exploited reefs of Spermonde Archipelago, Indonesia. Front. Mar. Sci. 2018, 5, 80. [Google Scholar] [CrossRef]
- Ruggeri, P.; Splendiani, A.; Bonanomi, S.; Arneri, E.; Cingolani, N.; Santojanni, A.; Belardinelli, A.; Giovannotti, M.; Caputo, V. Temporal genetic variation as revealed by a microsatellite analysis of European sardine (Sardina pilchardus) archived samples. Can. J. Fish. Aquat. Sci. 2012, 69, 1698–1709. [Google Scholar] [CrossRef]
- Hempel, G. Egg Production and Egg Mortality in Herring; Rapports et Procès-Verbaux des Réunions; Conseil Permanent International pour L’Exploration de la mer: Kolkata, India, 1971; Volume 160, pp. 8–11. Available online: https://www.digishelf.de/piresolver?id=404306500 (accessed on 30 August 2023).
- Collie, J.; Hiddink, J.G.; van Kooten, T.; Rijnsdorp, A.D.; Kaiser, M.J.; Jennings, S.; Hilborn, R. Indirect effects of bottom fishing on the productivity of marine fish. Fish Fish. 2017, 18, 619–637. [Google Scholar] [CrossRef]
- Tillotson, M.D.; Quinn, T.P. Selection on the timing of migration and breeding: A neglected aspect of fishing-induced evolution and trait change. Fish Fish. 2018, 19, 170–181. [Google Scholar] [CrossRef]
- Erisman, B.E.; Allen, L.G.; Claisse, J.T.; Pondella, D.J.; Miller, E.F.; Murray, J.H. The illusion of plenty: Hyperstability masks collapses in two recreational fisheries that target fish spawning aggregations. Can. J. Fish. Aquat. Sci. 2011, 68, 1705–1716. [Google Scholar] [CrossRef]
- Van Overzee, H.M.J.; Rijnsdorp, A.D. Effects of fishing during the spawning period: Implications for sustainable management. Rev. Fish Biol. Fish. 2015, 25, 65–83. [Google Scholar] [CrossRef]
- McCusker, M.R.; Bentzen, P. Positive relationships between genetic diversity and abundance in fishes. Mol. Ecol. 2010, 19, 4852–4862. [Google Scholar] [CrossRef]
- Chambers, R.C.; Leggett, W.C. Maternal influences on variation in egg sizes in temperate marine fishes. Am. Zool. 1996, 36, 180–196. [Google Scholar] [CrossRef]
- Pianka, E.R. On r- and K-Selection. Am. Nat. 1970, 104, 592–597. [Google Scholar] [CrossRef]
- Oosthuizen, E.; Daan, N. Egg fecundity and maturity of North Sea cod, Gadus morhua. Neth. J. Sea Res. 1974, 8, 378–397. [Google Scholar] [CrossRef]
- Hard, J.J.; Gross, M.R.; Heino, M.; Hilborn, R.; Kope, R.G.; Law, R.; Reynolds, J.D. Evolutionary consequences of fishing and their implications for salmon. Evol. Appl. 2008, 1, 388–408. [Google Scholar] [CrossRef]
- Smith, V.E. The Taking of Immature Salmon in the Waters of the State of Washington; State of Washington, Department of Fisheries: Olympia, WA, USA, 1920.
- Allan, J.D.; Abell, R.; Hogan, Z.; Revenga, C.; Taylor, B.W.; Welcomme, R.L.; Winemiller, K. Overfishing of inland waters. BioScience 2005, 55, 1041–1051. [Google Scholar] [CrossRef]
- Morita, K. Earlier migration timing of salmonids: An adaptation to climate change or maladaptation to the fishery? Can. J. Fish. Aquat. Sci. 2019, 76, 475–479. [Google Scholar] [CrossRef]
- Petit-Marty, N.; Liu, M.; Tan, I.Z.; Chung, A.; Terrasa, B.; Guijarro, B.; Ordines, F.; Ramírez-Amaro, S.; Massutí, E.; Schunter, C. Declining population sizes and loss of genetic diversity in commercial fishes: A simple method for a first diagnostic. Front. Mar. Sci. 2022, 9, 872537. [Google Scholar] [CrossRef]
- Biro, P.A.; Post, J.R. Rapid depletion of genotypes with fast growth and bold personality traits from harvested fish populations. Proc. Natl. Acad. Sci. USA 2008, 105, 2919–2922. [Google Scholar] [CrossRef] [PubMed]
- Sutter, D.A.H.; Suski, C.D.; Philipp, D.P.; Klefoth, T.; Wahl, D.H.; Kersten, P.; Cooke, S.J.; Arlinghaus, R. Recreational fishing selectively captures individuals with the highest fitness potential. Proc. Natl. Acad. Sci. USA 2012, 109, 20960–20965. [Google Scholar] [CrossRef] [PubMed]
- Monk, C.T.; Bekkevold, D.; Klefoth, T.; Pagel, T.; Palmer, M.; Arlinghaus, R. The battle between harvest and natural selection creates small and shy fish. Proc. Natl. Acad. Sci. USA 2021, 118, e2009451118. [Google Scholar] [CrossRef]
- Solmundsson, J.; Karlsson, H.; Palsson, J. Sexual differences in spawning behaviour and catchability of plaice (Pleuronectes platessa) west of Iceland. Fish. Res. 2003, 61, 57–71. [Google Scholar] [CrossRef]
- Free, C.M.; Thorson, J.T.; Pinsky, M.L.; Oken, K.L.; Wiedenmann, J.; Jensen, O.P. Impacts of historical warming on marine fisheries production. Science 2019, 363, 979–983. [Google Scholar] [CrossRef]
- Little, A.G.; Loughland, I.; Seebacher, F. What do warming waters mean for fish physiology and fisheries? J. Fish Biol. 2020, 97, 328–340. [Google Scholar] [CrossRef]
- Breitburg, D. Effects of hypoxia, and the balance between hypoxia and enrichment, on coastal fishes and fisheries. Estuaries 2002, 25, 767–781. [Google Scholar] [CrossRef]
- Diaz, R.J. Overview of hypoxia around the world. J. Environ. Qual. 2001, 30, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Le Quesne, W.J.F.; Pinnegar, J.K. The potential impacts of ocean acidification: Scaling from physiology to fisheries. Fish Fish. 2012, 13, 333–344. [Google Scholar] [CrossRef]
- Deegan, L.A.; Buchsbaum, R. The Decline of Fisheries Resources in New England: Evaluating the Impact of Overfishing, Contamination, and Habitat Degradation; MIT Sea Grant College Program: Cambridge, MA, USA, 2005. [Google Scholar]
- Pavlova, A.; Beheregaray, L.B.; Coleman, R.; Gilligan, D.; Harrisson, K.A.; Ingram, B.A.; Kearns, J.; Lamb, A.M.; Lintermans, M.; Lyon, J.; et al. Severe consequences of habitat fragmentation on genetic diversity of an endangered Australian freshwater fish: A call for assisted gene flow. Evol. Appl. 2017, 10, 531–550. [Google Scholar] [CrossRef] [PubMed]
- Kininmonth, S.; Blenckner, T.; Niiranen, S.; Watson, J.; Orio, A.; Casini, M.; Neuenfeldt, S.; Bartolino, V.; Hansson, M. Is diversity the missing link in coastal fisheries management? Diversity 2022, 14, 90. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sadler, D.E.; Watts, P.C.; Uusi-Heikkilä, S. The Riddle of How Fisheries Influence Genetic Diversity. Fishes 2023, 8, 510. https://doi.org/10.3390/fishes8100510
Sadler DE, Watts PC, Uusi-Heikkilä S. The Riddle of How Fisheries Influence Genetic Diversity. Fishes. 2023; 8(10):510. https://doi.org/10.3390/fishes8100510
Chicago/Turabian StyleSadler, Daniel E., Phillip C. Watts, and Silva Uusi-Heikkilä. 2023. "The Riddle of How Fisheries Influence Genetic Diversity" Fishes 8, no. 10: 510. https://doi.org/10.3390/fishes8100510
APA StyleSadler, D. E., Watts, P. C., & Uusi-Heikkilä, S. (2023). The Riddle of How Fisheries Influence Genetic Diversity. Fishes, 8(10), 510. https://doi.org/10.3390/fishes8100510





