Comparative Transcriptome Analysis Identifies MAPK Signaling Pathway Associated with Regulating Ovarian Lipid Metabolism during Vitellogenesis in the Mud Crab, Scylla paramamosain
Abstract
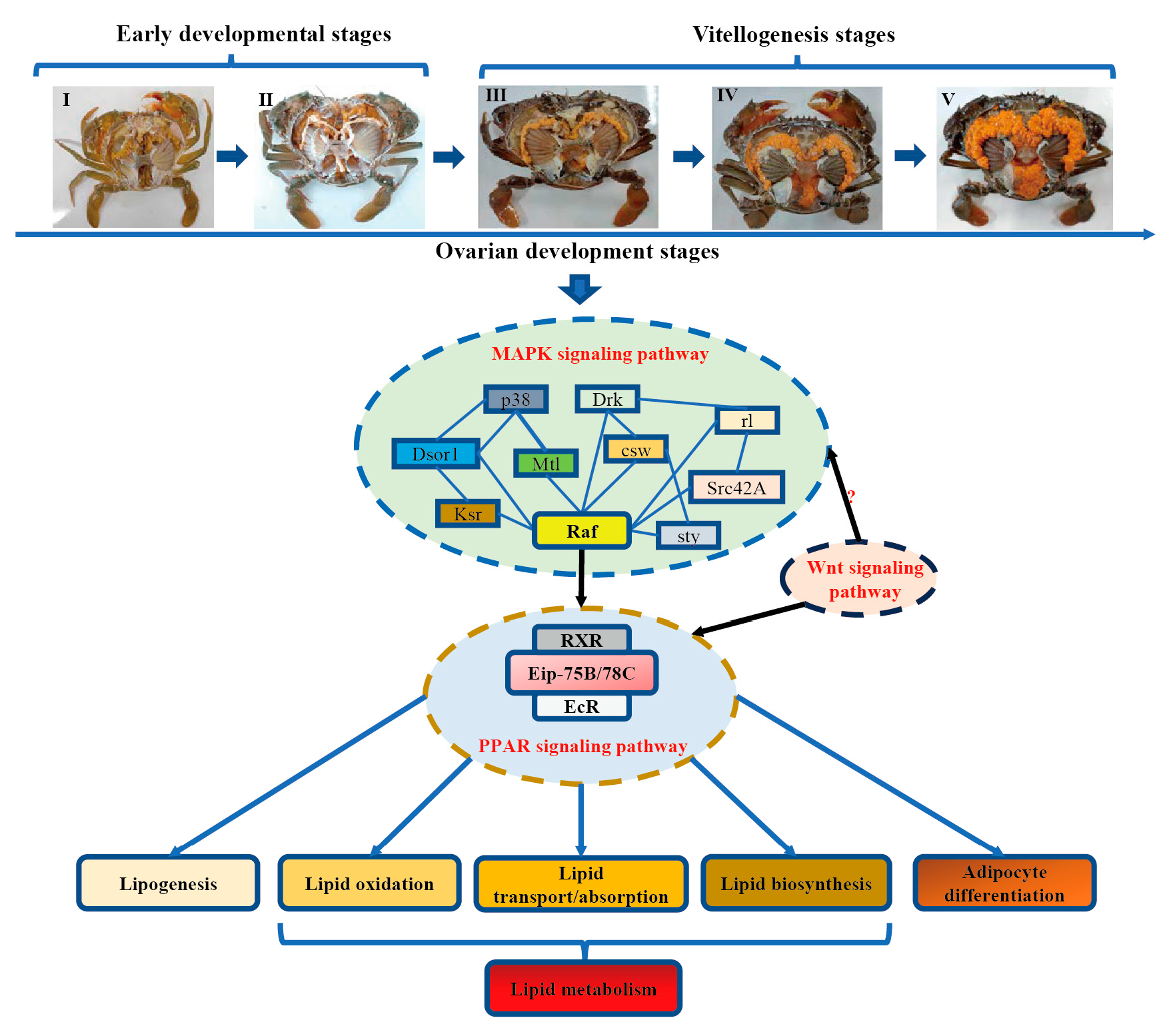
:1. Introduction
2. Materials and Methods
2.1. Animals and Sampling
2.2. Library Constructing and Sequencing
2.3. De Novo Assembly, Clustering Analysis, and Functional Annotation
2.4. Analysis of DEGs and Functional Enrichment
2.5. Analysis of Functional Genes and Signaling Pathways Related to Ovarian Lipid Metabolism
2.6. Analysis of PPI Networks
2.7. Confirmation of DEGs from Transcriptome Data
2.8. Data Transformation and Statistical Analysis
3. Results
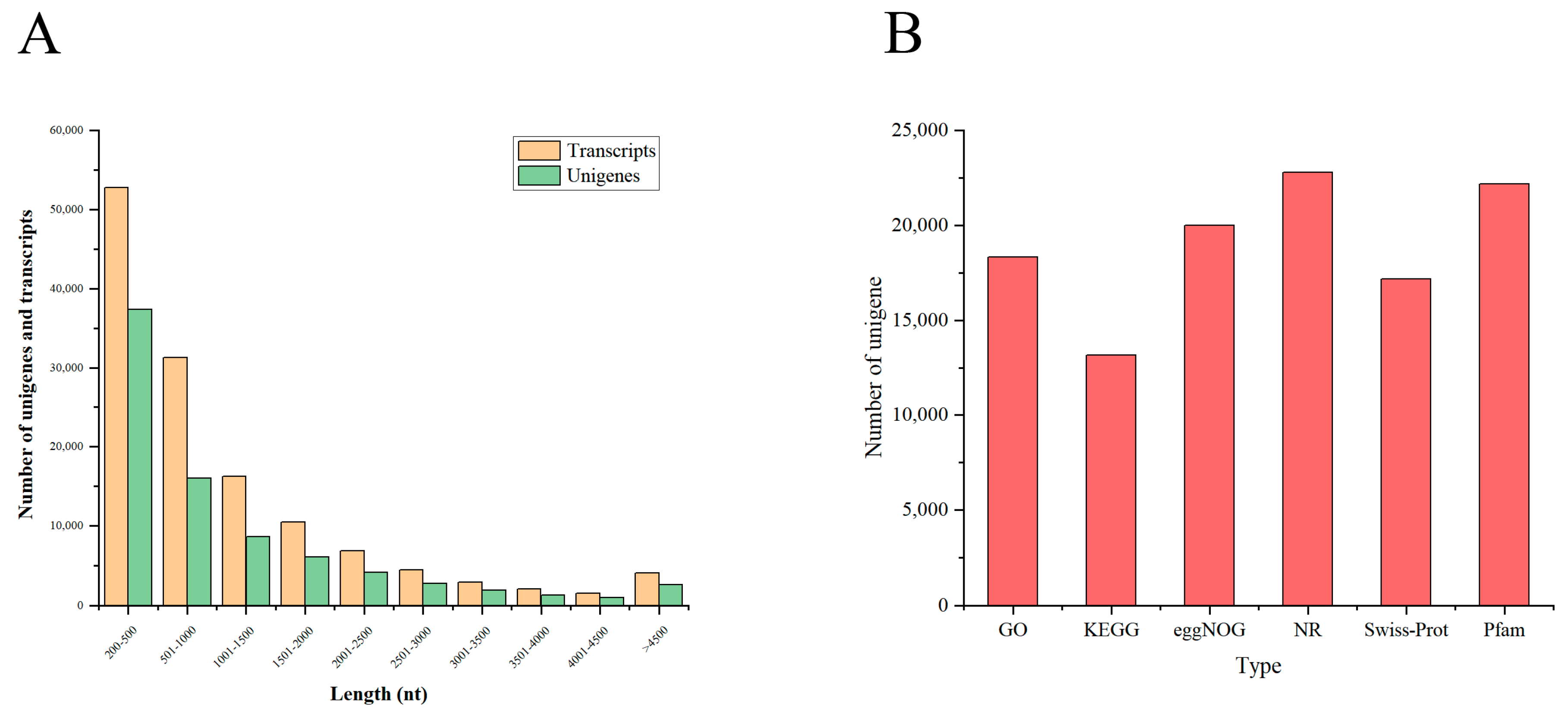
3.1. Assembly and Quality Control of Transcriptome Data
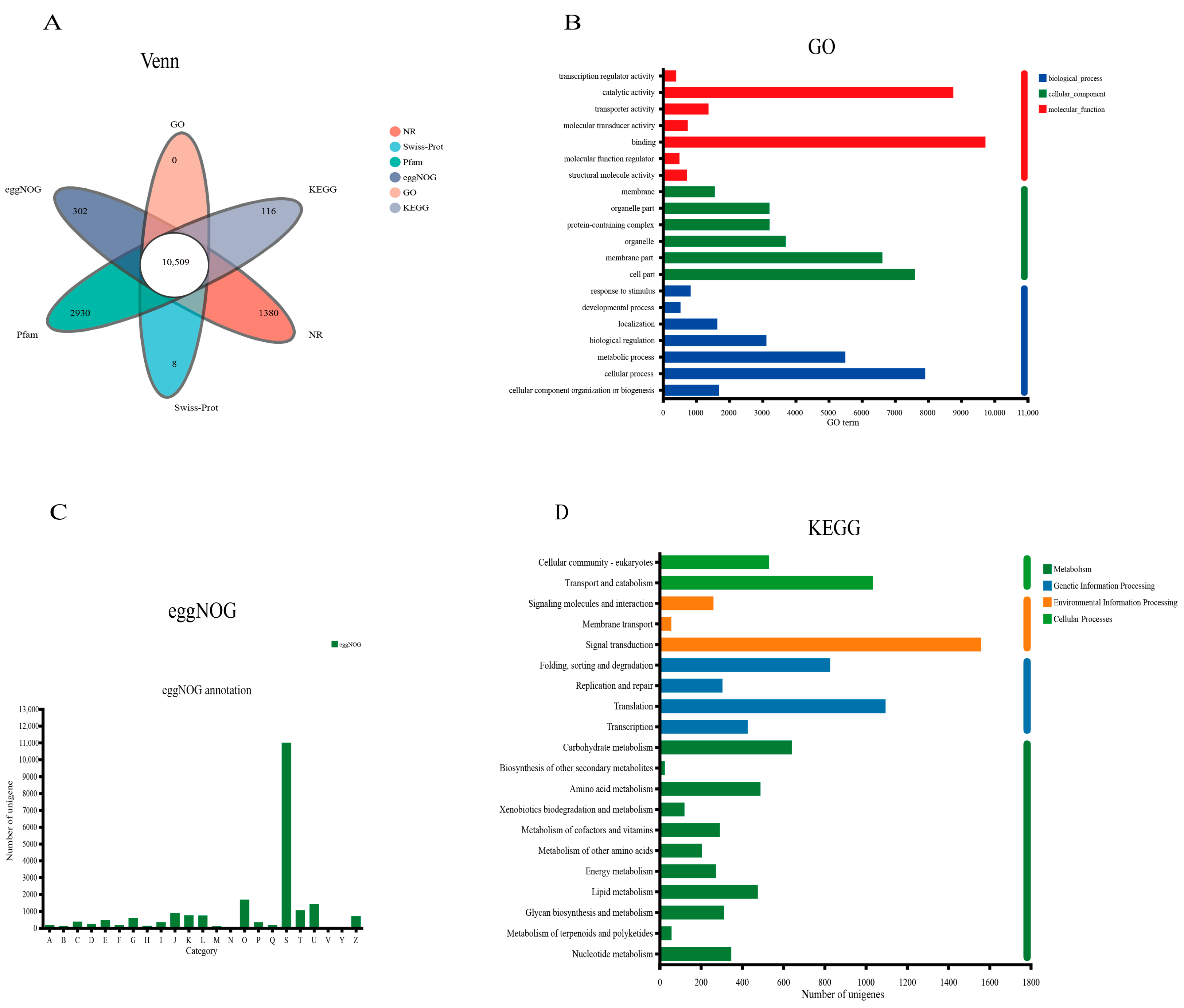
3.2. Overall Functional Annotation
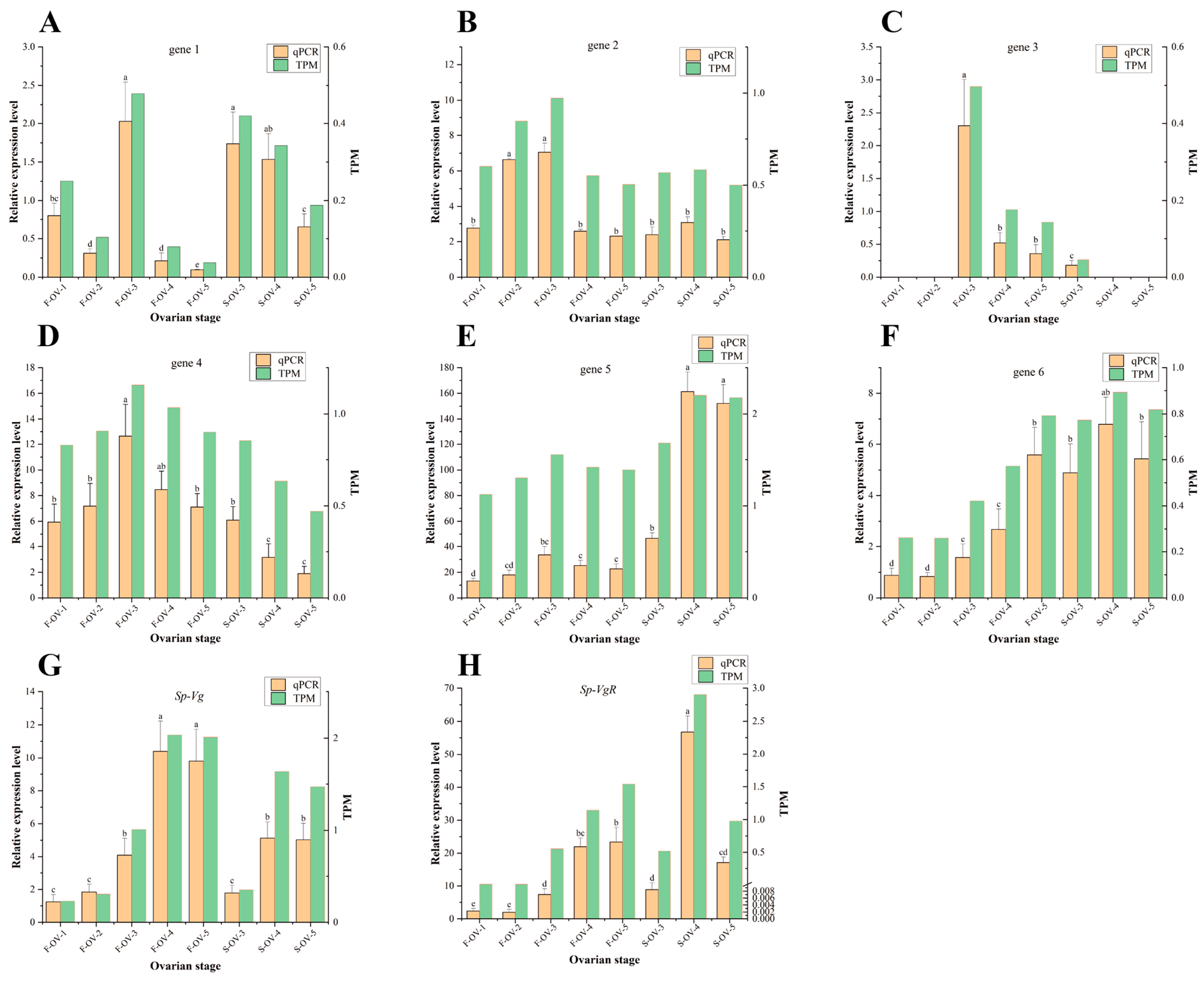
3.3. qRT-PCR Validation of Transcriptomic Sequencing Data
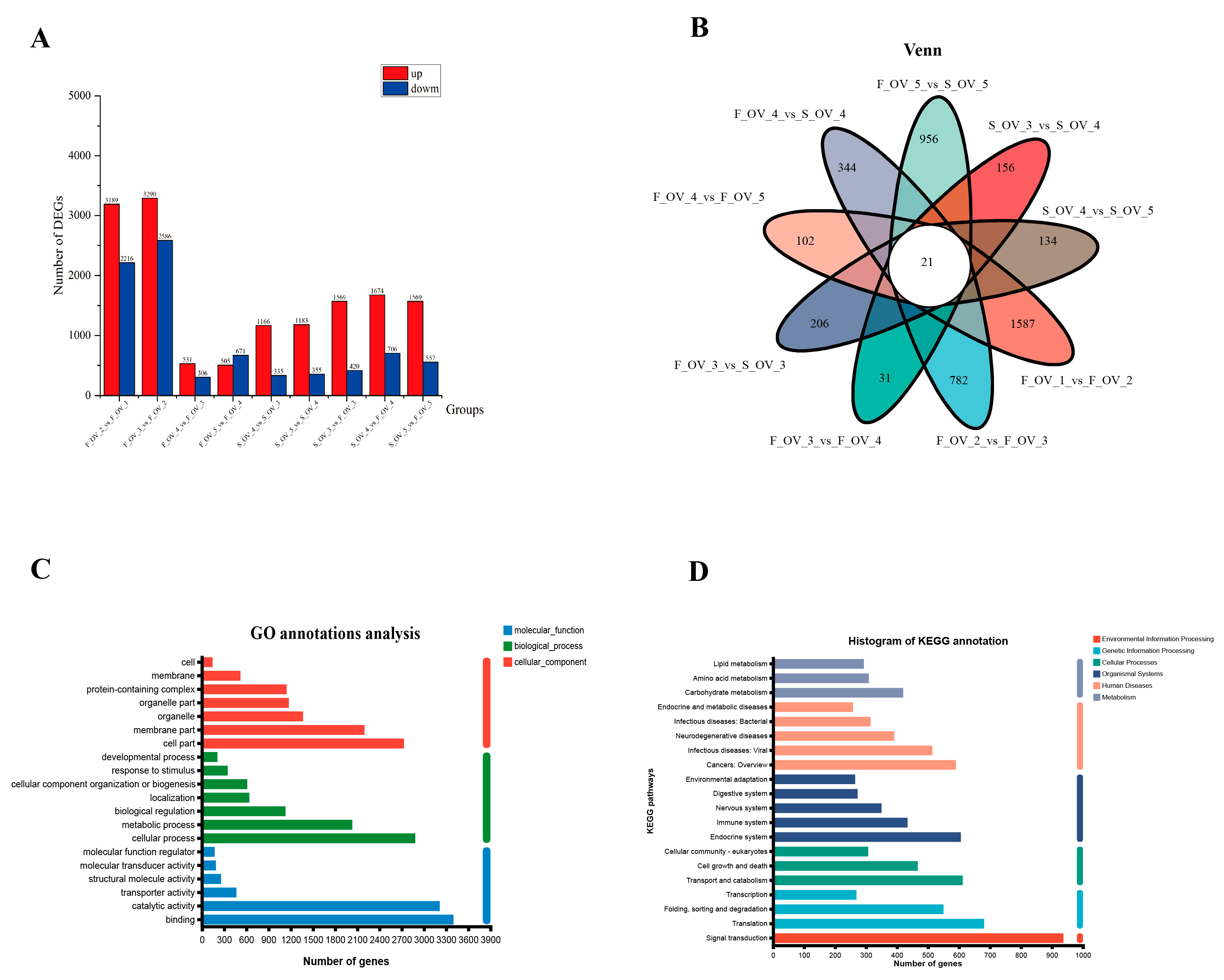
3.4. Identification and Function Analysis of DEGs
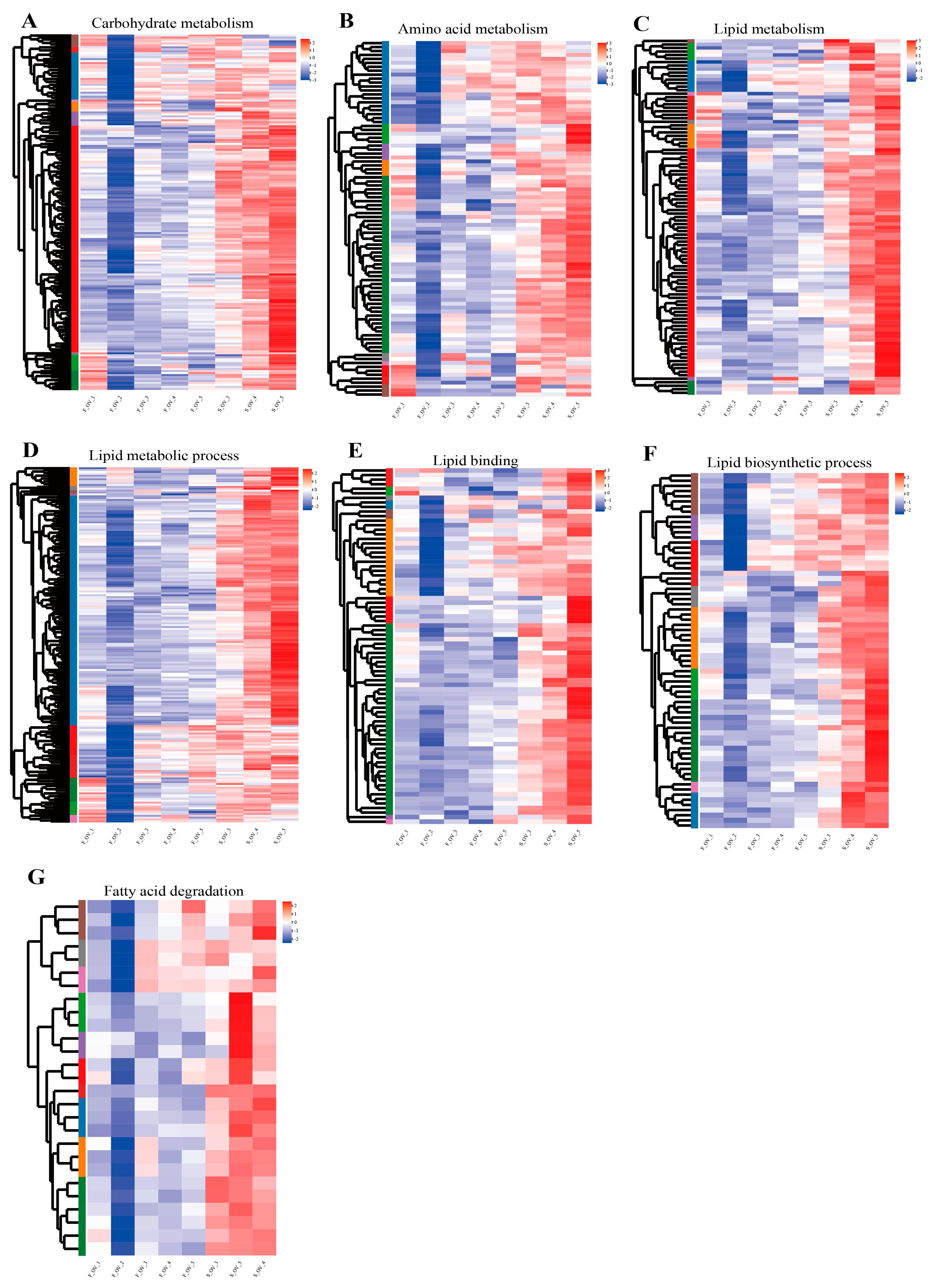
3.5. Expression Analysis of Genes Related to Lipid Metabolism
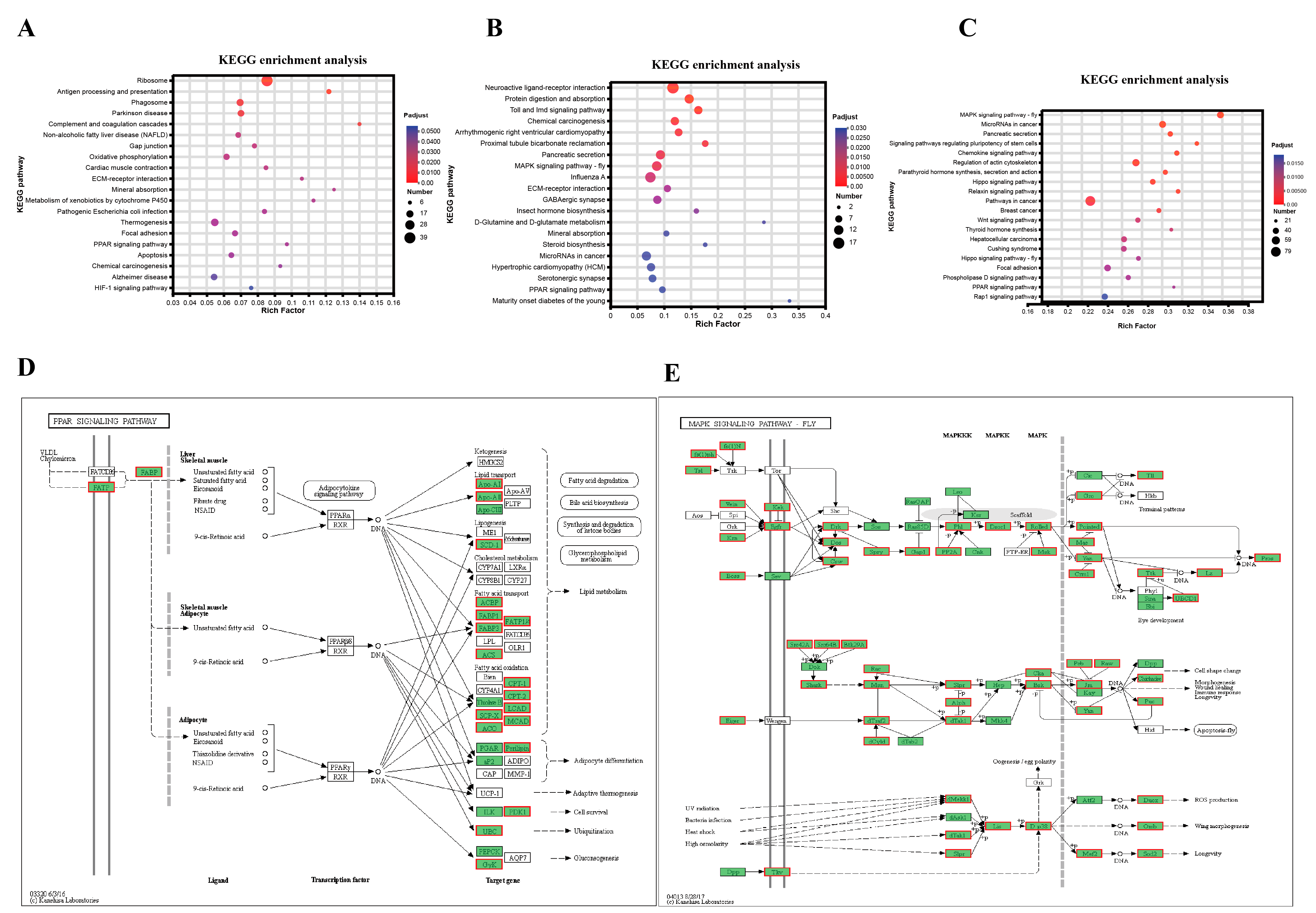
3.6. Identification of Candidate Signaling Pathways and Genes Related to Regulation of Lipid Metabolism
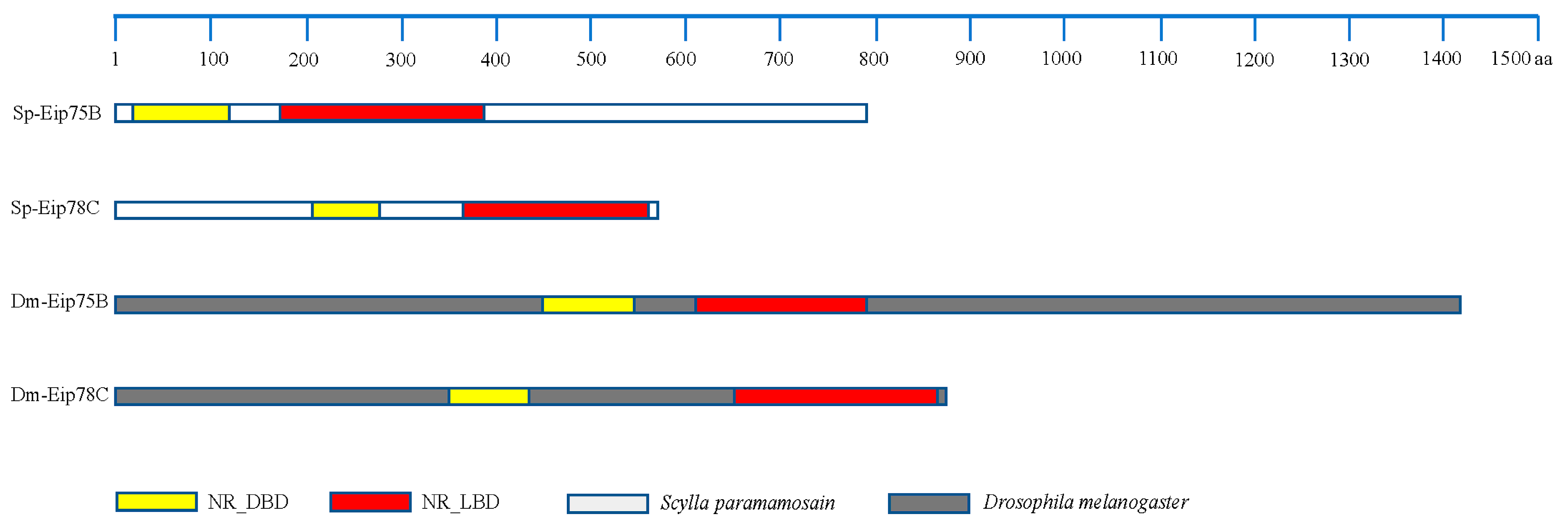
3.7. Identification of the Key Genes, Notably PPARγ-Homologous, Involved in Mediating Lipid Metabolism
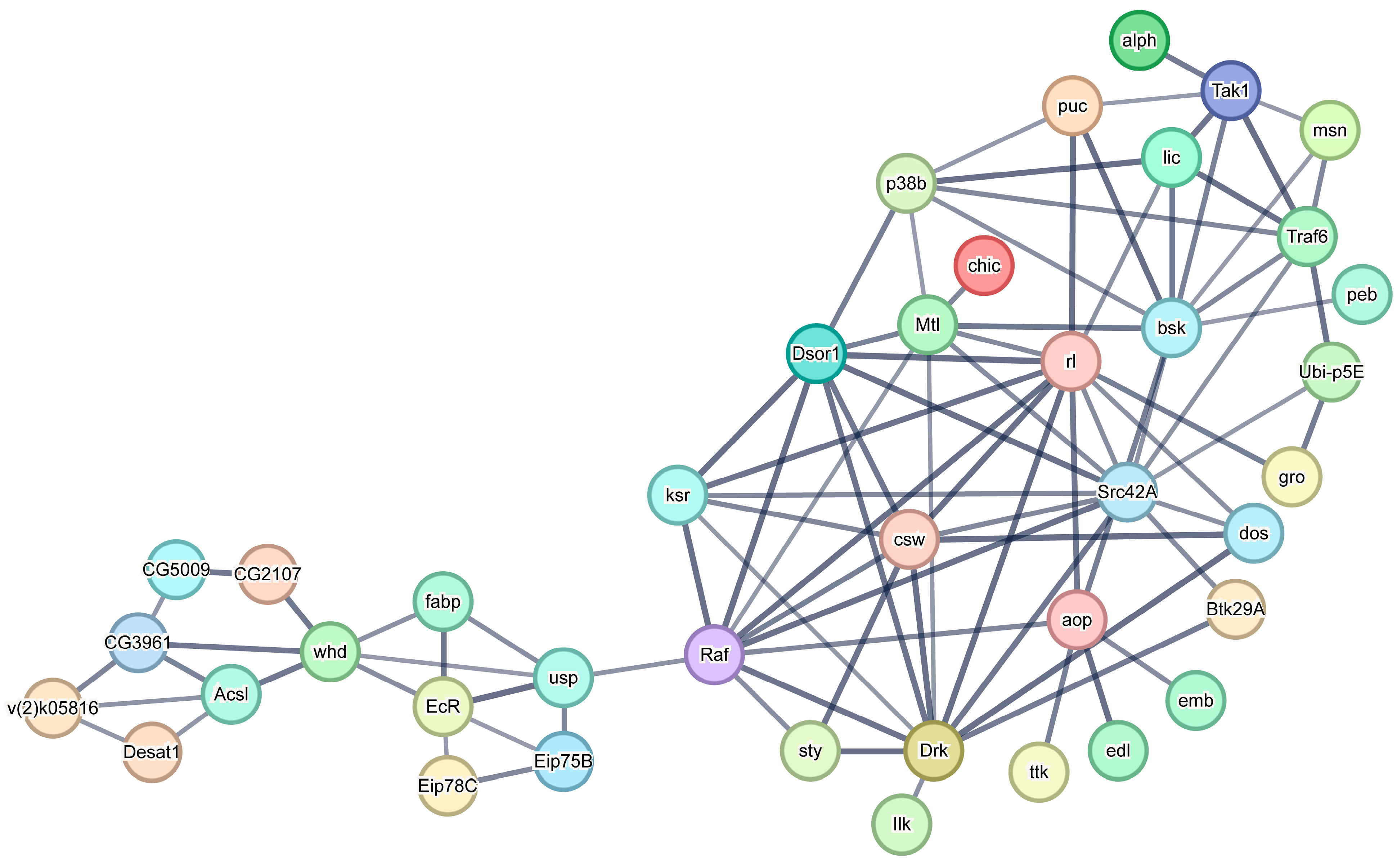
3.8. Construction of a Protein-Protein Interaction Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Le Vay, L. Ecology and Management of Mud Crab Scylla spp. Asian Fish. Sci. 2001, 14, 101–112. [Google Scholar] [CrossRef]
- Islam, M.S.; Kodama, K.; Kurokora, H. Ovarian Development of the Mud Crab Scylla paramamosain in a Tropical Mangrove Swamps, Thailand. J. Sci. Res. 2010, 2, 380–389. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.; Lu, J.; Wu, Q.; Waiho, K.; Aweya, J.J.; Fazhan, H.; Zhang, Y.; Li, S.; Zheng, H.; Lin, F.; et al. Comparative analysis of growth performance between female and male mud crab Scylla paramamosain crablets: Evidences from a four-month successive growth experiment. Aquaculture 2019, 505, 351–362. [Google Scholar] [CrossRef]
- Wan, H.; Zhong, J.; Zhang, Z.; Zou, P.; Wang, Y. Comparative Transcriptome Reveals the Potential Modulation Mechanisms of Spfoxl-2 Affecting Ovarian Development of Scylla paramamosain. Mar. Biotechnol. 2022, 24, 125–135. [Google Scholar] [CrossRef]
- Angell, C. Summary of the proceedings of the seminar on mud crab culture and trade. In Seminar on Mud Crab Culture and Trade, Bay of Bengal Program; Angell, C.A., Ed.; BOBP: Madras, India, 1992; pp. 1–2. [Google Scholar]
- Gu, Z.; He, L. Histological and cytological observation on the development cycle of crab (Eriocheir sinensis) ovary. Oceanol. Limnol. Sin. 1997, 28, 138–145. [Google Scholar]
- Wu, X.; Liu, M.; Pan, J.; Chen, H.; Zeng, C.; Cheng, Y. The ovarian development pattern of pond-reared Chinese mitten crab, Eriocheir sinensis H. Milne-Edwards, 1853. Crustaceana 2017, 90, 449–470. [Google Scholar] [CrossRef]
- Huang, X.; Ye, H.; Huang, H.; Yang, Y.; Gong, J. An insulin-like androgenic gland hormone gene in the mud crab, Scylla paramamosain, extensively expressed and involved in the processes of growth and female reproduction. Gen. Comp. Endocrinol. 2014, 204, 229–238. [Google Scholar] [CrossRef]
- Wu, Q.; Waiho, K.; Huang, Z.; Li, S.; Zheng, H.; Zhang, Y.; Ikhwanuddin, M.; Lin, F.; Ma, H. Growth performance and biochemical composition dynamics of ovary, hepatopancreas and muscle tissues at different ovarian maturation stages of female mud crab, Scylla paramamosain. Aquaculture 2019, 515, 734560. [Google Scholar] [CrossRef]
- Liu, Z.; Wu, X.; Cheng, Y.; Zeng, C.; Yang, X. Ovarian re-maturation following the first spawning in the Chinese mitten crab, Eriocheir sinensis (H. Milne-Edwards). Aquac. Res. 2010, 42, 417–426. [Google Scholar] [CrossRef]
- Wu, X.; Cheng, Y.; Zeng, C.; Wang, C.; Cui, Z. Reproductive performance and offspring quality of the first and the second brood of female swimming crab, Portunus trituberculatus. Aquaculture 2010, 303, 94–100. [Google Scholar] [CrossRef]
- Ren, Y.; Wang, W.; Liu, Z.; Luo, M.; Fu, Y.; Zhang, F.; Ma, C.; Zhao, M.; Chen, W.; Jiang, K.; et al. Insight of vitellogenesis patterns: A comparative analysis of the differences between the primary and secondary vitellogenesis period in the ovary, hepatopancreas, and muscle of mud crab, Scylla paramamosain. Front. Genet. 2022, 13, 965070. [Google Scholar] [CrossRef] [PubMed]
- Valle, D. Vitellogenesis in insects and other groups: A review. Mem. Inst. Oswaldo Cruz. 1993, 88, 1–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramoniam, T. Mechanisms and control of vitellogenesis in crustaceans. Fish. Sci. 2010, 77, 1–21. [Google Scholar] [CrossRef]
- Harrison, K.E. The role of nutrition in maturation, reproduction and embryonic development of decapod crustacean: A review. J. Shellfish Res. 1990, 9, 1–28. [Google Scholar]
- Xu, X.; Ji, W.; Castell, J.; O’Dor, R. Essential fatty acid requirement of the Chinese prawn, Penaeus chinensis. Aquaculture 1994, 127, 29–40. [Google Scholar] [CrossRef]
- Li, S.; Lin, S.; Liu, L. In Studies on Lipid Classes & Fatty Acid Compositions during Ovarian Development of Mud Crab, Scylla serrata (Forskal). J. Xiamen Univ. Nat. Sci. 1994, 33, 109–115. [Google Scholar]
- Alava, V.R.; Quinitio, E.T.; de Pedro, J.B.; Priolo, F.M.P.; Orozco, Z.G.A.; Wille, M. Lipids and fatty acids in wild and pond-reared mud crab Scylla serrata (Forsskål) during ovarian maturation and spawning. Aquac. Res. 2007, 38, 1468–1477. [Google Scholar] [CrossRef] [Green Version]
- Ghazali, A.; Azra, M.N.; Noordin, N.M.; Abol-Munafi, A.B.; Ikhwanuddin, M. Ovarian morphological development and fatty acids profile of mud crab (Scylla olivacea) fed with various diets. Aquaculture 2017, 468, 45–52. [Google Scholar] [CrossRef]
- Djunaidah, I.S.; Wille, M.; Kontara, E.; Sorgeloos, P. Reproductive performance and offspring quality in mud crab (Scylla paramamosain) broodstock fed different diets. Aquac. Int. 2003, 11, 3–15. [Google Scholar] [CrossRef]
- Wu, X.; Cheng, Y.; Zeng, C.; Wang, C.; Yang, X. Reproductive performance and offspring quality of wild-caught and pond-reared swimming crab Portunus trituberculatus broodstock. Aquaculture 2010, 301, 78–84. [Google Scholar] [CrossRef]
- Gusarova, V.; Brodsky, J.L.; Fisher, E.A. Apolipoprotein B100 Exit from the Endoplasmic Reticulum (ER) Is COPII-dependent, and Its Lipidation to Very Low Density Lipoprotein Occurs Post-ER. J. Biol. Chem. 2003, 278, 48051–48058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Z.; Qi, C.; Han, F.; Chen, X.; Qin, C.; Wang, C.; Wang, X.; Qin, J.; Chen, L. Selecting suitable phospholipid source for female Eriocheir sinensis in pre-reproductive phase. Aquaculture 2020, 528, 735610. [Google Scholar] [CrossRef]
- Zeng, X.; Wan, H.; Zhong, J.; Feng, Y.; Zhang, Z.; Wang, Y. Large lipid transfer proteins in hepatopancreas of the mud crab Scylla paramamosain. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 38, 100801. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Zheng, J.; Chen, C.; Wu, K.; Lin, F.; Ning, L.; Rong, H.; Chen, C.; Xiao, F.; Zhang, H.; et al. Differences in lipid accumulation and mobilization in the hepatopancreas and ovary of female mud crab (Scylla paramamosain, Estampador, 1949) during ovarian development. Aquaculture 2023, 564, 739046. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, S.; Qiao, H.; Xiong, Y.; Fu, H.; Zhang, W.; Gong, Y.; Jin, S.; Wu, Y. Transcriptome analysis of five ovarian stages reveals gonad maturation in female Macrobrachium nipponense. BMC Genom. 2021, 22, 1–11. [Google Scholar] [CrossRef]
- Zhong, Y.; Zhao, W.; Tang, Z.; Huang, L.; Zhu, X.; Liang, X.; Yan, A.; Lu, Z.; Yu, Y.; Tang, D.; et al. Comparative transcriptomic analysis of the different developmental stages of ovary in red swamp crayfish Procambarus clarkii. BMC Genom. 2021, 22, 1–20. [Google Scholar] [CrossRef]
- Wouters, R.; Molina, C.; Lavens, P.; Calderón, J. Lipid composition and vitamin content of wild female Litopenaeus vannamei in different stages of sexual maturation. Aquaculture 2001, 198, 307–323. [Google Scholar] [CrossRef] [Green Version]
- Zmora, N.; Trant, J.; Zohar, Y.; Chung, J.S. Molt-inhibiting hormone stimulates vitellogenesis at advanced ovarian developmental stages in the female blue crab, Callinectes sapidus 1: An ovarian stage dependent involvement. Saline Syst. 2009, 5, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Heras, H.; Gonzalez-Baró, M.R.; Pollero, R.J. Lipid and fatty acid composition and energy partitioning during embryo development in the shrimp Macrobrachium borellii. Lipids 2000, 35, 645–651. [Google Scholar] [CrossRef]
- Chen, J.-S.; Sappington, T.; Raikhel, A.S. Extensive Sequence Conservation Among Insect, Nematode, and Vertebrate Vitellogenins Reveals Ancient Common Ancestry. J. Mol. Evol. 1997, 44, 440–451. [Google Scholar] [CrossRef]
- Wei, H.; Ren, Z.; Tang, L.; Yao, H.; Li, X.; Wang, C.; Mu, C.; Shi, C.; Wang, H. JNK signaling pathway regulates the development of ovaries and synthesis of vitellogenin (Vg) in the swimming crab Portunus trituberculatus. Cell Stress Chaperones 2020, 25, 441–453. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Nan, G. The Mitogen-Activated Protein Kinase (MAPK) Signaling Pathway as a Discovery Target in Stroke. J. Mol. Neurosci. 2016, 59, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Rosette, C.; Karin, M. Ultraviolet Light and Osmotic Stress: Activation of the JNK Cascade Through Multiple Growth Factor and Cytokine Receptors. Science 1996, 274, 1194–1197. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shi, C.; Kong, M.; Mu, C.; Wei, H.; Wang, C. Cloning and expression of a transcription factor activator protein-1 member identified from the swimming crab Portunus trituberculatus. Cell Stress Chaperones 2018, 23, 1275–1282. [Google Scholar] [CrossRef]
- Zhao, J.C.; Wang, Y.L.; Li, Q.; Zhu, M.; Sun, W.J.; Wu, T.M.; Wang, Q.; He, L. Molecular cloning and characterization of p38 gene in the Chinese Mitten Crab, Eriocheir sinensis. Aquac. Res. 2014, 47, 1353–1363. [Google Scholar] [CrossRef]
- Ma, A.; Wang, Y.; Zou, Z.; Fu, M.; Lin, P.; Zhang, Z. Erk2 in Ovarian Development of Green Mud Crab Scylla paramamosain. DNA Cell Biol. 2012, 31, 1233–1244. [Google Scholar] [CrossRef] [Green Version]
- Feng, Q.-M.; Liu, M.-M.; Cheng, Y.-X.; Wu, X.-G. Comparative proteomics elucidates the dynamics of ovarian development in the Chinese mitten crab Eriocheir sinensis. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 40, 100878. [Google Scholar] [CrossRef]
- Fu, C.; Liu, L.; Li, F. Acetate alters the process of lipid metabolism in rabbits. Animal 2018, 12, 1895–1902. [Google Scholar] [CrossRef]
- Engelman, J.A.; Berg, A.H.; Lewis, R.Y.; Lin, A.; Lisanti, M.P.; Scherer, P.E. Constitutively Active Mitogen-activated Protein Kinase Kinase 6 (MKK6) or Salicylate Induces Spontaneous 3T3-L1 Adipogenesis. J. Biol. Chem. 1999, 274, 35630–35638. [Google Scholar] [CrossRef] [Green Version]
- Lei, Y.; Li, F.; Mortimer, M.; Li, Z.; Peng, B.-X.; Li, M.; Guo, L.-H.; Zhuang, G. Antibiotics disrupt lipid metabolism in zebrafish (Danio rerio) larvae and 3T3-L1 preadipocytes. Sci. Total. Environ. 2023, 858, 159755. [Google Scholar] [CrossRef]
- Du, J.; Xiang, X.; Xu, D.; Zhang, J.; Fang, W.; Xu, W.; Mai, K.; Ai, Q. FXR, a Key Regulator of Lipid Metabolism, Is Inhibited by ER Stress-Mediated Activation of JNK and p38 MAPK in Large Yellow Croakers (Larimichthys crocea) Fed High Fat Diets. Nutrients 2021, 13, 4343. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yu, Z.; Gao, P.; Yin, D. Multigenerational effects of perfluorooctanoic acid on lipid metabolism of Caenorhabditis elegans and its potential mechanism. Sci. Total. Environ. 2019, 703, 134762. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, S.; Poidevin, M.; Lemaitre, B. The Drosophila MAPK p38c Regulates Oxidative Stress and Lipid Homeostasis in the Intestine. PLoS Genet. 2014, 10, e1004659. [Google Scholar] [CrossRef] [PubMed]
- Nidhina, P.A.H.; Poulose, N.; Gopalakrishnapillai, A. Vanillin induces adipocyte differentiation in 3T3-L1 cells by activating extracellular signal regulated kinase 42/44. Life Sci. 2011, 88, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, A.S.; Shen, W.-J.; Muliro, K.; Patel, S.; Souza, S.C.; Roth, R.A.; Kraemer, F.B. Stimulation of Lipolysis and Hormone-sensitive Lipase via the Extracellular Signal-regulated Kinase Pathway. J. Biol. Chem. 2001, 276, 45456–45461. [Google Scholar] [CrossRef] [Green Version]
- Desvergne, B.; Wahli, W. Peroxisome Proliferator-Activated Receptors: Nuclear Control of Metabolism. Endocr. Rev. 1999, 20, 649–688. [Google Scholar] [CrossRef] [Green Version]
- Berger, J.; Moller, D.E. The Mechanisms of Action of PPARs. Annu. Rev. Med. 2002, 53, 409–435. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.-H.; Olson, P.; Evans, R.M. Minireview: Lipid Metabolism, Metabolic Diseases, and Peroxisome Proliferator-Activated Receptors. Endocrinology 2003, 144, 2201–2207. [Google Scholar] [CrossRef] [Green Version]
- Desvergne, B.; Michalik, L.; Wahli, W. Transcriptional Regulation of Metabolism. Physiol. Rev. 2006, 86, 465–514. [Google Scholar] [CrossRef]
- Müller-Brüsselbach, S.; Ebrahimsade, S.; Jäkel, J.; Eckhardt, J.; Rapp, U.; Peters, J.; Moll, R.; Müller, R. Growth of transgenic RAF-induced lung adenomas is increased in mice with a disrupted PPARβ/δ gene. Int. J. Oncol. 2007, 31, 607–611. [Google Scholar] [CrossRef]
- Viswakarma, N.; Jia, Y.; Bai, L.; Vluggens, A.; Borensztajn, J.; Xu, J.; Reddy, J.K. Coactivators in PPAR-Regulated Gene Expression. PPAR Res. 2010, 2010, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chawla, A.; Repa, J.J.; Evans, R.M.; Mangelsdorf, D.J. Nuclear Receptors and Lipid Physiology: Opening the X-Files. Science 2001, 294, 1866–1870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bookout, A.L.; Jeong, Y.; Downes, M.; Yu, R.T.; Evans, R.M.; Mangelsdorf, D.J. Anatomical Profiling of Nuclear Receptor Expression Reveals a Hierarchical Transcriptional Network. Cell 2006, 126, 789–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamauchi, T.; Kamon, J.; Waki, H.; Murakami, K.; Motojima, K.; Komeda, K.; Ide, T.; Kubota, N.; Terauchi, Y.; Tobe, K.; et al. The Mechanisms by Which Both Heterozygous Peroxisome Proliferator-activated Receptor γ (PPARγ) Deficiency and PPARγ Agonist Improve Insulin Resistance. J. Biol. Chem. 2001, 276, 41245–41254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forman, B.M.; Tontonoz, P.; Chen, J.; Brun, R.P.; Spiegelman, B.M.; Evans, R. 15-Deoxy-Δ12,14-Prostaglandin J2 is a ligand for the adipocyte determination factor PPARγ. Cell 1995, 83, 803–812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonet, M.L.; Ribot, J.; Palou, A. Lipid metabolism in mammalian tissues and its control by retinoic acid. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2012, 1821, 177–189. [Google Scholar] [CrossRef]
- Blommer, J.; Fischer, M.C.; Olszewski, A.R.; Katzenberger, R.J.; Ganetzky, B.; Wassarman, D.A. Ketogenic diet reduces early mortality following traumatic brain injury in drosophila via the PPARγ ortholog Eip75B. PLoS ONE 2021, 16, e0258873. [Google Scholar] [CrossRef]
- Hong, J.-W.; Park, K.W. Further understanding of fat biology: Lessons from a fat fly. Exp. Mol. Med. 2010, 42, 12–20. [Google Scholar] [CrossRef] [Green Version]
- King-Jones, K.; Thummel, C.S. Nuclear receptors—A perspective from Drosophila. Nat. Rev. Genet. 2005, 6, 311–323. [Google Scholar] [CrossRef]
- Zipper, L.; Jassmann, D.; Burgmer, S.; Görlich, B.; Reiff, T. Ecdysone steroid hormone remote controls intestinal stem cell fate decisions via the PPARγ-homolog Eip75B in Drosophila. Elife 2020, 9, e55795. [Google Scholar] [CrossRef]
- Yao, T.-P.; Segraves, W.A.; Oro, A.E.; McKeown, M.; Evans, R.M. Drosophila ultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell 1992, 71, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H.E.; Stunnenberg, H.G.; Stewart, A.F. Heterodimerization of the Drosophila ecdysone receptor with retinoid X receptor and ultraspiracle. Nature 1993, 362, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Palanker, L.; Necakov, A.S.; Sampson, H.M.; Ni, R.; Hu, C.; Thummel, C.S.; Krause, H.M. Dynamic regulation of Drosophila nuclear receptor activity in vivo. Development 2006, 133, 3549–3562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwema, T.; Chaumot, A.; Studer, R.A.; Robinson-Rechavi, M.; Billas, I.M.; Moras, D.; Laudet, V.; Bonneton, F. Structural and Evolutionary Innovation of the Heterodimerization Interface between USP and the Ecdysone Receptor ECR in Insects. Mol. Biol. Evol. 2009, 26, 753–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Techa, S.; Chung, J.S. Ecdysone and retinoid-X receptors of the blue crab, Callinectes sapidus: Cloning and their expression patterns in eyestalks and Y-organs during the molt cycle. Gene 2013, 527, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, P.M.; Durica, D.; Washington, T. RXR isoforms and endogenous retinoids in the fiddler crab, Uca pugilator. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2008, 151, 602–614. [Google Scholar] [CrossRef]
- Gong, J.; Huang, C.; Shu, L.; Bao, C.; Huang, H.; Ye, H.; Zeng, C.; Li, S. The retinoid X receptor from mud crab: New insights into its roles in ovarian development and related signaling pathway. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Girish, B.P.; Swetha, C.H.; Reddy, P.S. Expression of RXR, EcR, E75 and VtG mRNA levels in the hepatopancreas and ovary of the freshwater edible crab, Oziothelphusa senex senex (Fabricius, 1798) during different vitellogenic stages. Sci. Nat. 2015, 102, 20. [Google Scholar] [CrossRef]
- McCubrey, J.A.; Steelman, L.S.; Chappell, W.H.; Abrams, S.L.; Wong, E.W.T.; Chang, F.; Lehmann, B.; Terrian, D.M.; Milella, M.; Tafuri, A.; et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2007, 1773, 1263–1284. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Liu, W.-Z.; Liu, T.; Feng, X.; Yang, N.; Zhou, H.-F. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J. Recept. Signal Transduct. 2015, 35, 600–604. [Google Scholar] [CrossRef]
- Blenis, J. Growth-regulated signal transduction by the MAP kinases and RSKs. Cancer Cells 1991, 3, 445–449. [Google Scholar] [PubMed]
- Bruder, J.T.; Heidecker, G.; Rapp, U.R. Serum-, TPA-, and Ras-induced expression from Ap-1/Ets-driven promoters requires Raf-1 kinase. Genes Dev. 1992, 6, 545–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabian, J.R.; Morrison, D.K.; Daar, I. Requirement for Raf and MAP kinase function during the meiotic maturation of Xenopus oocytes. Int. J. Biochem. Cell Biol. 1993, 122, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Grunt, T.W. Interacting Cancer Machineries: Cell Signaling, Lipid Metabolism, and Epigenetics. Trends Endocrinol. Metab. 2018, 29, 86–98. [Google Scholar] [CrossRef]
- Yang, S.; Park, Y.; Kim, J.I.; Lee, Y.; Lee, T.; Jang, B. LY3009120, a pan-Raf kinase inhibitor, inhibits adipogenesis of 3T3-L1 cells by controlling the expression and phosphorylation of C/EBP-α, PPAR-γ, STAT-3, FAS, ACC, perilipin A, and AMPK. Int. J. Mol. Med. 2018, 42, 3477–3484. [Google Scholar] [CrossRef] [Green Version]
- Krey, G.; Braissant, O.; L’Horset, F.; Kalkhoven, E.; Perroud, M.; Parker, M.G.; Wahli, W. Fatty Acids, Eicosanoids, and Hypolipidemic Agents Identified as Ligands of Peroxisome Proliferator-Activated Receptors by Coactivator-Dependent Receptor Ligand Assay. Mol. Endocrinol. 1997, 11, 779–791. [Google Scholar] [CrossRef]
- Hashimoto, T.; Cook, W.S.; Qi, C.; Yeldandi, A.V.; Reddy, J.K.; Rao, M.S. Defect in Peroxisome Proliferator-activated Receptor α-inducible Fatty Acid Oxidation Determines the Severity of Hepatic Steatosis in Response to Fasting. J. Biol. Chem. 2000, 275, 28918–28928. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-X.; Lee, C.-H.; Tiep, S.; Yu, R.T.; Ham, J.; Kang, H.; Evans, R.M. Peroxisome-Proliferator-Activated Receptor δ Activates Fat Metabolism to Prevent Obesity. Cell 2003, 113, 159–170. [Google Scholar] [CrossRef] [Green Version]
- Rosen, E.D.; Spiegelman, B.M. PPARγ: A Nuclear Regulator of Metabolism, Differentiation, and Cell Growth. J. Biol. Chem. 2001, 276, 37731–37734. [Google Scholar] [CrossRef] [Green Version]
- Guo, D.; Liu, W.; Zhang, X.; Zhao, M.; Zhu, B.; Hou, T.; He, H. Duck Egg White–Derived Peptide VSEE (Val-Ser-Glu-Glu) Regulates Bone and Lipid Metabolisms by Wnt/β-Catenin Signaling Pathway and Intestinal Microbiota. Mol. Nutr. Food Res. 2019, 63, e1900525. [Google Scholar] [CrossRef]
- Takada, I.; Kouzmenko, A.P.; Kato, S. Wnt and PPARγ signaling in osteoblastogenesis and adipogenesis. Nat. Rev. Rheumatol. 2009, 5, 442–447. [Google Scholar] [CrossRef] [PubMed]
- Mulholland, D.J.; Dedhar, S.; Coetzee, G.A.; Nelson, C.C. Interaction of Nuclear Receptors with the Wnt/β-Catenin/Tcf Signaling Axis: Wnt You Like to Know? Endocr. Rev. 2005, 26, 898–915. [Google Scholar] [CrossRef] [PubMed]
| Sample ID | Developmental Period (F/S) | Developmental Stage |
|---|---|---|
| F_OV_1 | First ovarian maturation period (FOMP) | Stage I |
| F_OV_2 | First ovarian maturation period (FOMP) | Stage II |
| F_OV_3 | First ovarian maturation period (FOMP) | Stage III |
| F_OV_4 | First ovarian maturation period (FOMP) | Stage IV |
| F_OV_5 | First ovarian maturation period (FOMP) | Stage V |
| S_OV_3 | Second ovarian maturation period (SOMP) | Stage III |
| S_OV_4 | Second ovarian maturation period (SOMP) | Stage IV |
| S_OV_5 | Second ovarian maturation period (SOMP) | Stage V |
| Type | Unigene | Transcript |
|---|---|---|
| Total number | 81,893 | 132,744 |
| Total base | 92,837,704 | 154,178,017 |
| Largest length (bp) | 20,317 | 20,317 |
| Smallest length (bp) | 201 | 201 |
| Average length (bp) | 1133.65 | 1161.47 |
| N50 length (bp) | 2111 | 2758 |
| Fragment mapped percent (%) | 71.497 | 83.346 |
| GC percent (%) | 46.62 | 46.75 |
| BUSCO (%) | 96.6 | 98.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, Y.; Wang, W.; Fu, Y.; Liu, Z.; Zhao, M.; Xu, L.; Zhan, T.; Huang, T.; Luo, M.; Chen, W.; et al. Comparative Transcriptome Analysis Identifies MAPK Signaling Pathway Associated with Regulating Ovarian Lipid Metabolism during Vitellogenesis in the Mud Crab, Scylla paramamosain. Fishes 2023, 8, 145. https://doi.org/10.3390/fishes8030145
Ren Y, Wang W, Fu Y, Liu Z, Zhao M, Xu L, Zhan T, Huang T, Luo M, Chen W, et al. Comparative Transcriptome Analysis Identifies MAPK Signaling Pathway Associated with Regulating Ovarian Lipid Metabolism during Vitellogenesis in the Mud Crab, Scylla paramamosain. Fishes. 2023; 8(3):145. https://doi.org/10.3390/fishes8030145
Chicago/Turabian StyleRen, Yuanhao, Wei Wang, Yin Fu, Zhiqiang Liu, Ming Zhao, Likun Xu, Tianyong Zhan, Ting Huang, Minghao Luo, Wei Chen, and et al. 2023. "Comparative Transcriptome Analysis Identifies MAPK Signaling Pathway Associated with Regulating Ovarian Lipid Metabolism during Vitellogenesis in the Mud Crab, Scylla paramamosain" Fishes 8, no. 3: 145. https://doi.org/10.3390/fishes8030145
APA StyleRen, Y., Wang, W., Fu, Y., Liu, Z., Zhao, M., Xu, L., Zhan, T., Huang, T., Luo, M., Chen, W., Ma, C., Zhang, F., Jiang, K., & Ma, L. (2023). Comparative Transcriptome Analysis Identifies MAPK Signaling Pathway Associated with Regulating Ovarian Lipid Metabolism during Vitellogenesis in the Mud Crab, Scylla paramamosain. Fishes, 8(3), 145. https://doi.org/10.3390/fishes8030145






