Skin and Blood Microbial Signatures of Sedentary and Migratory Trout (Salmo trutta) of the Kerguelen Islands
Abstract
1. Introduction
2. Materials and Methods
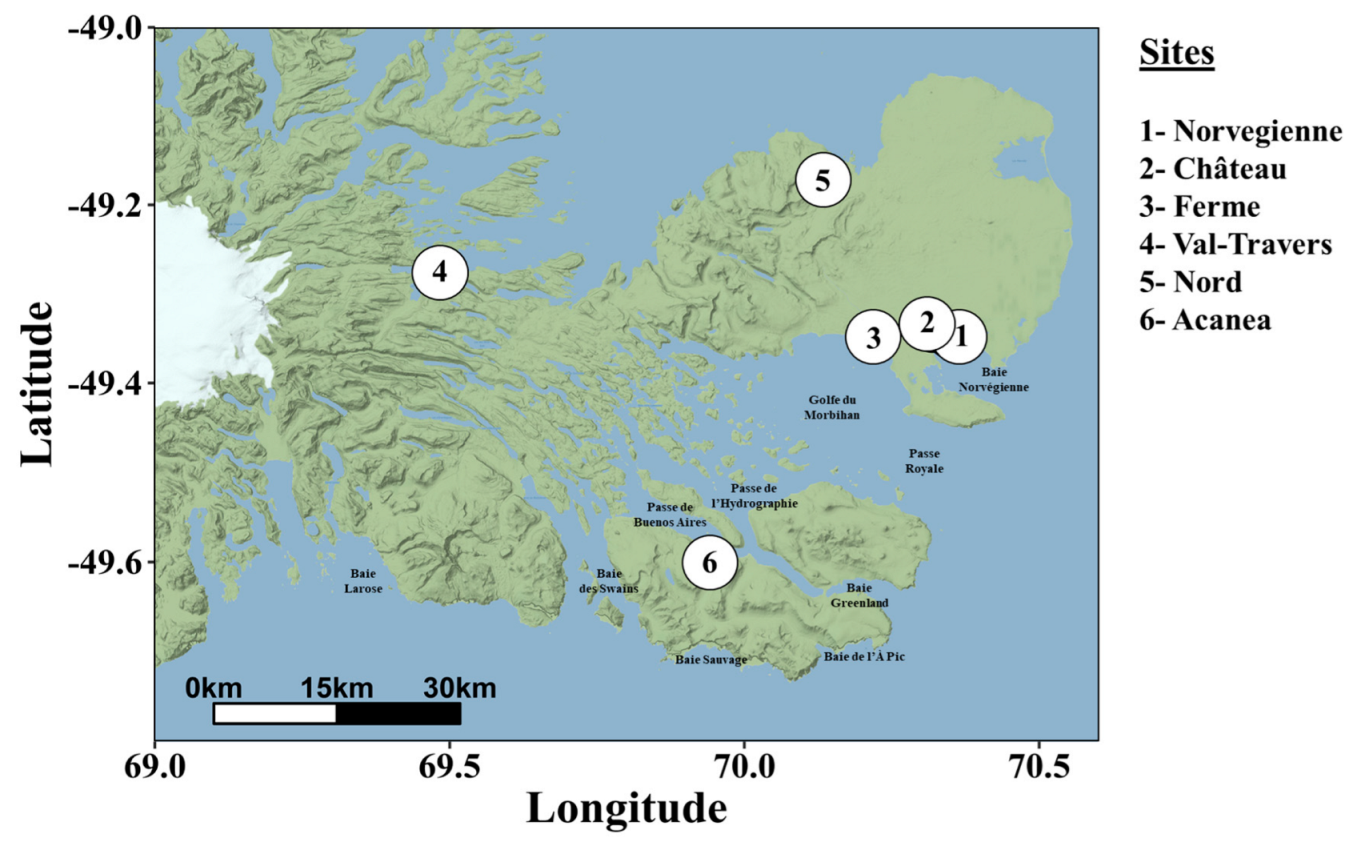
2.1. Sample Collection
2.2. DNA Extraction, Preprocessing, and Sequencing
2.3. 16S rRNA Data Processing
2.4. Statistical Analysis
3. Results
3.1. General View of the Mucosal and Circulating Bacterial Microbiome
3.2. Comparative Analysis between Sedentary and Migratory S. trutta
3.3. Site-Specific Variations
3.4. Functional Analysis of the CmDNA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, H.; Ma, Y.; Liu, Z.; Li, J.; Li, X.; Yang, F.; Qiu, M. Circulating microbiome DNA: An emerging paradigm for cancer liquid biopsy. Cancer Lett. 2021, 521, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic—Implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Cabillon, N.A.R.; Lazado, C.C. Mucosal barrier functions of fish under changing environmental conditions. Fishes 2019, 4, 2. [Google Scholar] [CrossRef]
- Lowrey, L.; Woodhams, D.C.; Tacchi, L.; Salinas, I. Topographical mapping of the rainbow trout (Oncorhynchus mykiss) microbiome reveals a diverse bacterial community with antifungal properties in the skin. Appl. Environ. Microbiol. 2015, 81, 6915–6925. [Google Scholar] [CrossRef] [PubMed]
- Tarnecki, A.M.; Brennan, N.P.; Schloesser, R.W.; Rhody, N.R. Shifts in the skin-associated microbiota of hatchery-reared common snook Centropomus undecimalis during acclimation to the wild. Microb. Ecol. 2019, 77, 770–781. [Google Scholar] [CrossRef]
- Uren Webster, T.M.; Consuegra, S.; Hitchings, M.; Garcia de Leaniz, C. Interpopulation variation in the Atlantic Salmon microbiome reflects environmental and genetic diversity. Appl. Environ. Microbiol. 2018, 84, e00691-18. [Google Scholar] [CrossRef]
- Uren Webster, T.M.; Rodriguez-Barreto, D.; Castaldo, G.; Gough, P.; Consuegra, S.; de Leaniz, C.G. Environmental plasticity and colonisation history in the Atlantic salmon microbiome: A translocation experiment. Mol. Ecol. 2020, 29, 886–898. [Google Scholar] [CrossRef]
- Dehler, C.E.; Secombes, C.J.; Martin, S.A. Seawater transfer alters the intestinal microbiota profiles of Atlantic salmon (Salmo salar L.). Sci. Rep. 2017, 7, 13877. [Google Scholar] [CrossRef]
- Rudi, K.; Angell, I.L.; Pope, P.B.; Vik, J.O.; Sandve, S.R.; Snipen, L.G. Stable core gut microbiota across the freshwater-to-saltwater transition for farmed Atlantic salmon. Appl. Environ. Microbiol. 2018, 84, e01974-17. [Google Scholar] [CrossRef]
- Whittle, E.; Leonard, M.O.; Harrison, R.; Gant, T.W.; Tonge, D.P. Multi-method characterization of the human circulating microbiome. Front. Microbiol. 2019, 9, 3266. [Google Scholar] [CrossRef]
- Mandel, P.; Metais, P. Nuclear Acids in Human Blood Plasma. C. R. Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar] [PubMed]
- Blauwkamp, T.A.; Thair, S.; Rosen, M.J.; Blair, L.; Lindner, M.S.; Vilfan, I.D.; Kawli, T.; Christians, F.C.; Venkatasubrahmanyam, S.; Wall, G.D.; et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat. Microbiol. 2019, 4, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.K.; Blauwkamp, T.A.; Kertesz, M.; Bercovici, S.; Truong, C.; Banaei, N. Liquid biopsy for infectious diseases: Sequencing of cell-free plasma to detect pathogen DNA in patients with invasive fungal disease. Diagn. Microbiol. Infect. Dis. 2018, 92, 210–213. [Google Scholar] [CrossRef]
- Kowarsky, M.; Camunas-Soler, J.; Kertesz, M.; De Vlaminck, I.; Koh, W.; Pan, W.; Martin, L.; Neff, N.F.; Okamoto, J.; Wong, R.J.; et al. Numerous uncharacterized and highly divergent microbes which colonize humans are revealed by circulating cell-free DNA. Proc. Natl. Acad. Sci. USA 2017, 114, 9623–9628. [Google Scholar] [CrossRef] [PubMed]
- Amar, J.; Lange, C.; Payros, G.; Garret, C.; Chabo, C.; Lantieri, O.; Courtney, M.; Marre, M.; Charles, M.A.; Balkau, B.; et al. Blood microbiota dysbiosis is associated with the onset of cardiovascular events in a large general population: The D.E.S.I.R. study. PLoS ONE 2013, 8, e54461. [Google Scholar] [CrossRef]
- Cho, E.J.; Leem, S.; Kim, S.A.; Yang, J.; Lee, Y.B.; Kim, S.S.; Cheong, J.Y.; Cho, S.W.; Kim, J.W.; Kim, S.-M.; et al. Circulating microbiota-based metagenomic signature for detection of hepatocellular carcinoma. Sci. Rep. 2019, 9, 7536. [Google Scholar] [CrossRef]
- Huang, C.C.; Du, M.; Wang, L. Bioinformatics analysis for circulating cell-free DNA in cancer. Cancers 2019, 11, 805. [Google Scholar] [CrossRef]
- Paisse, S.; Valle, C.; Servant, F.; Courtney, M.; Burcelin, R.; Amar, J.; Lelouvier, B. Comprehensive description of blood microbiome from healthy donors assessed by 16S targeted metagenomic sequencing. Transfusion 2016, 56, 1138–1147. [Google Scholar] [CrossRef]
- Potgieter, M.; Bester, J.; Kell, D.B.; Pretorius, E. The dormant blood microbiome in chronic, inflammatory diseases. FEMS Microbiol. Rev. 2015, 39, 567–591. [Google Scholar] [CrossRef]
- Auguste, M.; Lasa, A.; Pallavicini, A.; Gualdi, S.; Vezzulli, L.; Canesi, L. Exposure to TiO2 nanoparticles induces shifts in the microbiota composition of Mytilus galloprovincialis hemolymph. Sci. Total Environ. 2019, 670, 129–137. [Google Scholar] [CrossRef]
- Lokmer, A.; Wegner, M.K. Hemolymph microbiome of Pacific oysters in response to temperature, temperature stress and infection. ISME J. 2015, 9, 670–682. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, L.G.E.; Leray, M.; O’Dea, A.; Yuen, B.; Peixoto, R.S.; Pereira, T.J.; Bik, H.M.; Coil, D.A.; Duffy, J.E.; Herre, E.A.; et al. Host-associated microbiomes drive structure and function of marine ecosystems. PLoS Biol. 2019, 17, e3000533. [Google Scholar] [CrossRef] [PubMed]
- Rynkiewicz, E.C.; Hemmerich, C.; Rusch, D.B.; Fuqua, C.; Clay, K. Concordance of 605 bacterial communities of two tick species and blood of their shared rodent host. Mol. Ecol. 2015, 24, 2566–2579. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.J.; Cunha, F.; Vieira-Neto, A.; Bicalho, R.C.; Lima, S.; Bicalho, M.L.; Galvão, K.N. Blood as a route of transmission of uterine pathogens from the gut to the uterus in cows. Microbiome 2017, 5, 109. [Google Scholar] [CrossRef]
- Scarsella, E.; Sandri, M.; Monego, S.D.; Licastro, D.; Stefanon, B. Blood Microbiome: A New Marker of Gut Microbial Population in Dogs? Vet. Sci. 2020, 7, 198. [Google Scholar] [CrossRef]
- Hyun, H.; Lee, M.S.; Park, I.; Ko, H.S.; Yun, S.; Jang, D.H.; Kim, S.; Kim, H.; Kang, J.H.; Lee, J.H.; et al. Analysis of Porcine Model of Fecal-Induced Peritonitis Reveals the Tropism of Blood Microbiome. Front. Cell. Infect. Microbiol. 2021, 11, 676650. [Google Scholar] [CrossRef]
- Scarsella, E.; Zecconi, A.; Cintio, M.; Stefanon, B. Characterization of microbiome on feces, blood and milk in dairy cows with different milk leucocyte pattern. Animals 2021, 11, 1463. [Google Scholar] [CrossRef] [PubMed]
- Vientós-Plotts, A.I.; Ericsson, A.C.; Rindt, H.; Reinero, C.R. Blood cultures and blood microbiota analysis as surrogates for bronchoalveolar lavage fluid analysis in dogs with bacterial pneumonia. BMC Vet. Res. 2021, 17, 129. [Google Scholar] [CrossRef]
- Tilahun, Y.; Pinango, J.Q.; Johnson, F.; Lett, C.; Smith, K.; Gipson, T.; McCallum, M.; Hoyt, P.; Tritt, A.; Yadav, A.; et al. Transcript and blood-microbiome analysis towards a blood diagnostic tool for goats affected by Haemonchus contortus. Sci. Rep. 2022, 12, 5362. [Google Scholar] [CrossRef]
- Tarnecki, A.M.; Patterson, W.F.; Arias, C.R. Microbiota of wild-caught red snapper Lutjanus campechanus. BMC Microbiol. 2016, 16, 245. [Google Scholar] [CrossRef]
- Kelly, C.; Salinas, I. Under pressure: Interactions between commensal microbiota and the teleost immune system. Front. Immunol. 2017, 8, 559. [Google Scholar] [CrossRef]
- Pratte, Z.A.; Besson, M.; Hollman, R.D.; Stewart, F.J. The gills of reef fish support a distinct microbiome influenced by host-specific factors. Appl. Environ. Microbiol. 2018, 84, e00063-18. [Google Scholar] [CrossRef] [PubMed]
- Féral, J.-P.; Saucède, T.; Poulin, E.; Marschal, C.; Marty, G.; Roca, J.C.; Motreuil, S.; Beurier, J.-P. PROTEKER: Implementation of a submarine observatory at the Kerguelen islands (Southern Ocean). Underw. Technol. 2016, 34, 3–10. [Google Scholar] [CrossRef]
- Meredith, M.; Sommerkorn, M.; Cassotta, S.; Derksen, C.; Ekaykin, A.; Hollowed, A.; Kofinas, G.; Mackintosh, A.; Melbourne-Thomas, J.; Muelbert, M.M.C.; et al. Polar Regions. In IPCC Special Report on the Ocean and Cryosphere in a Changing Climate; Pörtner, H.-O., Roberts, D.C., Masson-Delmotte, V., Zhai, P., Eds.; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2019; pp. 203–320. [Google Scholar]
- Lecomte, F.; Beall, E.; Chat, J.; Davaine, P.; Gaudin, P. The complete history of salmonid introductions in the Kerguelen Islands, Southern Ocean. Polar Biol. 2013, 36, 457–475. [Google Scholar] [CrossRef]
- Dodson, J.J.; Aubin-Horth, N.; Thériault, V.; Páez, D.J. The evolutionary ecology of alternative migratory tactics in salmonid fishes. Biol. Rev. 2013, 88, 602–625. [Google Scholar] [CrossRef]
- Labonne, J.; Vignon, M.; Prévost, E.; Lecomte, F.; Dodson, J.J. Invasion Dynamics of a Fish-Free Landscape by Brown Trout (Salmo trutta). PLoS ONE 2013, 8, e71052. [Google Scholar] [CrossRef]
- Marandel, L.; Gaudin, P.; Guéraud, F.; Glise, S.; Herman, A.; Plagues-Juan, E.; Véron, V.; Panserat, S.; Labonne, J. A reassessment of the carnivorous status of salmonids: Hepatic glucokinase is expressed in wild fish in Kerguelen Islands. Sci. Total Environ. 2018, 612, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Caza, F.; Joly de Boissel, P.G.; Villemur, R.; Betoulle, S.; St-Pierre, Y. Liquid biopsies for omics-based analysis in sentinel mussels. PLoS ONE 2019, 14, e0223525. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Wang, L.-Y.; Ke, W.J.; Sun, X.-B.; Liu, J.-F.; Gu, J.-D.; Mu, B.-Z. Comparison of bacterial community in aqueous and oil phases of water-flooded petroleum reservoirs using pyrosequencing and clone library approaches. Appl. Microbiol. Biotechnol. 2014, 98, 4209–4221. [Google Scholar] [CrossRef]
- Vilo, C.; Dong, Q. Evaluation of the RDP classifier accuracy using 16S rRNA gene variable regions. Metagenomics 2012, 1, 104303. [Google Scholar] [CrossRef]
- Hung, Y.M.; Lyu, W.N.; Tsai, M.L.; Liu, C.L.; Lai, L.C.; Tsai, M.H.; Chuang, E.Y. To compare the performance of prokaryotic taxonomy classifiers using curated 16S full-length rRNA sequences. Comput. Biol. Med. 2022, 145, 105416. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; MicrobiomeMarker: Microbiome Biomarker Analysis. R Package Version 0.0. 1.9000. Available online: https://github.com/yiluheihei/microbiomeMarker (accessed on 23 October 2022).
- Ssekagiri, A.; Sloan, W.; Ijaz, U.Z.; MicrobiomeSeq: An R Package for Analysis of Microbial Communities in an Environmental Context. ISCB Africa ASBCB Conference. Available online: https://github.com/umerijaz/microbiomeSeq (accessed on 22 February 2019).
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Solymos, P.; Stevens, M.H.; Wagner, H.; Vegan: Community Ecology Package. R Package Version 2.5-7. Available online: https://cran.r-project.org/package=vegan (accessed on 11 October 2022).
- Narayan, N.R.; Weinmaier, T.; Laserna-Mendieta, E.J.; Claesson, M.J.; Shanahan, F.; Dabbagh, K.; Iwai, S.; DeSantis, T.Z. Piphillin predicts metagenomic composition and dynamics from DADA2-corrected 16S rDNA sequences. BMC Genom. 2020, 21, 56. [Google Scholar]
- Cao, S.; Zhang, W.; Ding, W.; Wang, M.; Fan, S.; Yang, B.; Mcminn, A.; Wang, M.; Xie, B.-B.; Qin, Q.L.; et al. Structure and function of the Arctic and Antarctic marine microbiota as revealed by metagenomics. Microbiome 2020, 8, 47. [Google Scholar] [CrossRef]
- Hedaoo, M.; Gore, D.; Fadnavis, S.; Dange, M.; Soni, M.A.; Kopulwar, A.P. Bioinformatics approach in speciation of oil degrading uncultured bacterium and its frequency recording. J. Pharm. Res. 2018, 12, 628–635. [Google Scholar]
- Koner, S.; Chen, J.S.; Hsu, B.M.; Tan, C.W.; Fan, C.W.; Chen, T.H.; Hussain, B.; Nagarajan, V. Assessment of Carbon Substrate Catabolism Pattern and Functional Metabolic Pathway for Microbiota of Limestone Caves. Microorganisms 2021, 9, 1789. [Google Scholar] [CrossRef]
- Peng, W.; Huang, J.; Yang, J.; Zhang, Z.; Yu, R.; Fayyaz, S.; Zhang, S.; Qin, Y.-H. Integrated 16S rRNA sequencing, metagenomics, and metabolomics to characterize gut microbial composition, function, and fecal metabolic phenotype in non-obese type 2 diabetic Goto-Kakizaki rats. Front. Microbiol. 2020, 10, 3141. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, M.A. Alterations in lipid metabolism accompanying smoltification and seawater adaptation of salmonid fish. Aquaculture 1989, 82, 191–203. [Google Scholar] [CrossRef]
- Lokesh, J.; Kiron, V. Transition from freshwater to seawater reshapes the skin-associated microbiota of Atlantic salmon. Sci. Rep. 2016, 6, 19707. [Google Scholar] [CrossRef] [PubMed]
- Fogarty, C.; Burgess, C.M.; Cotter, P.D.; Cabrera-Rubio, R.; Whyte, P.; Smyth, C.; Bolton, D.J. Diversity and composition of the gut microbiota of Atlantic salmon (Salmo salar) farmed in Irish waters. J. Appl. Microbiol. 2019, 127, 648–657. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.; Abdelhafiz, Y.A.A.; Abreu, H.; Silva, J.; Valente, L.M.; Viswanath, K. Gracilaria gracilis and Nannochloropsis oceanica, singly or in combination, in diets alter the intestinal microbiota of European seabass (Dicentrarchus labrax). Front. Mar. Sci. 2022, 9, 1001942. [Google Scholar] [CrossRef]
- Ferchiou, S.; Caza, F.; Villemur, R.; Betoulle, S.; St-Pierre, Y. Species- and site-specific circulating bacterial DNA in Subantarctic sentinel mussels Aulacomya atra and Mytilus platensis. Sci. Rep. 2022, 12, 9547. [Google Scholar] [CrossRef]
- Chiu, C.Y.; Miller, S.A. Clinical metagenomics. Nat. Rev. Genet. 2019, 20, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Chiu, K.-P.; Yu, A.L. Application of cell-free DNA sequencing in characterization of bloodborne microbes and the study of microbe-disease interactions. PeerJ 2019, 7, e7426. [Google Scholar] [CrossRef]
- Guo, W.; Zhang, Y.; Guo, S.; Mei, Z.; Liao, H.; Dong, H.; Wu, K.; Ye, H.; Zhang, Y.; Zhu, Y.; et al. Tumor microbiome contributes to an aggressive phenotype in the basal-like subtype of pancreatic cancer. Commun. Biol. 2021, 4, 1019. [Google Scholar] [CrossRef]
- Velmurugan, G.; Dinakaran, V.; Rajendhran, J.; Swaminathan, K. Blood microbiota and circulating microbial metabolites in diabetes and cardiovascular disease. Trends Endocrinol. Metab. 2020, 31, 835–847. [Google Scholar] [CrossRef]
- Onarheim, A.M.; Wiik, R.; Burghardt, J.; Stackebrandt, E. Characterization and identification of two Vibrio species indigenous to the intestine of fish in cold sea water; description of Vibrio iliopiscarius sp. nov. Syst. Appl. Microbiol. 1994, 17, 370–379. [Google Scholar] [CrossRef]
- Delghandi, M.R.; El-Matbouli, M.; Menanteau-Ledouble, S. Renibacterium salmoninarum—The causative agent of bacterial kidney disease in salmonid fish. Pathogens 2020, 9, 845. [Google Scholar] [CrossRef]
- Caraguel, J.M.; Guerri, O.; Davaine, P. Pacage marin du saumon à Kerguelen. In Rapport de Campagne ARMOR 1992; INRA: Paris, France, 1993. [Google Scholar]
- Eldøy, S.H.; Davidsen, J.; Vignon, M.; Power, M. The biology and feeding ecology of Arctic charr in the Kerguelen Islands. J. Fish Biol. 2020, 98, 526–536. [Google Scholar] [CrossRef] [PubMed]
- Austin, B.; Austin, D.A. Vibrionaceae representatives. In Bacterial Fish Pathogens; Austin, B., Austin, D.A., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 357–411. [Google Scholar]
- Klemetsen, T.; Karlsen, C.R.; Willassen, N.P. Phylogenetic revision of the genus Aliivibrio: Intra-and inter-species variance among clusters suggest a wider diversity of species. Front. Microbiol. 2021, 12, 626759. [Google Scholar] [CrossRef] [PubMed]
- Davidsen, J.G.; Bordeleau, X.; Eldøy, S.H.; Whoriskey, F.; Power, M.; Crossin, G.T.; Buhariwalla, C.; Gaudin, P. Marine habitat use and feeding ecology of introduced anadromous brown trout at the colonization front of the sub-Antarctic Kerguelen archipelago. Sci. Rep. 2021, 11, 11917. [Google Scholar] [CrossRef]
- Castillo, D.J.; Rifkin, R.F.; Cowan, D.A.; Potgieter, M. The healthy human blood microbiome: Fact or fiction? Front. Cell. Infect. Microbiol. 2019, 9, 148. [Google Scholar] [CrossRef] [PubMed]
- Schierwagen, R.; Alvarez-Silva, C.; Madsen, M.S.A.; Kolbe, C.C.; Meyer, C.; Thomas, D.; Uschner, F.E.; Magdaleno, F.; Jansen, C.; Pohlmann, A.; et al. Circulating microbiome in blood of different circulatory compartments. Gut 2019, 68, 578–580. [Google Scholar] [CrossRef] [PubMed]
- Schierwagen, R.; Alvarez-Silva, C.; Servant, F.; Trebicka, J.; Lelouvier, B.; Arumugam, M. Trust is good, control is better: Technical considerations in blood microbiome analysis. Gut 2020, 69, 1362–1363. [Google Scholar] [CrossRef] [PubMed]
- Labonne, J.; Kaeuffer, R.; Gueraud, F.; Zhou, M.; Manicki, A.; Hendry, A.P. From the bare minimum: Genetics and selection in populations founded by only a few parents. Evol. Ecol. Res. 2016, 17, 21–34. [Google Scholar]
- Frey, F.A.; Weis, D.; Yang, H.J.; Nicolaysen, K.; Leyrit, H.; Giret, A. Temporal geochemical trends in Kerguelen Archipelago basalts: Evidence of decreasing magma supply from the Kerguelen plume. Chem. Geol. 2000, 164, 61–80. [Google Scholar] [CrossRef]
- Snyder, M.W.; Kircher, M.; Hill, A.J.; Daza, R.M.; Shendure, J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissue origin. Cell 2016, 164, 57–68. [Google Scholar] [CrossRef]
- Abril, M.K.; Barnett, A.S.; Wegermann, K.; Fountain, E.; Strand, A.; Heyman, B.M.; Blough, B.A.; Swaminathan, A.C.; Sharma-Kuinkel, B.; Ruffin, F.; et al. Diagnosis of Capnocytophaga canimorsus sepsis by next generation whole-genome sequencing. Open Forum Infect. Dis. 2016, 3, ofw144. [Google Scholar] [CrossRef]
- Miller, R.R.; Montoya, V.; Gardy, J.L.; Patrick, D.M.; Tang, P. Metagenomics for pathogen detection in public health. Genome Med. 2013, 5, 81. [Google Scholar] [CrossRef] [PubMed]
- Poore, G.D.; Kopylova, E.; Zhu, Q.; Carpenter, C.; Fraraccio, S.; Wandro, S.; Kosciolek, T.; Janssen, S.; Metcalf, J.; Song, S.J.; et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020, 579, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Cardona-Ospina, J.A.; Villalba-Miranda, M.F.; Palechor-Ocampo, L.A.; Mancilla, L.I.; Sepúlveda-Arias, J.C. A systematic review of FTA cards® as a tool for viral RNA preservation in fieldwork: Are they safe and effective? Prev. Vet. Med. 2019, 172, 104772. [Google Scholar] [CrossRef] [PubMed]
- Cortes, A.L.; Montiel, E.R.; Gimeno, I.M. Validation of Marek’s disease diagnosis and monitoring of Marek’s disease vaccines from samples collected in FTA® Cards. Avian Dis. 2009, 54, 510–516. [Google Scholar] [CrossRef]
- Moscoso, H.; Raybon, E.O.; Thayer, S.G.; Hofacre, C.L. Molecular detection and serotyping of infectious bronchitis virus from FTA® filter paper. Avian Dis. 2005, 49, 24–29. [Google Scholar] [CrossRef]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferchiou, S.; Caza, F.; Villemur, R.; Labonne, J.; St-Pierre, Y. Skin and Blood Microbial Signatures of Sedentary and Migratory Trout (Salmo trutta) of the Kerguelen Islands. Fishes 2023, 8, 174. https://doi.org/10.3390/fishes8040174
Ferchiou S, Caza F, Villemur R, Labonne J, St-Pierre Y. Skin and Blood Microbial Signatures of Sedentary and Migratory Trout (Salmo trutta) of the Kerguelen Islands. Fishes. 2023; 8(4):174. https://doi.org/10.3390/fishes8040174
Chicago/Turabian StyleFerchiou, Sophia, France Caza, Richard Villemur, Jacques Labonne, and Yves St-Pierre. 2023. "Skin and Blood Microbial Signatures of Sedentary and Migratory Trout (Salmo trutta) of the Kerguelen Islands" Fishes 8, no. 4: 174. https://doi.org/10.3390/fishes8040174
APA StyleFerchiou, S., Caza, F., Villemur, R., Labonne, J., & St-Pierre, Y. (2023). Skin and Blood Microbial Signatures of Sedentary and Migratory Trout (Salmo trutta) of the Kerguelen Islands. Fishes, 8(4), 174. https://doi.org/10.3390/fishes8040174







