Genetic Diversity and Population Structure Analysis of Chinese Mitten Crab (Eriocheir sinensis) in the Yangtze and Liaohe Rivers
Abstract
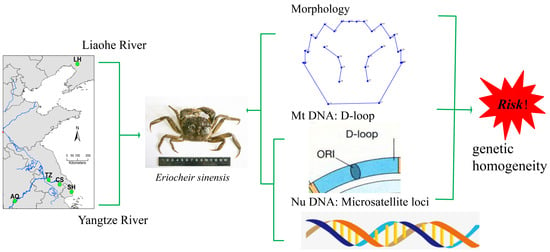
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Collection
2.3. DNA Extraction, Microsatellite, and D-Loop Fragment Amplification
2.4. Carapace Morphology
2.5. Molecular Genetic Analysis
3. Result
3.1. Geometric Morphometrics
3.2. Population Genetic Diversity
3.3. Population Structure
4. Discussion
4.1. Geometric Morphological Analysis
4.2. Genetic Diversity
4.3. Genetic Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sui, L.; Zhang, F.; Wang, X.; Bossier, P.; Sorgeloos, P.; Hänfling, B. Genetic Diversity and Population Structure of the Chinese Mitten Crab Eriocheir sinensis in Its Native Range. Mar. Biol. 2009, 156, 1573–1583. (In Chinese) [Google Scholar] [CrossRef]
- Wang, H.; Feng, G.; Zhang, Y. Studies on Genetic Diversity of Chinese Mitten Crab Eriocheir sinensis of Yangtze River System Based on Mitochondrial DNA Control Region. J. Phys. Conf. Ser. 2020, 1549, 032010. [Google Scholar] [CrossRef]
- Chang, Y.; Liang, L.; Ma, H.; He, J.; Sun, X. Microsatellite Analysis of Genetic Diversity and Population Structure of Chinese Mitten Crab (Eriocheir sinensis). J. Genet. Genom. 2008, 35, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xu, P.; Zhou, G.; Li, X.; Lu, Q.; Liu, X.; Zhou, J.; Wang, C. Genetic Improvement and Breeding Practices for Chinese Mitten Crab, Eriocheir sinensis. J. World Aquac. Soc. 2018, 49, 292–301. [Google Scholar] [CrossRef]
- Deng, Y.; Xu, Y.; Xu, Z.; Yang, Y.; Bai, R.; Ge, J.; Pan, J. Progress in Genetic Breeding of Chinese Mitten Crab. J. Aquac. 2017, 38, 39–42. (In Chinese) [Google Scholar]
- Li, J.; Chen, H.; Geng, X.; Dong, X.; Sun, J. Genetic Diversity Analysis of Wild and Cultured Population of Eriocheir sinensis from Different Water Systems. Prog. Fish. Sci. 2019, 40, 105–113. (In Chinese) [Google Scholar] [CrossRef]
- Espinoza-Donoso, S.; Angulo-Bedoya, M.; Lemic, D.; Benítez, H.A. Assessing the Influence of Allometry on Sexual and Non-Sexual Traits: An Example in Cicindelidia trifasciata (Coleoptera: Cicindelinae) Using Geometric Morphometrics. Zool. Anz. 2020, 287, 61–66. [Google Scholar] [CrossRef]
- Benítez, H.A.; Briones, R.; Jerez, V. Intra and Inter-Population Morphological Variation of Shape and Size of the Chilean Magnificent Beetle, Ceroglossus Chilensis in the Baker River Basin, Chilean Patagonia. J. Insect Sci. 2011, 11, 1536–2442. [Google Scholar] [CrossRef]
- Kaliontzopoulou, A.; Carretero, M.A.; Llorente, G.A. Intraspecific Ecomorphological Variation: Linear and Geometric Morphometrics Reveal Habitat-Related Patterns within Podarcis bocagei Wall Lizards. J. Evol. Biol. 2010, 23, 1234–1244. [Google Scholar] [CrossRef]
- Tozetto, L.; Lattke, J.E. Revealing Male Genital Morphology in the Giant Ant Genus Dinoponera with Geometric Morphometrics. Arthropod Struct. Dev. 2020, 57, 100943. [Google Scholar] [CrossRef]
- dos Santos, C.F.; Souza dos Santos, P.D.; Marques, D.M.; da-Costa, T.; Blochtein, B. Geometric Morphometrics of the Forewing Shape and Size Discriminate Plebeia Species (Hymenoptera: Apidae) Nesting in Different Substrates. Syst. Entomol. 2019, 44, 787–796. [Google Scholar] [CrossRef]
- Trallero, L.; Farré, M.; Phillips, R.A.; Navarro, J. Geometric Morphometrics Reveal Interspecific and Sexual Differences in Bill Morphology in Four Sympatric Planktivorous Petrels. J. Zool. 2019, 307, 167–177. [Google Scholar] [CrossRef]
- Walker, J.A. Ecological Morphology of Lacustrine Threespine Stickleback Gasterosteus Aculeatus L. (Gasterosteidae) Body Shape. Biol. J. Linn. Soc. 1997, 61, 3–50. [Google Scholar] [CrossRef]
- Ponton, D.; Carassou, L.; Raillard, S.; Borsa, P. Geometric Morphometrics as a Tool for Identifying Emperor fish (Lethrinidae) Larvae and Juveniles. J. Fish Biol. 2013, 83, 14–27. [Google Scholar] [CrossRef]
- Gemilang Simanjuntak, R.; Eprilurahman, R.; Selatan, M.J.T.; Utara, S.; Yogyakarta, I. Geometric Morphometrics Analysis of Chelae and Carapace of the Freshwater Prawn Macrobrachium Bate, 1868. Biog. J. Ilm. Biol. 2019, 7, 58–66. [Google Scholar] [CrossRef]
- Torres, M.V.; Giri, F.; Collins, P.A. Geometric Morphometric Analysis of the Freshwater Prawn Macrobrachium borellii (Decapoda: Palaemonidae) at a Microgeographical Scale in a Floodplain System. Ecol. Res. 2014, 29, 959–968. [Google Scholar] [CrossRef]
- Zheng, C.; Jiang, T.; Luo, R.; Chen, X.; Liu, H.; Yang, J. Geometric Morphometric Analysis of the Chinese Mitten Crab Eriocheir sinensis: A Potential Approach for Geographical Origin Authentication. N. Am. J. Fish. Manag. 2021, 41, 891–903. [Google Scholar] [CrossRef]
- Brian, J.V.; Fernandes, T.; Ladle, R.J.; Todd, P.A. Patterns of Morphological and Genetic Variability in UK Populations of the Shore Crab, Carcinus maenas Linnaeus, 1758 (Crustacea: Decapoda: Brachyura). J. Exp. Mar. Bio. Ecol. 2006, 329, 47–54. [Google Scholar] [CrossRef]
- Binashikhbubkr, K.; Adam Malik, A.; Al-Misned, F.; Mahboob, S.; Naim, D.M. Geometric Morphometric Discrimination between Seven Populations of Kawakawa Euthynnus Affinis (Cantor, 1849) from Peninsular Malaysia. J. King Saud Univ.-Sci. 2022, 34, 101863. [Google Scholar] [CrossRef]
- Grover, A.; Sharma, P.C. Development and Use of Molecular Markers: Past and Present. Crit. Rev. Biotechnol. 2016, 36, 290–302. [Google Scholar] [CrossRef]
- Zhang, W.; Jiang, S.; Salumy, K.R.; Xuan, Z.; Xiong, Y.; Jin, S.; Gong, Y.; Wu, Y.; Qiao, H.; Fu, H. Comparison of Genetic Diversity and Population Structure of Eight Macrobrachium nipponense Populations in China Based on D-Loop Sequences. Aquac. Rep. 2022, 23, 101086. [Google Scholar] [CrossRef]
- Sun, N.; Zhu, D.M.; Li, Q.; Wang, G.Y.; Chen, J.; Zheng, F.; Li, P.; Sun, Y.H. Genetic Diversity Analysis of Topmouth culter (Culter Alburnus) Based on Microsatellites and D-Loop Sequences. Environ. Biol. Fishes 2021, 104, 213–228. [Google Scholar] [CrossRef]
- Gallagher, J.; Finarelli, J.A.; Jonasson, J.P.; Carlsson, J. Mitochondrial D-Loop DNA Analyses of Norway Lobster (Nephrops Norvegicus) Reveals Genetic Isolation between Atlantic and East Mediterranean Populations. J. Mar. Biol. Assoc. UK 2019, 99, 933–940. [Google Scholar] [CrossRef]
- Duan, B.; Liu, W.; Li, S.; Yu, Y.; Guan, Y.; Mu, S.; Li, Z.; Ji, X.; Kang, X. Microsatellite Analysis of Genetic Diversity in Wild and Cultivated Portunus trituberculatus in Bohai Bay. Mol. Biol. Rep. 2022, 49, 2543–2551. [Google Scholar] [CrossRef]
- Guo, X.Z.; Chen, H.M.; Wang, A.B.; Qian, X.Q. Population Genetic Structure of the Yellow Catfish (Pelteobagrus fulvidraco) in China Inferred from Microsatellite Analyses: Implications for Fisheries Management and Breeding. J. World Aquac. Soc. 2022, 53, 174–191. [Google Scholar] [CrossRef]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; de Freitas Munhoz, C. Microsatellite Markers: What They Mean and Why They Are so Useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef]
- Wirgin, I.; Maceda, L.; Stabile, J.; Waldman, J. Genetic Population Structure of Summer Flounder Paralichthys Dentatus Using Microsatellite DNA Analysis. Fish. Res. 2022, 250, 106270. [Google Scholar] [CrossRef]
- Wang, Y.J.; Lu, J.H.; Liu, Z.; Zhang, J.P. Genetic Diversity of Gymnocypris chilianensis (Cypriniformes, Cyprinidae) Unveiled by the Mitochondrial DNA D-Loop Region. Mitochondrial. DNA Part B Resour. 2021, 6, 1292–1297. [Google Scholar] [CrossRef]
- Hansen, M.M.; Kenchington, E.; Nielsen, E.E. Assigning Individual Fish to Populations Using Microsatellite DNA Markers. Fish Fish. 2001, 2, 93–112. [Google Scholar] [CrossRef]
- Ma, H.; Ma, C.; Ma, L.; Cui, H. Novel Polymorphic Microsatellite Markers in Scylla paramamosain and Cross-Species Amplification in Related Crab Species. J. Crustac. Biol. 2010, 30, 441–444. [Google Scholar] [CrossRef]
- Liu, Y.G.; Guo, Y.H.; Hao, J.; Liu, L.X. Genetic Diversity of Swimming Crab (Portunus trituberculatus) Populations from Shandong Peninsula as Assessed by Microsatellite Markers. Biochem. Syst. Ecol. 2012, 41, 91–97. [Google Scholar] [CrossRef]
- Švara, V.; Norf, H.; Luckenbach, T.; Brack, W.; Michalski, S.G. Isolation and Characterization of Eleven Novel Microsatellite Markers for Fine-Scale Population Genetic Analyses of Gammarus Pulex (Crustacea: Amphipoda). Mol. Biol. Rep. 2019, 46, 6609–6615. [Google Scholar] [CrossRef]
- González-Castellano, I.; González-López, J.; González-Tizón, A.M.; Martínez-Lage, A. Genetic Diversity and Population Structure of the Rockpool Shrimp Palaemon Elegans Based on Microsatellites: Evidence for a Cryptic Species and Differentiation across the Atlantic–Mediterranean Transition. Sci. Rep. 2020, 10, 10784. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Zhang, C.; Li, Q.; Zheng, H.; Cheng, Y.; Wu, X. Genetic Diversity Analysis of Wild and Cultured Eriocheir sinensis Populations from the Yangtze River, Yellow River, and Liaohe River Based on the Mitochondrial D-Loop Gene. J. Fish. Sci. China 2019, 26, 436–444. (In Chinese) [Google Scholar] [CrossRef]
- Xiao, Q. DNA Molecular Markers Discovery of Chinese Mitten Crab Eriocheir sinensis and Its Application. Master’s Thesis, Shanghai Ocean University, Shanghai, China, 2018. (In Chinese). [Google Scholar]
- Klingenberg, C.P. MorphoJ: An Integrated Software Package for Geometric Morphometrics. Mol. Ecol. Resour. 2011, 11, 353–357. [Google Scholar] [CrossRef]
- IBM SPSS. Corp Ibm SPSS Statistics for Windows, Version 26.0; IBM Corp: Armonk, NY, USA, 2018. [Google Scholar]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, Chapter 2, 1–22. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinform. Appl. Note 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. MICRO-CHECKER: Software for Identifying and Correcting Genotyping Errors in Microsatellite Data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT (Version 2.9.3.2): A Program to Estimate and Test Gene Diversities and Fixation Indices. Available online: http//www.unil.ch/izea/software/fstat.html (accessed on 22 June 2022).
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software Structure: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Pritchard, J.K. Documentation for Structure Software: Version 2.2. J. Pediatr. Surg. 2009, 41, 55–63. [Google Scholar]
- Baum, B.R. PHYLIP: Phylogeny Inference Package. Version 3.2. Joel Felsenstein. Q. Rev. Biol. 1989, 64, 539–541. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenALEx 6.5: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research-an Update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Cheng, Y.; Pan, J.; Li, X.; Wu, X. Landmark-Based Morphometric Identification of Wild Eriocheir sinensis with Geographically Different Origins. J. Fish. Sci. China 2019, 26, 1116–1125. (In Chinese) [Google Scholar]
- Lu, Y.; Wu, X.; He, J.; Wang, C.; Li, X.; Liu, N.; Wang, Y.; Cheng, Y. Comparative Studies of the Morphology and Biochemical Composition of Wild Juvenile Chinese Mitten Crabs from the Yangtze River, Yellow River and Liaohe River Systems. J. Fish. Sci. China 2016, 23, 382–395. (In Chinese) [Google Scholar]
- Li, Y.; Li, S.; Wang, C.; Li, C.; Zhao, J. Establishment and Application of Morphological Discrimination Model for Juveniles Eriocheir sinensis from Liaohe, Yangtze and Oujiang Rivers. J. Fish. China 2001, 25, 120–126. (In Chinese) [Google Scholar]
- Zu, L.; Yu, Q.; Jiang, X.; Cheng, Y.; Wu, X. Edible Yield and Proximate Composition Analysis of Wild Male Adult Mitten Crab, Eriocheir Sp. in Northern China. Fish. Sci. 2021, 40, 20–28. [Google Scholar]
- Zhang, W.; Lv, J.; Ti, X.; Zhao, X.Y.; Jin, L.; Wu, J.; Gao, B.; Liu, P. Verification of an Efficient Genetic Sex Identification Method in Portunus trituberculatus: Applied to Study Sex Ratios in Full-Sib Families. Aquaculture 2021, 543, 736976. [Google Scholar] [CrossRef]
- Wang, W.; Ma, C.; Chen, W.; Jin, Z.; Zhao, M.; Zhang, F.; Liu, Z.; Ma, L. Population Genetic Diversity of Mud Crab (Scylla paramamosain) from Southeast Coastal Regions of China Based on Mitochondrial COI Gene Sequence. Gene 2020, 751, 144763. [Google Scholar] [CrossRef]
- Duan, B.; Mu, S.; Guan, Y.; Li, S.; Yu, Y.; Liu, W.; Li, Z.; Ji, X.; Kang, X. Genetic Diversity and Population Structure of the Swimming Crab (Portunus trituberculatus) in China Seas Determined by Genotyping-by-Sequencing (GBS). Aquaculture 2022, 555, 738233. [Google Scholar] [CrossRef]
- Wang, S.; Luo, L.; Zhang, R.; Guo, K.; Xu, W.; Zhao, Z. Population Genetic Diversity and Differentiation of Mitten Crab, Genus Eriocheir, Based on Microsatellite Markers. Fishes 2022, 7, 182. (In Chinese) [Google Scholar] [CrossRef]
- Grant, W.S.; Bowen, B.W. Shallow Population Histories in Deep Evolutionary Lineages of Marine Fishes: Insights from Sardines and Anchovies and Lessons for Conservation. J. Hered. 1998, 89, 415–426. [Google Scholar] [CrossRef]
- Qiu, G.; Xu, Q.; Wang, L.; Fan, Z.; Chen, X. Molecular Taxonomy and Phylogeny of Four Species of Eriocheir (Decapoda: Brachyura: Grapsidae). Acta Zool. Sin. 2001, 47, 640–647. [Google Scholar]
- Raabová, J.; Münzbergová, Z.; Fischer, M. Ecological Rather than Geographic or Genetic Distance Affects Local Adaptation of the Rare Perennial Herb, Aster Amellus. Biol. Conserv. 2007, 139, 348–357. [Google Scholar] [CrossRef]
- Xu, J.; Chu, K.H. Genome Scan of the Mitten Crab Eriocheir Sensu Stricto in East Asia: Population Differentiation, Hybridization and Adaptive Speciation. Mol. Phylogenet. Evol. 2012, 64, 118–129. [Google Scholar] [CrossRef]
- Guo, E.; Cui, Z.; Wu, D.; Hui, M.; Liu, Y.; Wang, H. Genetic Structure and Diversity of Portunus Trituberculatus in Chinese Population Revealed by Microsatellite Markers. Biochem. Syst. Ecol. 2013, 50, 313–321. [Google Scholar] [CrossRef]
- Wright, S. The Interpretation of Population Structure by F-Statistics with Special Regard to Systems of Mating. Evolution 1965, 19, 395–420. [Google Scholar] [CrossRef]
- Marko, P.B.; Hart, M.W. The Complex Analytical Landscape of Gene Flow Inference. Trends Ecol. Evol. 2011, 26, 448–456. [Google Scholar] [CrossRef]
- Hey, J. Recent Advances in Assessing Gene Flow between Diverging Populations and Species. Curr. Opin. Genet. Dev. 2006, 16, 592–596. [Google Scholar] [CrossRef]
- Chen, W.; Li, C.; Chen, F.; Li, Y.; Yang, J.; Li, J.; Li, X. Phylogeographic Analyses of a Migratory Freshwater Fish (Megalobrama terminalis) Reveal a Shallow Genetic Structure and Pronounced Effects of Sea-Level Changes. Gene 2020, 737, 144478. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.; Shu, J.; Xu, S.; Li, H.; Chen, G. Effects of Sample Size and Sex Ratio on Various Genetic Diversity Measures with Microsatellite Markers. Genet. Breed. 2007, 43, 6–9. (In Chinese) [Google Scholar]
- Xiao, Q.; Liu, Q.; Wu, X.; Wang, H.; Dong, P.; Liu, H.; Cheng, Y. Genetic Diversity Analysis of Wild and Cultured Megalopa Population of Eriocheir sinensis from Yangtze River. Genom. Appl. Biol. 2017, 36, 1935–1945. (In Chinese) [Google Scholar] [CrossRef]
- Fraser, D.J.; Cook, A.M.; Eddington, J.D.; Bentzen, P.; Hutchings, J.A. Mixed Evidence for Reduced Local Adaptation in Wild Salmon Resulting from Interbreeding with Escaped Farmed Salmon: Complexities in Hybrid Fitness. Evol. Appl. 2008, 1, 501–512. [Google Scholar] [CrossRef]
- Gu, Q.; Wu, H.; Zhou, C.; Cao, X.; Wang, W.; Wen, Y.; Luo, F. Genetic Structure and Phylogeographic Relationships of the Bellamya Complex: A Nascent Aquacultural Snail in the Pearl River Basin, China. Aquac. Res. 2020, 51, 1323–1335. [Google Scholar] [CrossRef]
- Frandsen, H.R.; Figueroa, D.F.; George, J.A. Mitochondrial Genomes and Genetic Structure of the Kemp’s Ridley Sea Turtle (Lepidochelys kempii). Ecol. Evol. 2020, 10, 249–262. [Google Scholar] [CrossRef]





| Population | Liaohe River | Yangtze River |
|---|---|---|
| Liaohe River | 54 (100.0%) | 0 |
| Yangtze River | 10 (5.8%) | 162 (94.2%) |
| Locus | CX140 | CX287 | CX256 | CX416 | CX003 | CX254 | CX400 | CX427 | Mean | |
|---|---|---|---|---|---|---|---|---|---|---|
| LH | Ho | 0.433 | 0.666 | 0.966 | 0.344 | 0.4 | 0.6 | 0.533 | 0.633 | 0.571 |
| He | 0.397 | 0.765 | 0.957 | 0.434 | 0.467 | 0.816 | 0.68 | 0.837 | 0.669 | |
| AR | 3.609 | 6.305 | 15.335 | 4.22 | 4.032 | 6.898 | 4.597 | 6.845 | 6.48 | |
| HWE | 1 | 0.246 | 0.227 | 0.185 | 0.198 | 0.001 | 0.101 | 0.023 | ||
| CS | Ho | 0.25 | 0.666 | 0.909 | 0.75 | 0.666 | 0.583 | 0.583 | 0.416 | 0.602 |
| He | 0.358 | 0.67 | 0.982 | 0.666 | 0.586 | 0.731 | 0.586 | 0.847 | 0.678 | |
| AR | 2.917 | 4.83 | 19 | 4.83 | 4 | 5.83 | 4 | 6.833 | 6.53 | |
| HWE | 0.403 | 1 | 0.138 | 0.627 | 1 | 0.34 | 0.621 | 0.001 | ||
| AQ | Ho | 0.533 | 0.7 | 0.966 | 0.333 | 1 | 0.633 | 0.566 | 0.566 | 0.662 |
| He | 0.437 | 0.788 | 0.937 | 0.375 | 0.524 | 0.769 | 0.533 | 0.866 | 0.653 | |
| AR | 3.315 | 6.376 | 13.296 | 3.69 | 2.367 | 6.536 | 4.109 | 7.727 | 5.927 | |
| HWE | 0.735 | 0.156 | 0.014 | 0.295 | 0 | 0.076 | 0.688 | 0 | ||
| TZ | Ho | 0.4 | 0.633 | 0.933 | 0.433 | 1 | 0.8 | 0.533 | 0.566 | 0.662 |
| He | 0.345 | 0.777 | 0.939 | 0.363 | 0.508 | 0.861 | 0.596 | 0.856 | 0.655 | |
| AR | 2.875 | 6.771 | 14.137 | 3.094 | 2 | 7.7 | 3.698 | 7.635 | 5.988 | |
| HWE | 1 | 0.033 | 0.827 | 0.775 | 0 | 0.485 | 0.234 | 0.001 | ||
| SH | Ho | 0.633 | 0.766 | 1 | 0.433 | 1 | 0.633 | 0.567 | 0.667 | 0.712 |
| He | 0.493 | 0.771 | 0.927 | 0.37 | 0.54 | 0.847 | 0.65 | 0.799 | 0.674 | |
| AR | 3.639 | 6.786 | 12.478 | 3.196 | 2.733 | 7.18 | 4.703 | 6.292 | 5.875 | |
| HWE | 0.537 | 0.589 | 0.239 | 1 | 0 | 0.057 | 0.223 | 0.169 |
| Population | Number of Haplotypes (n) | Haplotype Diversity (Hd) | Nucleotide Diversity (Pi) | Variable Sites |
|---|---|---|---|---|
| LH | 6 | 1.000 ± 0.096 | 0.02009 ± 0.00360 | 24 |
| AQ | 6 | 1.000 ± 0.096 | 0.00624 ± 0.00948 | 9 |
| CS | 4 | 1.000 ± 0.177 | 0.00948 ± 0.00225 | 9 |
| TZ | 5 | 0.933 ± 0.122 | 0.01276 ± 0.00374 | 18 |
| SH | 5 | 1.000 ± 0.126 | 0.00778 ± 0.00253 | 9 |
| Haplotype | CS | LH | AQ | TZ | SH | Total |
|---|---|---|---|---|---|---|
| Hap_1 | 1 | 0 | 1 | 0 | 1 | 3 |
| Hap_2 | 1 | 0 | 0 | 0 | 0 | 1 |
| Hap_3 | 1 | 0 | 0 | 0 | 0 | 1 |
| Hap_4 | 1 | 0 | 0 | 0 | 0 | 1 |
| Hap_5 | 0 | 1 | 0 | 0 | 0 | 1 |
| Hap_6 | 0 | 1 | 0 | 0 | 0 | 1 |
| Hap_7 | 0 | 1 | 0 | 0 | 0 | 1 |
| Hap_8 | 0 | 1 | 0 | 0 | 0 | 1 |
| Hap_9 | 0 | 1 | 0 | 0 | 0 | 1 |
| Hap_10 | 0 | 1 | 0 | 0 | 0 | 1 |
| Hap_11 | 0 | 0 | 1 | 0 | 0 | 1 |
| Hap_12 | 0 | 0 | 1 | 0 | 0 | 1 |
| Hap_13 | 0 | 0 | 1 | 0 | 0 | 1 |
| Hap_14 | 0 | 0 | 1 | 0 | 0 | 1 |
| Hap_15 | 0 | 0 | 1 | 2 | 1 | 4 |
| Hap_16 | 0 | 0 | 0 | 1 | 0 | 1 |
| Hap_17 | 0 | 0 | 0 | 1 | 0 | 1 |
| Hap_18 | 0 | 0 | 0 | 1 | 0 | 1 |
| Hap_19 | 0 | 0 | 0 | 1 | 0 | 1 |
| Hap_20 | 0 | 0 | 0 | 0 | 1 | 1 |
| Hap_21 | 0 | 0 | 0 | 0 | 1 | 1 |
| Hap_22 | 0 | 0 | 0 | 0 | 1 | 1 |
| Source of Variation | d.f. | Sum of Components | Variance Components | Percentage of Variation | Fixation Index | p-Value | |
|---|---|---|---|---|---|---|---|
| (A) Microsatellites | Among populations | 4 | 24.586 | 0.0675 | 2.48 | ||
| Within populations | 259 | 686.342 | 2.650 | 97.52 | |||
| Total | 263 | 710.928 | 2.718 | 0.025 | 0.000 | ||
| (B) D-loop | Among groups | 1 | 0.595 | 0.021 | 4.15 | ||
| Among populations within groups | 3 | 1.238 | −0.015 | −3.08 | |||
| Within populations | 22 | 10.833 | 0.492 | 98.93 | |||
| Total | 26 | 12.667 | 0.498 | 0.011 | 0.892 | ||
| Population | LH | AQ | CS | TZ | SH | |
|---|---|---|---|---|---|---|
| (A) Microsatellites | LH | 0.000 | 0.054 | 0.000 | 0.000 | |
| AQ | 0.032 | 0.000 | 0.135 | 0.009 | ||
| CS | 0.018 | 0.048 | 0.000 | 0.000 | ||
| TZ | 0.036 | 0.003 | 0.057 | 0.793 | ||
| SH | 0.036 | 0.012 | 0.056 | −0.002 | ||
| (B) D-loop | LH | 0.036 | 0.063 | 0.135 | 0.027 | |
| AQ | 0.200 | 0.1387 | 0.928 | 0.829 | ||
| CS | 0.172 | 0.021 | 0.198 | 0.315 | ||
| TZ | 0.081 | −0.030 | 0.038 | 0.162 | ||
| SH | 0.202 | −0.046 | 0.033 | 0.044 |
| Population | LH | AQ | CS | TZ | SH |
|---|---|---|---|---|---|
| LH | 2.000 | 2.395 | 5.678 | 1.969 | |
| AQ | 7.751 | 23.692 | inf | inf | |
| CS | 13.088 | 5.022 | 12.800 | 14.500 | |
| TZ | 6.524 | 70.863 | 4.182 | 10.989 | |
| SH | 6.559 | 23.918 | 4.301 | inf |
| Population | Tajima’s D Test | p-Value | Fu’s Fs Test | p-Value |
|---|---|---|---|---|
| LH | −0.196 | 0.435 | −1.064 | 0.154 |
| AQ | −1.011 | 0.201 | −3.709 | 0.005 |
| CS | −0.069 | 0.603 | −0.715 | 0.163 |
| TZ | −1.260 | 0.095 | −0.055 | 0.415 |
| SH | −0.526 | 0.412 | −1.716 | 0.056 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, L.; Gao, J.; Yang, Y.; Nie, Z.; Liu, K.; Xu, G. Genetic Diversity and Population Structure Analysis of Chinese Mitten Crab (Eriocheir sinensis) in the Yangtze and Liaohe Rivers. Fishes 2023, 8, 253. https://doi.org/10.3390/fishes8050253
Zhou L, Gao J, Yang Y, Nie Z, Liu K, Xu G. Genetic Diversity and Population Structure Analysis of Chinese Mitten Crab (Eriocheir sinensis) in the Yangtze and Liaohe Rivers. Fishes. 2023; 8(5):253. https://doi.org/10.3390/fishes8050253
Chicago/Turabian StyleZhou, Lin, Jiancao Gao, Yanping Yang, Zhijuan Nie, Kai Liu, and Gangchun Xu. 2023. "Genetic Diversity and Population Structure Analysis of Chinese Mitten Crab (Eriocheir sinensis) in the Yangtze and Liaohe Rivers" Fishes 8, no. 5: 253. https://doi.org/10.3390/fishes8050253
APA StyleZhou, L., Gao, J., Yang, Y., Nie, Z., Liu, K., & Xu, G. (2023). Genetic Diversity and Population Structure Analysis of Chinese Mitten Crab (Eriocheir sinensis) in the Yangtze and Liaohe Rivers. Fishes, 8(5), 253. https://doi.org/10.3390/fishes8050253