Comparative Transcriptomic Analysis Reveals Adaptive Traits in Antarctic Scallop Adamussium colbecki
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling, Sequencing, and Assembly of A. colbecki Transcriptome
2.2. Molecular Data from Other Pectinids
2.3. Identification of Orthologous Sequences
2.4. Annotation of the Orthologous Sequence Dataset
2.5. Quantification of Gene Expression
2.6. Differential Gene Expression Analysis
2.7. GO Enrichment Analysis of URGs
2.8. Further Analysis of URGs
3. Results
3.1. Identification and Annotation of Orthologous Sequences
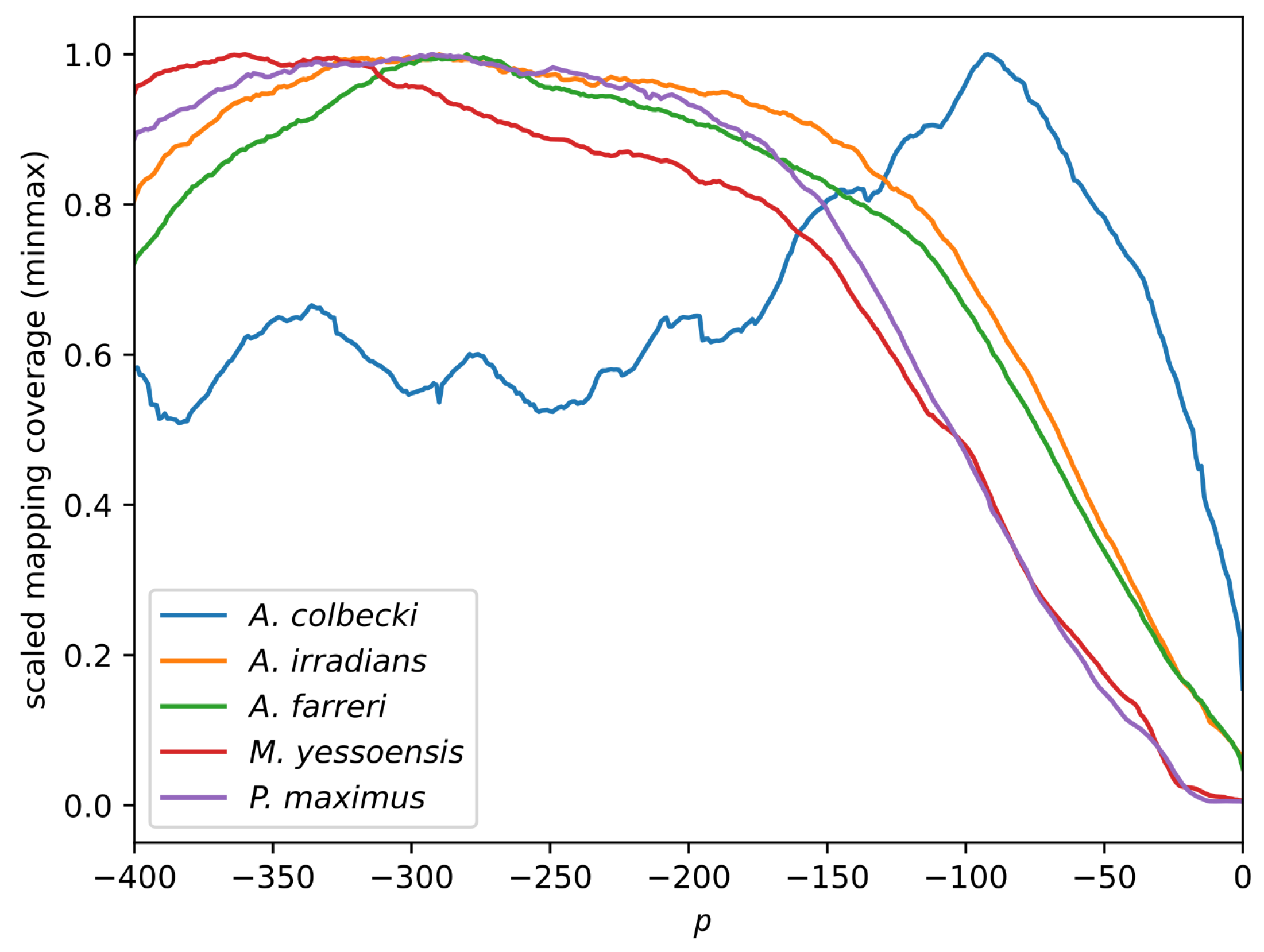
3.2. Read Mapping
3.3. Differential Expression and GO Enrichment Analysis
4. Discussion
4.1. Limitations of the Study
4.2. Multiple A. colbecki Tissues Display a Shared Transcriptomic Adaptation to Cold
4.3. The Antarctic Scallop Displays Improved Control of mRNA Transcription, Processing, and Turnover
4.4. Evidence of an Enhanced Control of Protein Production and Turnover
4.5. Optimization of Cytoskeletal Function at Low Temperatures
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MDPI | Multidisciplinary Digital Publishing Institute |
| DOAJ | Directory of open access journals |
| TLA | Three letter acronym |
| LD | Linear dichroism |
References
- Deacon, G.E.R. The Antarctic Circumpolar Ocean; Studies in Polar Research; Cambridge University Press: Cambridge, UK; New York, NY, USA, 1984. [Google Scholar]
- Hunt, B.M.; Hoefling, K.; Cheng, C.H.C. Annual Warming Episodes in Seawater Temperatures in McMurdo Sound in Relationship to Endogenous Ice in Notothenioid Fish. Antarct. Sci. 2003, 15, 333–338. [Google Scholar] [CrossRef]
- Dayton, P.K. Polar Benthos. In Polar Oceanography; Elsevier: Amsterdam, The Netherlands, 1990; pp. 631–685. [Google Scholar] [CrossRef]
- Dayton, P.K.; Mordida, B.J.; Bacon, F. Polar Marine Communities. Am. Zool. 1994, 34, 90–99. [Google Scholar] [CrossRef]
- Smith, E., VII. Mollusca. Report on the Collections of Natural History Made in the Antarctic Regions during the Voyage of the “Southern Cross”: 201–213, 1902. Available online: https://agris.fao.org/agris-search/search.do?recordID=GB2021468990 (accessed on 18 May 2023).
- Stockton, W. The Biology and Ecology of the Epifaunal Scallop Adamussium colbecki on the West Side of McMurdo Sound, Antarctica. Mar. Biol. 1984, 78, 171–178. [Google Scholar] [CrossRef]
- Guidetti, M.; Marcato, S.; Chiantore, M.; Patarnello, T.; Albertelli, G.; Cattaneo-Vietti, R. Exchange between Populations of Adamussiumcolbecki (Mollusca: Bivalvia) in the Ross Sea. Antarct. Sci. 2006, 18, 645–653. [Google Scholar] [CrossRef]
- Schiaparelli, S.; Linse, K. A Reassessment of the Distribution of the Common Antarctic Scallop Adamussium colbecki (Smith, 1902). Deep. Sea Res. Part II Top. Stud. Oceanogr. 2006, 53, 912–920. [Google Scholar] [CrossRef]
- Peck, L.S.; Morley, S.A.; Richard, J.; Clark, M.S. Acclimation and Thermal Tolerance in Antarctic Marine Ectotherms. J. Exp. Biol. 2014, 217, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Regoli, F.; Nigro, M.; Bertoli, E.; Principato, G.; Orlando, E. Defenses against Oxidative Stress in the Antarctic Scallop Adamussium colbecki and Effects of Acute Exposure to Metals. In Interactions and Adaptation Strategies of Marine Organisms; Springer: Berlin/Heidelberg, Germany, 1997; pp. 139–144. [Google Scholar]
- Chiantore, M.; Cattaneo-Vietti, R.; Berkman, P.A.; Nigro, M.; Vacchi, M.; Schiaparelli, S.; Albertelli, G. Antarctic Scallop (Adamussium colbecki ) Spatial Population Variability along the Victoria Land Coast, Antarctica. Polar Biol. 2001, 24, 139–143. [Google Scholar] [CrossRef]
- Chiantore, M.; Cattaneo-Vietti, R.; Povero, P.; Albertelli, G. The Population Structure and Ecology of the Antarctic Scallop Adamussium colbecki in Terra Nova Bay. In Ross Sea Ecology; Faranda, F.M., Guglielmo, L., Ianora, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2000; pp. 563–573. [Google Scholar] [CrossRef]
- Broach, K.H.; Miller, M.F.; Bowser, S.S. Bioturbation by the Common Antarctic Scallop (Adamussium colbecki) and Ophiuroid (Ophionotus victoriae) Under Multi-Year Sea Ice: Ecologic and Stratigraphic Implications. PALAIOS 2016, 31, 280–290. [Google Scholar] [CrossRef]
- Speiser, D.I.; Johnsen, S. Scallops Visually Respond to the Size and Speed of Virtual Particles. J. Exp. Biol. 2008, 211, 2066–2070. [Google Scholar] [CrossRef]
- Shumway, S.E.; Parsons, G.J. (Eds.) Scallops: Biology, Ecology and Aquaculture, 2nd ed.; Number 35 in Developments in Aquaculture and Fisheries Science; Elsevier: Amsterdam, The Netherlands, 2006. [Google Scholar]
- Denny, M.; Miller, L. Jet Propulsion in the Cold: Mechanics of Swimming in the Antarctic Scallop Adamussiumcolbecki. J. Exp. Biol. 2006, 209, 4503–4514. [Google Scholar] [CrossRef] [PubMed]
- Heilmayer, O.; Brey, T. Saving by Freezing? Metabolic Rates of Adamussium colbecki in a Latitudinal Context. Mar. Biol. 2003, 143, 477–484. [Google Scholar] [CrossRef]
- Norkko, J.; Norkko, A.; Thrush, S.; Cummings, V. Detecting Growth under Environmental Extremes: Spatial and Temporal Patterns in Nucleic Acid Ratios in Two Antarctic Bivalves. J. Exp. Mar. Biol. Ecol. 2005, 326, 144–156. [Google Scholar] [CrossRef]
- Wong, W.S.; Hauer, L.; Cziko, P.A.; Meister, K. Cryofouling Avoidance in the Antarctic Scallop Adamussium colbecki. Commun. Biol. 2022, 5, 1–8. [Google Scholar] [CrossRef]
- Daane, J.M.; Detrich, H.W. Adaptations and Diversity of Antarctic Fishes: A Genomic Perspective. Annu. Rev. Anim. Biosci. 2022, 10, 39–62. [Google Scholar] [CrossRef]
- Moro, G.; Buonocore, F.; Barucca, M.; Spazzali, F.; Canapa, A.; Pallavicini, A.; Scapigliati, G.; Gerdol, M. The First Transcriptomic Resource for the Antarctic Scallop Adamussium colbecki. Mar. Genom. 2019, 44, 61–64. [Google Scholar] [CrossRef]
- Greco, S.; Gaetano, A.S.; Manfrin, C.; Capanni, F.; Santovito, G.; Giulianini, P.G.; Pallavicini, A.; Gerdol, M. The Antarctic Scallop Adamussium colbecki (Smith 1902) is Unable to Transcriptomically Respond to Captivity and Moderate Thermal Stress. Stresses 2023. accepted for publication. [Google Scholar]
- Ren, J.; Shen, X.; Jiang, F.; Liu, B. The Mitochondrial Genomes of Two Scallops, Argopecten irradians and Chlamys farreri (Mollusca: Bivalvia): The Most Highly Rearranged Gene Order in the Family Pectinidae. J. Mol. Evol. 2010, 70, 57–68. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, J.; Jiao, W.; Li, J.; Xun, X.; Sun, Y.; Guo, X.; Huan, P.; Dong, B.; Zhang, L.; et al. Scallop Genome Provides Insights into Evolution of Bilaterian Karyotype and Development. Nat. Ecol. Evol. 2017, 1, 0120. [Google Scholar] [CrossRef]
- Li, Y.; Sun, X.; Hu, X.; Xun, X.; Zhang, J.; Guo, X.; Jiao, W.; Zhang, L.; Liu, W.; Wang, J.; et al. Scallop Genome Reveals Molecular Adaptations to Semi-Sessile Life and Neurotoxins. Nat. Commun. 2017, 8, 1721. [Google Scholar] [CrossRef]
- Zeng, Q.; Liu, J.; Wang, C.; Wang, H.; Zhang, L.; Hu, J.; Bao, L.; Wang, S. High-Quality Reannotation of the King Scallop Genome Reveals No ‘Gene-Rich’ Feature and Evolution of Toxin Resistance. Comput. Struct. Biotechnol. J. 2021, 19, 4954–4960. [Google Scholar] [CrossRef]
- Mao, J.; Huang, X.; Sun, H.; Jin, X.; Guan, W.; Xie, J.; Wang, Y.; Wang, X.; Yin, D.; Hao, Z.; et al. Transcriptome Analysis Provides Insight into Adaptive Mechanisms of Scallops under Environmental Stress. Front. Mar. Sci. 2022, 9, 971796. [Google Scholar] [CrossRef]
- Ventoso, P.; Pazos, A.J.; Blanco, J.; Pérez-Parallé, M.L.; Triviño, J.C.; Sánchez, J.L. Transcriptional Response in the Digestive Gland of the King Scallop (Pecten maximus) After the Injection of Domoic Acid. Toxins 2021, 13, 339. [Google Scholar] [CrossRef] [PubMed]
- Kenny, N.J.; McCarthy, S.A.; Dudchenko, O.; James, K.; Betteridge, E.; Corton, C.; Dolucan, J.; Mead, D.; Oliver, K.; Omer, A.D.; et al. The Gene-Rich Genome of the Scallop Pecten maximus. GigaScience 2020, 9, giaa037. [Google Scholar] [CrossRef]
- Sato, M.; Nagashima, K. Molecular Characterization of a Mitochondrial DNA Segment from the Japanese Scallop (Patinopecten yessoensis): Demonstration of a Region Showing Sequence Polymorphism in the Population. Mar. Biotechnol. 2001, 3, 370–379. [Google Scholar] [CrossRef]
- Chi, C.; Giri, S.S.; Jun, J.W.; Kim, H.J.; Yun, S.; Kim, S.W.; Kang, J.W.; Park, S.C. Detoxification, Apoptosis, and Immune Transcriptomic Responses of the Gill Tissue of Bay Scallop Following Exposure to the Algicide Thiazolidinedione 49. Biomolecules 2019, 9, 310. [Google Scholar] [CrossRef] [PubMed]
- Chi, C.; Giri, S.; Jun, J.; Kim, S.; Kim, H.; Kang, J.; Park, S. Detoxification- and Immune-Related Transcriptomic Analysis of Gills from Bay Scallops (emphArgopecten irradians) in Response to Algal Toxin Okadaic Acid. Toxins 2018, 10, 308. [Google Scholar] [CrossRef]
- Han, W.; Liu, L.; Wang, J.; Wei, H.; Li, Y.; Zhang, L.; Guo, Z.; Li, Y.; Liu, T.; Zeng, Q.; et al. Ancient Homomorphy of Molluscan Sex Chromosomes Sustained by Reversible Sex-Biased Genes and Sex Determiner Translocation. Nat. Ecol. Evol. 2022, 6, 1891–1906. [Google Scholar] [CrossRef]
- Malkowsky, Y.; Klussmann-Kolb, A. Phylogeny and Spatio-Temporal Distribution of European Pectinidae (Mollusca: Bivalvia). Syst. Biodivers. 2012, 10, 233–242. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Grigoryev, D.N.; Ma, S.F.; Irizarry, R.A.; Ye, S.; Quackenbush, J.; Garcia, J.G. Orthologous Gene-Expression Profiling in Multi-Species Models: Search for Candidate Genes. Genome Biol. 2004, 5, R34. [Google Scholar] [CrossRef]
- Ansaloni, F.; Gerdol, M.; Torboli, V.; Fornaini, N.R.; Greco, S.; Giulianini, P.G.; Coscia, M.R.; Miccoli, A.; Santovito, G.; Buonocore, F.; et al. Cold Adaptation in Antarctic Notothenioids: Comparative Transcriptomics Reveals Novel Insights in the Peculiar Role of Gills and Highlights Signatures of Cobalamin Deficiency. Int. J. Mol. Sci. 2021, 22, 1812. [Google Scholar] [CrossRef] [PubMed]
- Saxena, A.; Sharma, V.; Muthuirulan, P.; Neufeld, S.J.; Tran, M.P.; Gutierrez, H.L.; Chen, K.D.; Erberich, J.M.; Birmingham, A.; Capellini, T.D.; et al. Interspecies Transcriptomics Identify Genes That Underlie Disproportionate Foot Growth in Jerboas. Curr. Biol. 2022, 32, 289–303.e6. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.; Ali, R.A.; Heyland, A. Comparative Transcriptomics Analysis Pipeline for the Meta-Analysis of Phylogenetically Divergent Datasets (CoRMAP). BMC Bioinform. 2022, 23, 415. [Google Scholar] [CrossRef]
- Vercruysse, J.; Van Bel, M.; Osuna-Cruz, C.M.; Kulkarni, S.R.; Storme, V.; Nelissen, H.; Gonzalez, N.; Inzé, D.; Vandepoele, K. Comparative Transcriptomics Enables the Identification of Functional Orthologous Genes Involved in Early Leaf Growth. Plant Biotechnol. J. 2020, 18, 553–567. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Hrdlickova, R.; Toloue, M.; Tian, B. RNA-Seq Methods for Transcriptome Analysis. Wiley Interdiscip. Rev. RNA 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The Pfam Protein Families Database. Nucleic Acids Res. 2012, 40, D290–D301. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Ma, F.; Fuqua, B.K.; Hasin, Y.; Yukhtman, C.; Vulpe, C.D.; Lusis, A.J.; Pellegrini, M. A Comparison Between Whole Transcript and 3’ RNA Sequencing Methods Using Kapa and Lexogen Library Preparation Methods. BMC Genom. 2019, 20, 9. [Google Scholar] [CrossRef]
- Xiong, Y.; Soumillon, M.; Wu, J.; Hansen, J.; Hu, B.; van Hasselt, J.G.C.; Jayaraman, G.; Lim, R.; Bouhaddou, M.; Ornelas, L.; et al. A Comparison of mRNA Sequencing with Random Primed and 3′-Directed Libraries. Sci. Rep. 2017, 7, 14626. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. MSOR Connect. 2014. Available online: https://www.r-project.org/ (accessed on 1 January 2022).
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential Expression Analysis of Multifactor RNA-Seq Experiments with Respect to Biological Variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Bullard, J.H.; Purdom, E.; Hansen, K.D.; Dudoit, S. Evaluation of Statistical Methods for Normalization and Differential Expression in mRNA-Seq Experiments. BMC Bioinform. 2010, 11, 94. [Google Scholar] [CrossRef]
- Risso, D.; Ngai, J.; Speed, T.P.; Dudoit, S. Normalization of RNA-seq Data Using Factor Analysis of Control Genes or Samples. Nat. Biotechnol. 2014, 32, 896–902. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A Scaling Normalization Method for Differential Expression Analysis of RNA-seq Data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Timmons, J.A.; Szkop, K.J.; Gallagher, I.J. Multiple Sources of Bias Confound Functional Enrichment Analysis of Global -Omics Data. Genome Biol. 2015, 16, 186. [Google Scholar] [CrossRef]
- Falcon, S.; Gentleman, R. Hypergeometric Testing Used for Gene Set Enrichment Analysis. In Bioconductor Case Studies; Springer: Berlin/Heidelberg, Germany, 2008; pp. 207–220. [Google Scholar]
- The UniProt Consortium; Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bye-A-Jee, H.; et al. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING V10: Protein–Protein Interaction Networks, Integrated over the Tree of Life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Moll, P.; Ante, M.; Seitz, A.; Reda, T. QuantSeq 3′ mRNA Sequencing for RNA Quantification. Nat. Methods 2014, 11, i–iii. [Google Scholar] [CrossRef]
- Gerdol, M.; Fujii, Y.; Hasan, I.; Koike, T.; Shimojo, S.; Spazzali, F.; Yamamoto, K.; Ozeki, Y.; Pallavicini, A.; Fujita, H. The Purplish Bifurcated Mussel Mytilisepta virgata Gene Expression Atlas Reveals a Remarkable Tissue Functional Specialization. BMC Genom. 2017, 18, 590. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Christina Cheng, C.-H.; Zhang, J.; Cao, L.; Chen, L.; Zhou, L.; Jin, Y.; Ye, H.; Deng, C.; Dai, Z.; et al. Transcriptomic and Genomic Evolution under Constant Cold in Antarctic Notothenioid Fish. Proc. Natl. Acad. Sci. USA 2008, 105, 12944–12949. [Google Scholar] [CrossRef] [PubMed]
- Kristiansson, E.; Österlund, T.; Gunnarsson, L.; Arne, G.; Joakim Larsson, D.G.; Nerman, O. A Novel Method for Cross-Species Gene Expression Analysis. BMC Bioinform. 2013, 14, 70. [Google Scholar] [CrossRef]
- Beers, J.M.; Jayasundara, N. Antarctic Notothenioid Fish: What Are the Future Consequences of ’losses’ and ’Gains’ Acquired during Long-Term Evolution at Cold and Stable Temperatures? J. Exp. Biol. 2015, 218, 1834–1845. [Google Scholar] [CrossRef]
- Fletcher, G.L.; Hew, C.L.; Davies, P.L. Antifreeze Proteins of Teleost Fishes. Annu. Rev. Physiol. 2001, 63, 359–390. [Google Scholar] [CrossRef]
- Bista, I.; Wood, J.M.D.; Desvignes, T.; McCarthy, S.A.; Matschiner, M.; Ning, Z.; Tracey, A.; Torrance, J.; Sims, Y.; Chow, W.; et al. Genomics of Cold Adaptations in the Antarctic Notothenioid Fish Radiation. BiorXiv 2022, 2022.06.08.494096. [Google Scholar] [CrossRef]
- DeVries, A.R. Antifreeze Peptides and Glycopeptides in Cold-Water Fishes. Annu. Rev. Physiol. 1983, 45, 245–260. [Google Scholar] [CrossRef]
- Hofmann, G.E.; Buckley, B.A.; Airaksinen, S.; Keen, J.E.; Somero, G.N. Heat-Shock Protein Expression Is Absent in the Antarctic Fish Trematomus bernacchii (Family Nototheniidae). J. Exp. Biol. 2000, 203 Pt 15, 2331–2339. [Google Scholar] [CrossRef]
- Greco, S.; Agostino, E.D.; Manfrin, C.; Gaetano, A.S.; Furlanis, G.; Capanni, F.; Santovito, G.; Edomi, P.; Giulianini, P.G.; Gerdol, M.; et al. RNA-sequencing Indicates High Hemocyanin Expression as a Key Strategy for Cold Adaptation in the Antarctic Amphipod Eusirus cf. giganteus Clade G3. Biocell 2021, 45, 1611–1619. [Google Scholar] [CrossRef]
- Becskei, A.; Rahaman, S. The Life and Death of RNA across Temperatures. Comput. Struct. Biotechnol. J. 2022, 20, 4325–4336. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.W.F.; dos Santos, J.A.; Vianna, M.V.; Vieira, J.M.F.; Mallagutti, V.H.; Inforsato, F.J.; Wentzel, L.C.P.; Lario, L.D.; Rodrigues, A.; Pagnocca, F.C.; et al. Cold-Adapted Enzymes Produced by Fungi from Terrestrial and Marine Antarctic Environments. Crit. Rev. Biotechnol. 2018, 38, 600–619. [Google Scholar] [CrossRef] [PubMed]
- Marx, J.C.; Collins, T.; D’Amico, S.; Feller, G.; Gerday, C. Cold-Adapted Enzymes from Marine Antarctic Microorganisms. Mar. Biotechnol. 2007, 9, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.W.F.; Barato, M.B.; Nobre, F.S.; Polezel, D.A.; de Oliveira, T.B.; dos Santos, J.A.; Rodrigues, A.; Sette, L.D. Production of Cold-Adapted Enzymes by Filamentous Fungi from King George Island, Antarctica. Polar Biol. 2018, 41, 2511–2521. [Google Scholar] [CrossRef]
- Decker, C.J.; Parker, R. A Turnover Pathway for Both Stable and Unstable mRNAs in Yeast: Evidence for a Requirement for Deadenylation. Genes Develop. 1993, 7, 1632–1643. [Google Scholar] [CrossRef] [PubMed]
- Doidge, R.; Mittal, S.; Aslam, A.; Winkler, G.S. Deadenylation of Cytoplasmic mRNA by the Mammalian Ccr4–Not Complex. Biochem. Soc. Trans. 2012, 40, 896–901. [Google Scholar] [CrossRef]
- Collart, M.A. The Ccr4-Not Complex Is a Key Regulator of Eukaryotic Gene Expression. Wiley Interdiscip. Rev. RNA 2016, 7, 438–454. [Google Scholar] [CrossRef]
- Behm-Ansmant, I.; Gatfield, D.; Rehwinkel, J.; Hilgers, V.; Izaurralde, E. A Conserved Role for Cytoplasmic Poly(A)-Binding Protein 1 (PABPC1) in Nonsense-Mediated mRNA Decay. EMBO J. 2007, 26, 1591–1601. [Google Scholar] [CrossRef]
- Hanet, A.; Räsch, F.; Weber, R.; Ruscica, V.; Fauser, M.; Raisch, T.; Kuzuoğlu-Öztürk, D.; Chang, C.T.; Bhandari, D.; Igreja, C.; et al. HELZ Directly Interacts with CCR4–NOT and Causes Decay of Bound mRNAs. Life Sci. Alliance 2019, 2, e201900405. [Google Scholar] [CrossRef]
- Fukushima, M.; Hosoda, N.; Chifu, K.; Hoshino, S.I. TDP -43 Accelerates Deadenylation of Target mRNA s by Recruiting Caf1 Deadenylase. FEBS Lett. 2019, 593, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Olsen, M.T.; Webb, K.; Bennett, E.J.; Lykke-Andersen, J. Recruitment of the 4EHP-GYF2 Cap-Binding Complex to Tetraproline Motifs of Tristetraprolin Promotes Repression and Degradation of mRNAs with AU-rich Elements. RNA 2016, 22, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Braun, J.E.; Huntzinger, E.; Fauser, M.; Izaurralde, E. GW182 Proteins Directly Recruit Cytoplasmic Deadenylase Complexes to miRNA Targets. Mol. Cell 2011, 44, 120–133. [Google Scholar] [CrossRef]
- Sun, X.; Fan, G.; Su, L.; Wang, W.; Liang, Z.; Li, S.; Xin, H. Identification of Cold-Inducible microRNAs in Grapevine. Front. Plant Sci. 2015, 6, 595. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yu, Q.; Yang, Y.; Su, Y.; Ahmad, W.; Guo, J.; Gao, S.; Xu, L.; Que, Y. Identification of Cold-Related miRNAs in Sugarcane by Small RNA Sequencing and Functional Analysis of a Cold Inducible ScmiR393 to Cold Stress. Environ. Exp. Bot. 2018, 155, 464–476. [Google Scholar] [CrossRef]
- Pilotte, J.; Dupont-Versteegden, E.E.; Vanderklish, P.W. Widespread Regulation of miRNA Biogenesis at the Dicer Step by the Cold-Inducible RNA-Binding Protein, RBM3. PLoS ONE 2011, 6, e28446. [Google Scholar] [CrossRef]
- LLeonart, M. A New Generation of Proto-Oncogenes: Cold-inducible RNA Binding Proteins. Biochim. Biophys. Acta (BBA)—Rev. Cancer 2010, 1805, 43–52. [Google Scholar] [CrossRef]
- Neumann, A.; Meinke, S.; Goldammer, G.; Strauch, M.; Schubert, D.; Timmermann, B.; Heyd, F.; Preußner, M. Alternative Splicing Coupled mRNA Decay Shapes the Temperature-dependent Transcriptome. EMBO Rep. 2020, 21, e51369. [Google Scholar] [CrossRef]
- Kirstein, N.; Gomes Dos Santos, H.; Blumenthal, E.; Shiekhattar, R. The Integrator Complex at the Crossroad of Coding and Noncoding RNA. Curr. Opin. Cell Biol. 2021, 70, 37–43. [Google Scholar] [CrossRef]
- Jin, G.; Zhang, Z.; Wan, J.; Wu, X.; Liu, X.; Zhang, W. G3BP2: Structure and Function. Pharmacol. Res. 2022, 186, 106548. [Google Scholar] [CrossRef]
- Blencowe, B.J.; Issner, R.; Nickerson, J.A.; Sharp, P.A. A Coactivator of Pre-mRNA Splicing. Genes Dev. 1998, 12, 996–1009. [Google Scholar] [CrossRef]
- Sánchez-Hernández, N.; Boireau, S.; Schmidt, U.; Muñoz-Cobo, J.P.; Hernández-Munain, C.; Bertrand, E.; Suñé, C. The in Vivo Dynamics of TCERG1, a Factor That Couples Transcriptional Elongation with Splicing. RNA 2016, 22, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Tollervey, J.R.; Curk, T.; Rogelj, B.; Briese, M.; Cereda, M.; Kayikci, M.; König, J.; Hortobágyi, T.; Nishimura, A.L.; Župunski, V.; et al. Characterizing the RNA Targets and Position-Dependent Splicing Regulation by TDP-43. Nat. Neurosci. 2011, 14, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Seshaiah, P.; Miller, B.; Myat, M.M.; Andrew, D.J. Pasilla, the Drosophila Homologue of the Human Nova-1 and Nova-2 Proteins, Is Required for Normal Secretion in the Salivary Gland. Develop. Biol. 2001, 239, 309–322. [Google Scholar] [CrossRef]
- Storch, D.; Pörtner, H. The Protein Synthesis Machinery Operates at the Same Expense in Eurythermal and Cold Stenothermal Pectinids. Physiol. Biochem. Zool. PBZ 2003, 76, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Fraser, K.P.P.; Clarke, A.; Peck, L.S. Low-Temperature Protein Metabolism: Seasonal Changes in Protein Synthesis and RNA Dynamics in the Antarctic Limpet Nacellaconcinna Strebel 1908. J. Exp. Biol. 2002, 205, 3077–3086. [Google Scholar] [CrossRef]
- Carré, C.; Shiekhattar, R. Human GTPases Associate with RNA Polymerase II To Mediate Its Nuclear Import. Mol. Cell. Biol. 2011, 31, 3953–3962. [Google Scholar] [CrossRef]
- Gregory, G.D.; Vakoc, C.R.; Rozovskaia, T.; Zheng, X.; Patel, S.; Nakamura, T.; Canaani, E.; Blobel, G.A. Mammalian ASH1L Is a Histone Methyltransferase That Occupies the Transcribed Region of Active Genes. Mol. Cell. Biol. 2007, 27, 8466–8479. [Google Scholar] [CrossRef]
- Lee, S.; Roeder, R.G.; Lee, J.W. Chapter 10 Roles of Histone H3-Lysine 4 Methyltransferase Complexes in NR-Mediated Gene Transcription. In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2009; Volume 87, pp. 343–382. [Google Scholar] [CrossRef]
- Doyon, Y.; Selleck, W.; Lane, W.S.; Tan, S.; Côté, J. Structural and Functional Conservation of the NuA4 Histone Acetyltransferase Complex from Yeast to Humans. Mol. Cell. Biol. 2004, 24, 1884–1896. [Google Scholar] [CrossRef]
- Bilyk, K.T.; Cheng, C.H. Model of Gene Expression in Extreme Cold - Reference Transcriptome for the High-Antarctic Cryopelagic Notothenioid Fish Pagothenia borchgrevinki. BMC Genom. 2013, 14, 634. [Google Scholar] [CrossRef]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular Chaperones in Protein Folding and Proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Feller, G. Protein Folding at Extreme Temperatures: Current Issues. Semin. Cell Develop. Biol. 2018, 84, 129–137. [Google Scholar] [CrossRef]
- Lopez, C.F.; Darst, R.K.; Rossky, P.J. Mechanistic Elements of Protein Cold Denaturation. J. Phys. Chem. B 2008, 112, 5961–5967. [Google Scholar] [CrossRef] [PubMed]
- Dias, C.L.; Ala-Nissila, T.; Wong-ekkabut, J.; Vattulainen, I.; Grant, M.; Karttunen, M. The Hydrophobic Effect and Its Role in Cold Denaturation. Cryobiology 2010, 60, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Place, S.P.; Zippay, M.L.; Hofmann, G.E. Constitutive Roles for Inducible Genes: Evidence for the Alteration in Expression of the Inducible Hsp70 Gene in Antarctic Notothenioid Fishes. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2004, 287, R429–R436. [Google Scholar] [CrossRef] [PubMed]
- Buckley, B.A.; Place, S.P.; Hofmann, G.E. Regulation of Heat Shock Genes in Isolated Hepatocytes from an Antarctic Fish, Trematomus Bernacchii. J. Exp. Biol. 2004, 207 Pt 21, 3649–3656. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, H.W.; Lee, J.H.; Park, J.; Lee, H.; Kim, S.; Shin, S.C. Gene Family Expansions in Antarctic Winged Midge as a Strategy for Adaptation to Cold Environments. Sci. Rep. 2022, 12, 18263. [Google Scholar] [CrossRef]
- Todgham, A.E.; Hoaglund, E.A.; Hofmann, G.E. Is Cold the New Hot? Elevated Ubiquitin-Conjugated Protein Levels in Tissues of Antarctic Fish as Evidence for Cold-Denaturation of Proteins in Vivo. J. Comp. Physiol. B 2007, 177, 857–866. [Google Scholar] [CrossRef]
- Glickman, M.H.; Ciechanover, A. The Ubiquitin-Proteasome Proteolytic Pathway: Destruction for the Sake of Construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef]
- Gorbea, C.; Goellner, G.M.; Teter, K.; Holmes, R.K.; Rechsteiner, M. Characterization of Mammalian Ecm29, a 26 S Proteasome-associated Protein That Localizes to the Nucleus and Membrane Vesicles. J. Biol. Chem. 2004, 279, 54849–54861. [Google Scholar] [CrossRef]
- Ballar, P.; Pabuccuoglu, A.; Kose, F.A. Different P97/VCP Complexes Function in Retrotranslocation Step of Mammalian Er-associated Degradation (ERAD). Int. J. Biochem. Cell Biol. 2011, 43, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Antao, A.M.; Kaushal, K.; Das, S.; Singh, V.; Suresh, B.; Kim, K.S.; Ramakrishna, S. USP48 Governs Cell Cycle Progression by Regulating the Protein Level of Aurora B. Int. J. Mol. Sci. 2021, 22, 8508. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.B.; Shao, Y.M.; Miao, S.; Wang, L. The Diversity of the DnaJ/Hsp40 Family, the Crucial Partners for Hsp70 Chaperones. Cell. Mol. Life Sci. 2006, 63, 2560–2570. [Google Scholar] [CrossRef] [PubMed]
- Rospert, S.; Dubaquié, Y.; Gautschi, M. Nascent-Polypeptide-Associated Complex. Cell. Mol. Life Sci. 2002, 59, 1632–1639. [Google Scholar] [CrossRef]
- Skarie, J.M.; Link, B.A. The Primary Open-Angle Glaucoma Gene WDR36 Functions in Ribosomal RNA Processing and Interacts with the P53 Stress-Response Pathway. Hum. Mol. Genet. 2008, 17, 2474–2485. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.F.; Hsu, C.M.; Huang, Y.S. CPEB 2-dependent Translation of Long 3′- UTR Ucp1 mRNA Promotes Thermogenesis in Brown Adipose Tissue. EMBO J. 2018, 37, e99071. [Google Scholar] [CrossRef]
- Melki, R.; Carlier, M.F.; Pantaloni, D.; Timasheff, S.N. Cold Depolymerization of Microtubules to Double Rings: Geometric Stabilization of Assemblies. Biochemistry 1989, 28, 9143–9152. [Google Scholar] [CrossRef]
- Billger, M.; Wallin, M.; Williams, R.C.; Detrich, H.W. Dynamic Instability of Microtubules from Cold-Living Fishes. Cell Motil. Cytoskelet. 1994, 28, 327–332. [Google Scholar] [CrossRef]
- Detrich, H.W.; Johnson, K.A.; Marchese-Ragona, S.P. Polymerization of Antarctic Fish Tubulins at Low Temperatures: Energetic Aspects. Biochemistry 1989, 28, 10085–10093. [Google Scholar] [CrossRef]
- Williams, R.C.; Correia, J.J.; De Vries, A.L. Formation of Microtubules at Low Temperature by Tubulin from Antarctic Fish. Biochemistry 1985, 24, 2790–2798. [Google Scholar] [CrossRef]
- Detrich, H.W.; Parker, S.K.; Williams, R.C.; Nogales, E.; Downing, K.H. Cold Adaptation of Microtubule Assembly and Dynamics. J. Biol. Chem. 2000, 275, 37038–37047. [Google Scholar] [CrossRef] [PubMed]
- Marziale, F.; Pucciarelli, S.; Ballarini, P.; Melki, R.; Uzun, A.; Ilyin, V.A.; Detrich III, H.W.; Miceli, C. Different Roles of Two γ-tubulin Isotypes in the Cytoskeleton of the Antarctic Ciliate Euplotes focardii: Remodelling of Interaction Surfaces May Enhance Microtubule Nucleation at Low Temperature. FEBS J. 2008, 275, 5367–5382. [Google Scholar] [CrossRef] [PubMed]
- Willem, S.; Srahna, M.; Devos, N.; Gerday, C.; Loppes, R.; Matagne, R.F. Protein Adaptation to Low Temperatures: A Comparative Study of α-tubulin Sequences in Mesophilic and Psychrophilic Algae. Extremophiles 1999, 3, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Pucciarelli, S.; Ballarini, P.; Miceli, C. Cold-adapted Microtubules: Characterization of Tubulin Posttranslational Modifications in the Antarctic Ciliate Euplotes focardii. Cell Motil. Cytoskelet. 1997, 38, 329–340. [Google Scholar] [CrossRef]
- Parker, S.K.; Detrich, H.W. Evolution, Organization, and Expression of α-Tubulin Genes in the Antarctic Fish Notothenia coriiceps. J. Biol. Chem. 1998, 273, 34358–34369. [Google Scholar] [CrossRef]
- Tartaglia, L.J.; Shain, D.H. Cold-adapted Tubulins in the Glacier Ice Worm, Mesenchytraeus solifugus. Gene 2008, 423, 135–141. [Google Scholar] [CrossRef]
- Starling, G.P.; Phillips, B.A.; Ganesh, S.; King, J.S. Katnip is Needed to Maintain Microtubule Function and Lysosomal Delivery to Autophagosomes and Phagosomes. Mol. Biol. Cell 2023, 34, ar12. [Google Scholar] [CrossRef]
- Tögel, M.; Wiche, G.; Propst, F. Novel Features of the Light Chain of Microtubule-associated Protein MAP1B: Microtubule Stabilization, Self Interaction, Actin Filament Binding, and Regulation by the Heavy Chain. J. Cell Biol. 1998, 143, 695–707. [Google Scholar] [CrossRef]
- Thauvin-Robinet, C.; Lee, J.S.; Lopez, E.; Herranz-Pérez, V.; Shida, T.; Franco, B.; Jego, L.; Ye, F.; Pasquier, L.; Loget, P.; et al. The Oral-Facial-Digital Syndrome Gene C2CD3 Encodes a Positive Regulator of Centriole Elongation. Nat. Genet. 2014, 46, 905–911. [Google Scholar] [CrossRef]
- Smith, E.F. Hydin Seek: Finding a Function in Ciliary Motility. J. Cell Biol. 2007, 176, 403–404. [Google Scholar] [CrossRef]
- Kim, T.Y.; Vigil, D.; Der, C.J.; Juliano, R.L. Role of DLC-1, a Tumor Suppressor Protein with RhoGAP Activity, in Regulation of the Cytoskeleton and Cell Motility. Cancer Metastasis Rev. 2009, 28, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Rajgor, D.; Shanahan, C.M. Nesprins: From The Nuclear Envelope and Beyond. Expert Rev. Mol. Med. 2013, 15, e5. [Google Scholar] [CrossRef] [PubMed]
- Dalpé, G.; Leclerc, N.; Vallée, A.; Messer, A.; Mathieu, M.; De Repentigny, Y.; Kothary, R. Dystonin is Essential For Maintaining Neuronal Cytoskeleton Organization. Mol. Cell. Neurosci. 1998, 10, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Papini, D.; Langemeyer, L.; Abad, M.A.; Kerr, A.; Samejima, I.; Eyers, P.A.; Jeyaprakash, A.A.; Higgins, J.M.G.; Barr, F.A.; Earnshaw, W.C. TD-60 links RalA GTPase Function o the CPC in Mitosis. Nat. Commun. 2015, 6, 7678. [Google Scholar] [CrossRef] [PubMed]
- Ching, Y.P.; Wong, C.M.; Chan, S.F.; Leung, T.H.Y.; Ng, D.C.H.; Jin, D.Y.; Ng, I.O.l. Deleted in Liver Cancer (DLC) 2 Encodes a RhoGAP Protein with Growth Suppressor Function and Is Underexpressed in Hepatocellular Carcinoma. J. Biol. Chem. 2003, 278, 10824–10830. [Google Scholar] [CrossRef]
- Albertson, R.; Doe, C.Q. Dlg, Scrib and Lgl Regulate Neuroblast Cell Size and Mitotic Spindle Asymmetry. Nat. Cell Biol. 2003, 5, 166–170. [Google Scholar] [CrossRef]
- Peters, J.M.; Tedeschi, A.; Schmitz, J. The Cohesin Complex and its Roles in Chromosome Biology. Genes Dev. 2008, 22, 3089–3114. [Google Scholar] [CrossRef]
- Sigl, R.; Wandke, C.; Rauch, V.; Kirk, J.; Hunt, T.; Geley, S. Loss of the Mammalian APC/C Activator FZR1 Shortens G1 and Lengthens S Phase but has Little Effect on Exit From Mitosis. J. Cell Sci. 2009, 122, 4208–4217. [Google Scholar] [CrossRef]
- Bosco, E.E.; Mulloy, J.C.; Zheng, Y. Rac1 GTPase: A “Rac” of All Trades. Cell. Mol. Life Sci. 2009, 66, 370–374. [Google Scholar] [CrossRef]
- Roskoski, R. ERK1/2 MAP Kinases: Structure, Function, and Regulation. Pharmacol. Res. 2012, 66, 105–143. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Greco, S.; Voltarel, G.; Gaetano, A.S.; Manfrin, C.; Pallavicini, A.; Giulianini, P.G.; Gerdol, M. Comparative Transcriptomic Analysis Reveals Adaptive Traits in Antarctic Scallop Adamussium colbecki. Fishes 2023, 8, 276. https://doi.org/10.3390/fishes8060276
Greco S, Voltarel G, Gaetano AS, Manfrin C, Pallavicini A, Giulianini PG, Gerdol M. Comparative Transcriptomic Analysis Reveals Adaptive Traits in Antarctic Scallop Adamussium colbecki. Fishes. 2023; 8(6):276. https://doi.org/10.3390/fishes8060276
Chicago/Turabian StyleGreco, Samuele, Giacomo Voltarel, Anastasia Serena Gaetano, Chiara Manfrin, Alberto Pallavicini, Piero Giulio Giulianini, and Marco Gerdol. 2023. "Comparative Transcriptomic Analysis Reveals Adaptive Traits in Antarctic Scallop Adamussium colbecki" Fishes 8, no. 6: 276. https://doi.org/10.3390/fishes8060276








