High-Temperature Stress Induces Autophagy in Rainbow Trout Skeletal Muscle
Abstract
:1. Introduction
2. Materials and Methods
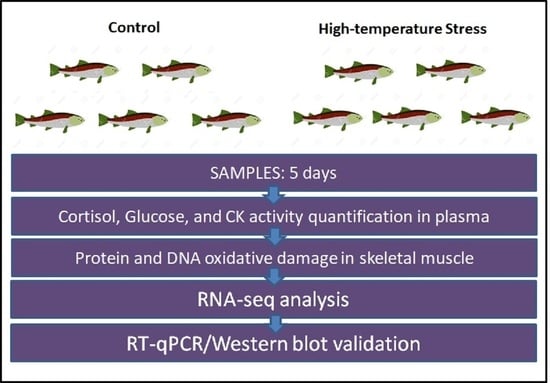
2.1. Experimental Thermal Stress Protocol
2.2. Cortisol, Creatine Kinase Activity, and Glucose Quantification in Blood Plasma
2.3. DNA and Protein Oxidative Damage in Skeletal Muscle
2.4. Skeletal Muscle RNA Extraction and Sequencing
2.5. RNA-Seq and GO Analysis
2.6. RNA-Seq Validation by Real-Time RT-qPCR
2.7. LC3 Western Blot Analysis
2.8. Statistical Analysis
3. Results
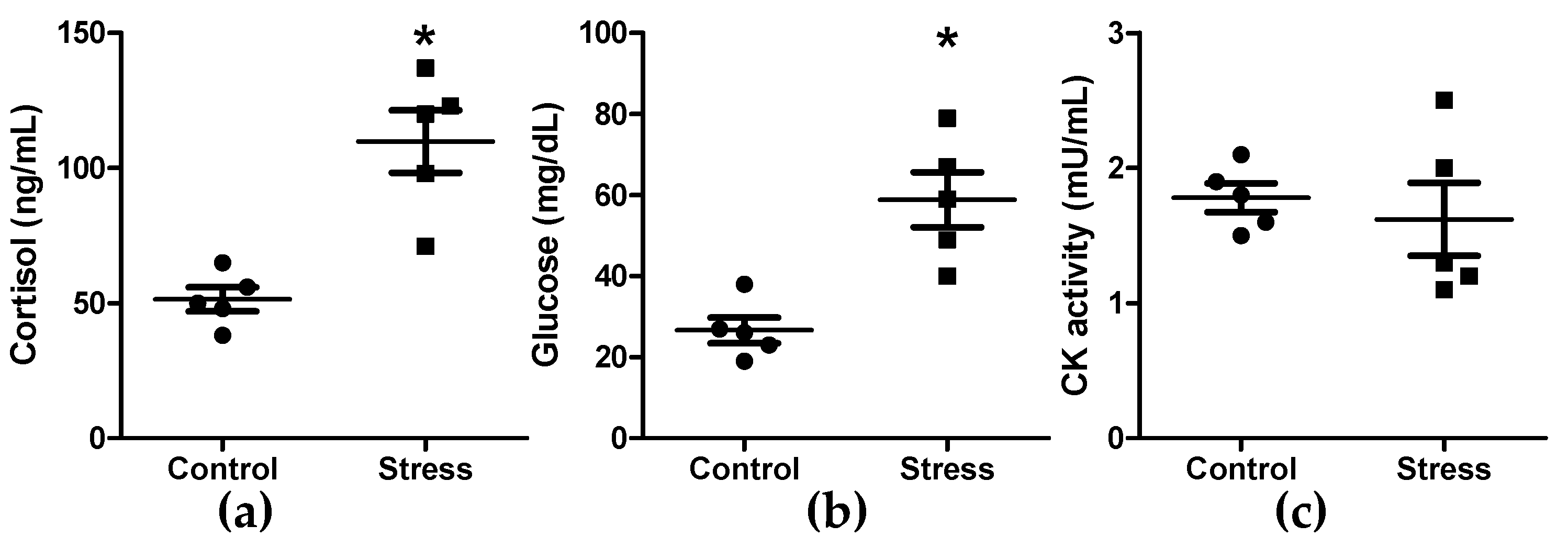
3.1. Cortisol, Glucose, and Creatine Kinase Activity Quantification in Plasma, and Oxidation in Skeletal Muscle Tissue
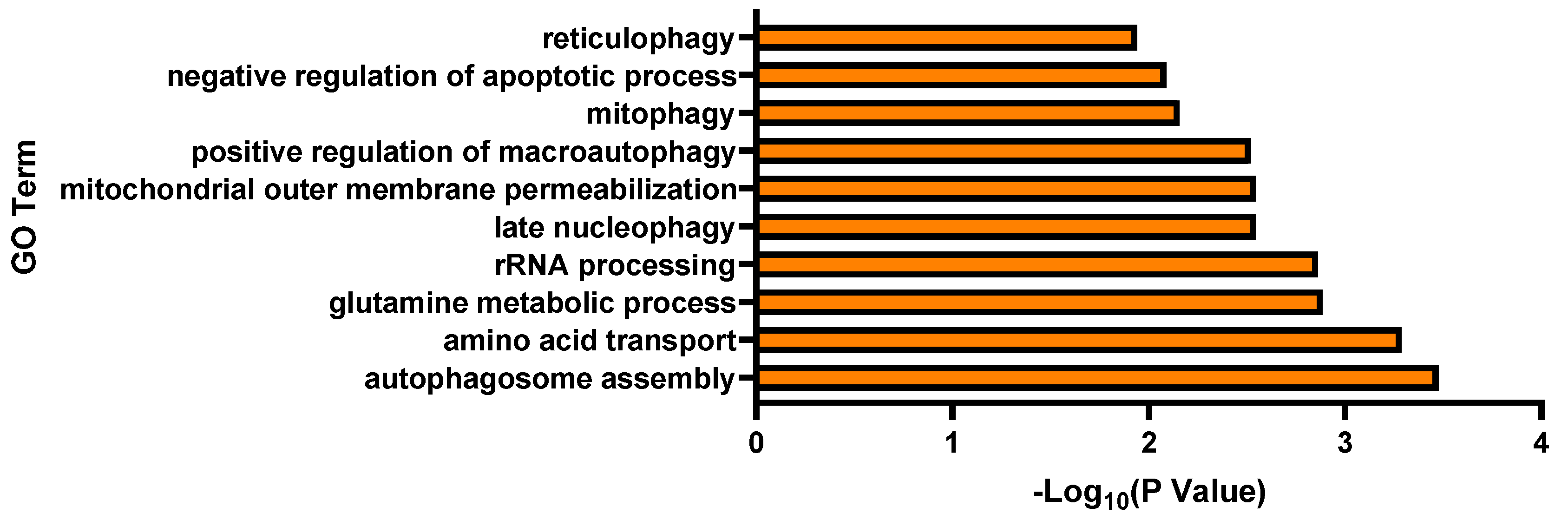
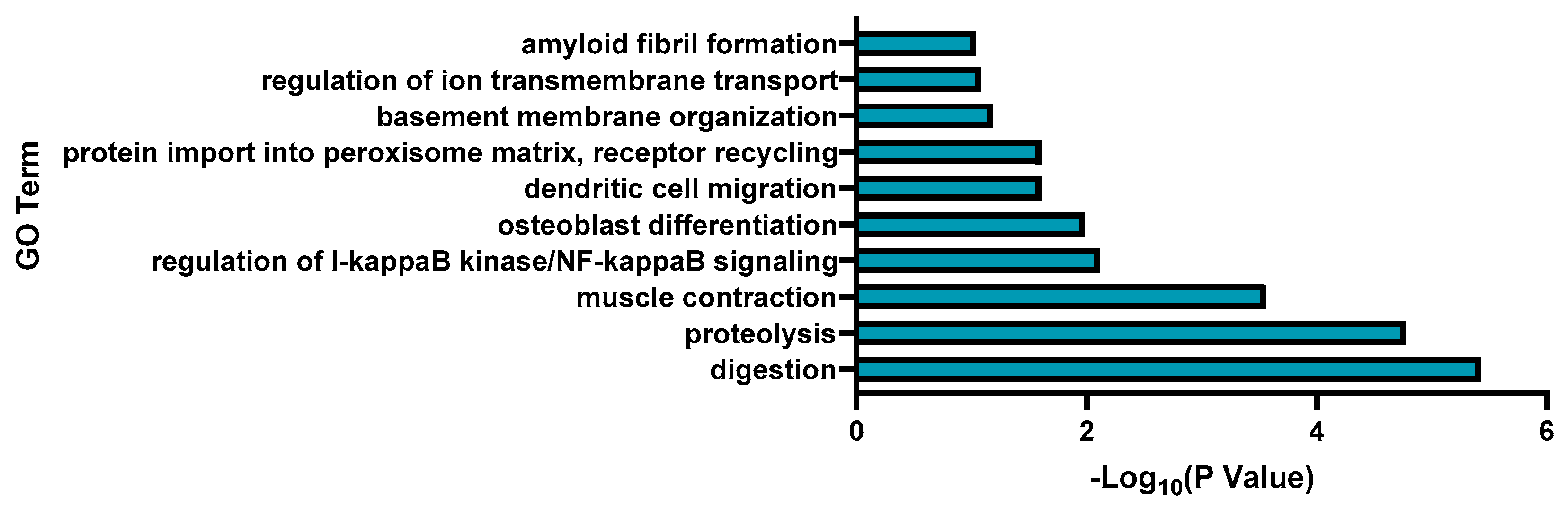
3.2. Transcriptomic Analysis and Pathway Enrichment Analysis
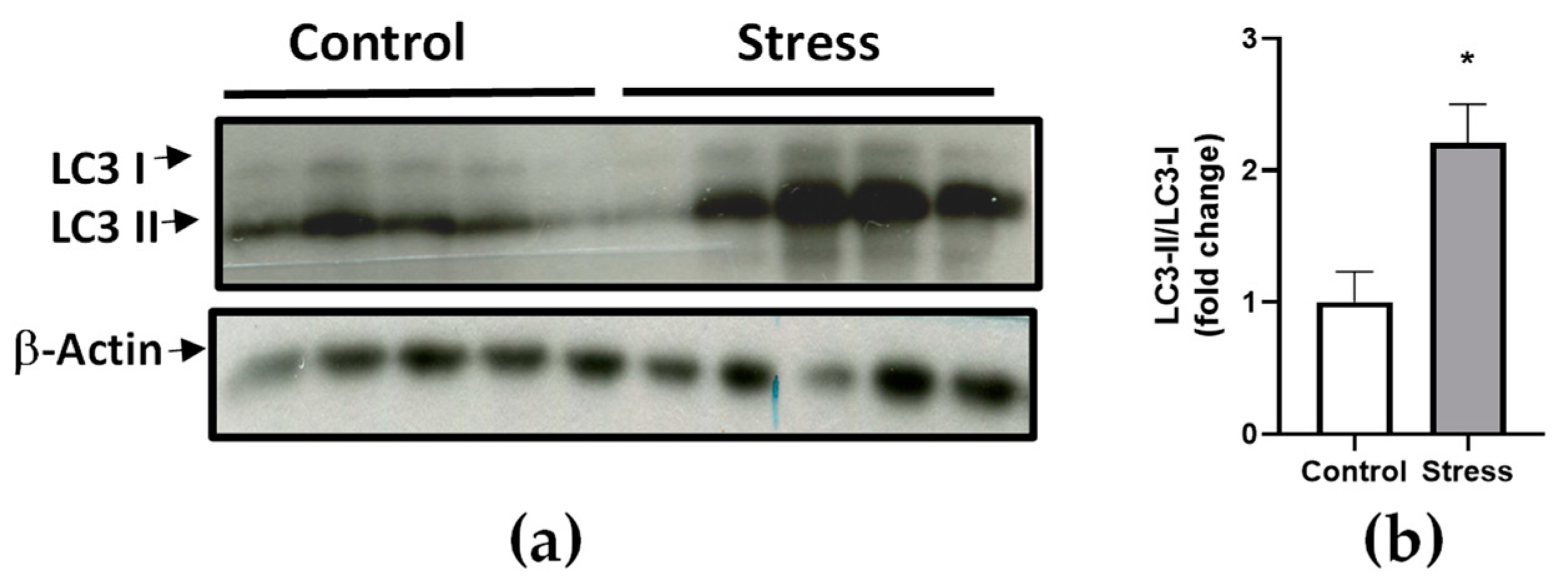
3.3. RNA-Seq Result Validation by Real-Time RT-qPCR and Western Blot
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abbass, K.; Qasim, M.Z.; Song, H.; Murshed, M.; Mahmood, H.; Younis, I. A Review of the Global Climate Change Impacts, Adaptation, and Sustainable Mitigation Measures. Environ. Sci. Pollut. Res. 2022, 29, 42539–42559. [Google Scholar] [CrossRef] [PubMed]
- Chophel, Y. Global Warming and Climate Change (GWCC) Realities. In The Nature, Causes, Effects and Mitigation of Climate Change on the Environment; Harris, S.A., Ed.; IntechOpen: Rijeka, Croatia, 2022; ISBN 978-1-83968-611-5. [Google Scholar]
- Garcia-Soto, C.; Cheng, L.; Caesar, L.; Schmidtko, S.; Jewett, E.B.; Cheripka, A.; Rigor, I.; Caballero, A.; Chiba, S.; Báez, J.C.; et al. An Overview of Ocean Climate Change Indicators: Sea Surface Temperature, Ocean Heat Content, Ocean PH, Dissolved Oxygen Concentration, Arctic Sea Ice Extent, Thickness and Volume, Sea Level and Strength of the AMOC (Atlantic Meridional Overturning Circulation). Front. Mar. Sci. 2021, 8, 642372. [Google Scholar] [CrossRef]
- Wang, B.; Luo, X.; Yang, Y.-M.; Sun, W.; Cane, M.A.; Cai, W.; Yeh, S.-W.; Liu, J. Historical Change of El Niño Properties Sheds Light on Future Changes of Extreme El Niño. Proc. Natl. Acad. Sci. USA 2019, 116, 22512–22517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alfonso, S.; Gesto, M.; Sadoul, B. Temperature Increase and Its Effects on Fish Stress Physiology in the Context of Global Warming. J. Fish. Biol. 2021, 98, 1496–1508. [Google Scholar] [CrossRef] [PubMed]
- Pankhurst, N.W. The Endocrinology of Stress in Fish: An Environmental Perspective. General. Comp. Endocrinol. 2011, 170, 265–275. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Yong, Y.; Ju, X. Effect of Heat Stress on Growth and Production Performance of Livestock and Poultry: Mechanism to Prevention. J. Therm. Biol. 2021, 99, 103019. [Google Scholar] [CrossRef]
- Faught, E.; Vijayan, M.M. Mechanisms of Cortisol Action in Fish Hepatocytes. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2016, 199, 136–145. [Google Scholar] [CrossRef]
- Manneken, J.D.; Dauer, M.V.P.; Currie, P.D. Dynamics of Muscle Growth and Regeneration: Lessons from the Teleost. Exp. Cell Res. 2022, 411, 112991. [Google Scholar] [CrossRef]
- Johnston, I.A.; Bower, N.I.; Macqueen, D.J. Growth and the Regulation of Myotomal Muscle Mass in Teleost Fish. J. Exp. Biol. 2011, 214, 1617–1628. [Google Scholar] [CrossRef] [Green Version]
- Sadoul, B.; Vijayan, M.M. Stress and Growth. In Fish Physiology; Elsevier: Amsterdam, The Netherlands, 2016; Volume 35, pp. 167–205. ISBN 978-0-12-802728-8. [Google Scholar]
- Dettleff, P.; Zuloaga, R.; Fuentes, M.; Gonzalez, P.; Aedo, J.; Estrada, J.M.; Molina, A.; Valdés, J.A. Physiological and Molecular Responses to Thermal Stress in Red Cusk-Eel (Genypterus chilensis) Juveniles Reveals Atrophy and Oxidative Damage in Skeletal Muscle. J. Therm. Biol. 2020, 94, 102750. [Google Scholar] [CrossRef]
- Dettleff, P.; Zuloaga, R.; Fuentes, M.; Gonzalez, P.; Aedo, J.; Estrada, J.M.; Molina, A.; Valdés, J.A. High-Temperature Stress Effect on the Red Cusk-Eel (Geypterus chilensis) Liver: Transcriptional Modulation and Oxidative Stress Damage. Biology 2022, 11, 990. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Chen, Y.; Liu, Y.; Wang, X. Autophagy in Muscle Regeneration: Potential Therapies for Myopathies. J. Cachexia Sarcopenia Muscle 2022, 13, 1673–1685. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Goh, K.Y.; Lee, W.X.; Choy, S.M.; Tang, H.-W. The Importance of MTORC1-Autophagy Axis for Skeletal Muscle Diseases. Int. J. Mol. Sci. 2022, 24, 297. [Google Scholar] [CrossRef]
- Chatzinikita, E.; Maridaki, M.; Palikaras, K.; Koutsilieris, M.; Philippou, A. The Role of Mitophagy in Skeletal Muscle Damage and Regeneration. Cells 2023, 12, 716. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; He, Y.; Wang, S.; Wang, Y.; Shan, P.; Li, P. Autophagy Regulation in Teleost Fish: A Double-Edged Sword. Aquaculture 2022, 558, 738369. [Google Scholar] [CrossRef]
- Rivas-Aravena, A.; Fuentes-Valenzuela, M.; Escobar-Aguirre, S.; Gallardo-Escarate, C.; Molina, A.; Valdés, J.A. Transcriptomic Response of Rainbow Trout (Oncorhynchus mykiss) Skeletal Muscle to Flavobacterium Psychrophilum. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 31, 100596. [Google Scholar] [CrossRef]
- Aedo, J.E.; Zuloaga, R.; Bastías-Molina, M.; Meneses, C.; Boltaña, S.; Molina, A.; Valdés, J.A. Early Transcriptomic Responses Associated with the Membrane-Initiated Action of Cortisol in the Skeletal Muscle of Rainbow Trout (Oncorhynchus mykiss). Physiol. Genom. 2019, 51, 596–606. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Gizińska, J.; Sojka, M. How Climate Change Affects River and Lake Water Temperature in Central-West Poland—A Case Study of the Warta River Catchment. Atmosphere 2023, 14, 330. [Google Scholar] [CrossRef]
- Yuan, D.; Wang, H.; Liu, X.; Wang, S.; Shi, J.; Cheng, X.; Gu, H.; Xiao, S.; Wang, Z. High Temperature Induced Metabolic Reprogramming and Lipid Remodeling in a High-Altitude Fish Species, Triplophysa Bleekeri. Front. Mar. Sci. 2022, 9, 1017142. [Google Scholar] [CrossRef]
- LeBlanc, S.; Middleton, S.; Gilmour, K.M.; Currie, S. Chronic Social Stress Impairs Thermal Tolerance in the Rainbow Trout (Oncorhynchus mykiss). J. Exp. Biol. 2011, 214, 1721–1731. [Google Scholar] [CrossRef] [Green Version]
- Basu, N.; Nakano, T.; Grau, E.G.; Iwama, G.K. The Effects of Cortisol on Heat Shock Protein 70 Levels in Two Fish Species. Gen. Comp. Endocrinol. 2001, 124, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Pal, A.K.; Sahu, N.P.; Jha, A.K.; Priya, P. Biochemical and Physiological Stress Responses to Heat Shock and Their Recovery in Labeo Rohita Fingerlings. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2015, 85, 485–490. [Google Scholar] [CrossRef]
- Uren Webster, T.M.; Rodriguez-Barreto, D.; Martin, S.A.M.; Van Oosterhout, C.; Orozco-terWengel, P.; Cable, J.; Hamilton, A.; Garcia De Leaniz, C.; Consuegra, S. Contrasting Effects of Acute and Chronic Stress on the Transcriptome, Epigenome, and Immune Response of Atlantic Salmon. Epigenetics 2018, 13, 1191–1207. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Casanova, J.C.; Afonso, L.O.B.; Johnson, S.C.; Currie, S.; Gamperl, A.K. The Stress and Metabolic Responses of Juvenile Atlantic Cod Gadus morhua L. to an Acute Thermal Challenge. J. Fish Biol. 2008, 72, 899–916. [Google Scholar] [CrossRef]
- Chadwick, J.G.; Nislow, K.H.; McCormick, S.D. Thermal Onset of Cellular and Endocrine Stress Responses Correspond to Ecological Limits in Brook Trout, an Iconic Cold-Water Fish. Conserv. Physiol. 2015, 3, cov017. [Google Scholar] [CrossRef]
- Aedo, J.E.; Zuloaga, R.; Aravena-Canales, D.; Molina, A.; Valdés, J.A. Role of Glucocorticoid and Mineralocorticoid Receptors in Rainbow Trout (Oncorhynchus mykiss) Skeletal Muscle: A Transcriptomic Perspective of Cortisol Action. Front. Physiol. 2023, 13, 1048008. [Google Scholar] [CrossRef]
- Khieokhajonkhet, A.; Sangphrom, S.; Aeksiri, N.; Tatsapong, P.; Wuthijaree, K.; Kaneko, G. Effects of Long-Term Exposure to High Temperature on Growth Performance, Chemical Composition, Hematological and Histological Changes, and Physiological Responses in Hybrid Catfish [♂Clarias Gariepinus (Burchell, 1822) ×♀C. Macrocephalus (Günther, 1864)]. J. Therm. Biol. 2022, 105, 103226. [Google Scholar] [CrossRef]
- Vo, T.T.M.; Amoroso, G.; Ventura, T.; Elizur, A. Histological and Transcriptomic Analysis of Muscular Atrophy Associated with Depleted Flesh Pigmentation in Atlantic Salmon (Salmo salar) Exposed to Elevated Seawater Temperatures. Sci. Rep. 2023, 13, 4218. [Google Scholar] [CrossRef]
- Feidantsis, K.; Georgoulis, I.; Zachariou, A.; Campaz, B.; Christoforou, M.; Pörtner, H.O.; Michaelidis, B. Energetic, Antioxidant, Inflammatory and Cell Death Responses in the Red Muscle of Thermally Stressed Sparus Aurata. J. Comp. Physiol. B 2020, 190, 403–418. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular Definitions of Autophagy and Related Processes. EMBO J. 2017, 36, 1811–1836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khandia, R.; Dadar, M.; Munjal, A.; Dhama, K.; Karthik, K.; Tiwari, R.; Yatoo, M.I.; Iqbal, H.M.N.; Singh, K.P.; Joshi, S.K.; et al. A Comprehensive Review of Autophagy and Its Various Roles in Infectious, Non-Infectious, and Lifestyle Diseases: Current Knowledge and Prospects for Disease Prevention, Novel Drug Design, and Therapy. Cells 2019, 8, 674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cassel, M.; de Paiva Camargo, M.; Oliveira de Jesus, L.W.; Borella, M.I. Involution Processes of Follicular Atresia and Post-Ovulatory Complex in a Characid Fish Ovary: A Study of Apoptosis and Autophagy Pathways. J. Mol. Hist. 2017, 48, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Godoi, F.G.A.; Forner-Piquer, I.; Randazzo, B.; Habibi, H.R.; Lo Nostro, F.L.; Moreira, R.G.; Carnevali, O. Effects of Di-Isononyl Phthalate (DiNP) on Follicular Atresia in Zebrafish Ovary. Front. Endocrinol. 2021, 12, 677853. [Google Scholar] [CrossRef]
- Cheng, Y.; Lai, F.; Wang, X.; Shang, D.; Zou, J.; Luo, M.; Xia, X.; Cheng, H.; Zhou, R. Srag Regulates Autophagy via Integrating into a Preexisting Autophagy Pathway in Testis. Mol. Biol. Evol. 2021, 38, 128–141. [Google Scholar] [CrossRef]
- Seiliez, I.; Belghit, I.; Gao, Y.; Skiba-Cassy, S.; Dias, K.; Cluzeaud, M.; Rémond, D.; Hafnaoui, N.; Salin, B.; Camougrand, N.; et al. Looking at the Metabolic Consequences of the Colchicine-Based in Vivo Autophagic Flux Assay. Autophagy 2016, 12, 343–356. [Google Scholar] [CrossRef] [Green Version]
- Dai, Z.; Cheng, J.; Bao, L.; Zhu, X.; Li, H.; Chen, X.; Zhang, Y.; Zhang, J.; Chu, W.; Pan, Y.; et al. Exposure to Waterborne Cadmium Induce Oxidative Stress, Autophagy and Mitochondrial Dysfunction in the Liver of Procypris Merus. Ecotoxicol. Environ. Saf. 2020, 204, 111051. [Google Scholar] [CrossRef]
- Masud, S.; Prajsnar, T.K.; Torraca, V.; Lamers, G.E.M.; Benning, M.; Van Der Vaart, M.; Meijer, A.H. Macrophages Target Salmonella by Lc3-Associated Phagocytosis in a Systemic Infection Model. Autophagy 2019, 15, 796–812. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Lu, Z.; Li, F.; Shi, F.; Zhan, F.; Zhao, L.; Li, Y.; Li, J.; Lin, L.; Qin, Z. Escherichia coli Induced Ferroptosis in Red Blood Cells of grass carp (Ctenopharyngodon Idella). Fish Shellfish Immunol. 2021, 112, 159–167. [Google Scholar] [CrossRef]
- Valenzuela, C.A.; Zuloaga, R.; Mercado, L.; Einarsdottir, I.E.; Björnsson, B.T.; Valdés, J.A.; Molina, A. Chronic Stress Inhibits Growth and Induces Proteolytic Mechanisms through Two Different Nonoverlapping Pathways in the Skeletal Muscle of a Teleost Fish. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2018, 314, R102–R113. [Google Scholar] [CrossRef] [PubMed]
- Aedo, J.E.; Maldonado, J.; Aballai, V.; Estrada, J.M.; Bastias-Molina, M.; Meneses, C.; Gallardo-Escarate, C.; Silva, H.; Molina, A.; Valdés, J.A. MRNA-Seq Reveals Skeletal Muscle Atrophy in Response to Handling Stress in a Marine Teleost, the Red Cusk-Eel (Genypterus chilensis). BMC Genom. 2015, 16, 1024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aedo, J.E.; Aravena-Canales, D.; Dettleff, P.; Fuentes-Valenzuela, M.; Zuloaga, R.; Rivas-Aravena, A.; Molina, A.; Valdés, J.A. RNA-Seq Analysis Reveals the Dynamic Regulation of Proteasomal and Autophagic Degradation Systems of Rainbow Trout (Oncorhynchus mykiss) Skeletal Muscle Challenged with Infectious Pancreatic Necrosis Virus (IPNV). Aquaculture 2022, 552, 738000. [Google Scholar] [CrossRef]
- Sun, C.-C.; Zhou, Z.-Q.; Chen, Z.-L.; Zhu, R.-K.; Yang, D.; Peng, X.-Y.; Zheng, L.; Tang, C.-F. Identification of Potentially Related Genes and Mechanisms Involved in Skeletal Muscle Atrophy Induced by Excessive Exercise in Zebrafish. Biology 2021, 10, 761. [Google Scholar] [CrossRef] [PubMed]
- Dhanasekaran, D.N.; Reddy, E.P. JNK-Signaling: A Multiplexing Hub in Programmed Cell Death. Genes. Cancer 2017, 8, 682–694. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Wen, H.; Qi, X.; Zhang, X.; Li, Y. Identification of Mapk Gene Family in Lateolabrax Maculatus and Their Expression Profiles in Response to Hypoxia and Salinity Challenges. Gene 2019, 684, 20–29. [Google Scholar] [CrossRef]
- Gao, A.; Jiang, J.; Xie, F.; Chen, L. Bnip3 in Mitophagy: Novel Insights and Potential Therapeutic Target for Diseases of Secondary Mitochondrial Dysfunction. Clin. Chim. Acta 2020, 506, 72–83. [Google Scholar] [CrossRef]
- Valenzuela, C.A.; Escobar, D.; Perez, L.; Zuloaga, R.; Estrada, J.M.; Mercado, L.; Valdés, J.A.; Molina, A. Transcriptional Dynamics of Immune, Growth and Stress Related Genes in Skeletal Muscle of the Fine Flounder (Paralichthys Adpersus) during Different Nutritional Statuses. Dev. Comp. Immunol. 2015, 53, 145–157. [Google Scholar] [CrossRef]
- Jin, M.; Klionsky, D.J. Transcriptional Regulation of ATG9 by the Pho23-Rpd3 Complex Modulates the Frequency of Autophagosome Formation. Autophagy 2014, 10, 1681–1682. [Google Scholar] [CrossRef] [Green Version]
- Donninger, H.; Schmidt, M.L.; Mezzanotte, J.; Barnoud, T.; Clark, G.J. Ras Signaling through RASSF Proteins. Semin. Cell Dev. Biol. 2016, 58, 86–95. [Google Scholar] [CrossRef] [Green Version]
- Yan, S.; Chen, R.; Wang, M.; Zha, J. Carbamazepine at Environmentally Relevant Concentrations Caused DNA Damage and Apoptosis in the Liver of Chinese Rare Minnows (Gobiocypris rarus) by the Ras/Raf/ERK/P53 Signaling Pathway. Environ. Pollut. 2021, 270, 116245. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, S.; Pearce, S.C.; Gabler, N.K.; Baumgard, L.H.; Rhoads, R.P.; Selsby, J.T. Short-Term Heat Stress Results in Increased Apoptotic Signaling and Autophagy in Oxidative Skeletal Muscle in Sus Scrofa. J. Therm. Biol. 2018, 72, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Hirunsai, M.; Srikuea, R. Autophagy-Lysosomal Signaling Responses to Heat Stress in Tenotomy-Induced Rat Skeletal Muscle Atrophy. Life Sci. 2021, 275, 119352. [Google Scholar] [CrossRef] [PubMed]
- Tamura, Y.; Kitaoka, Y.; Matsunaga, Y.; Hoshino, D.; Hatta, H. Daily Heat Stress Treatment Rescues Denervation-Activated Mitochondrial Clearance and Atrophy in Skeletal Muscle: Heat Stress Treatment in Denervated Skeletal Muscle. J. Physiol. 2015, 593, 2707–2720. [Google Scholar] [CrossRef] [Green Version]
- Summers, C.M.; Valentine, R.J. Acute Heat Exposure Alters Autophagy Signaling in C2C12 Myotubes. Front. Physiol. 2020, 10, 1521. [Google Scholar] [CrossRef]
- Yun, H.R.; Jo, Y.H.; Kim, J.; Shin, Y.; Kim, S.S.; Choi, T.G. Roles of Autophagy in Oxidative Stress. Int. J. Mol. Sci. 2020, 21, 3289. [Google Scholar] [CrossRef]
- Bolisetty, S.; Jaimes, E. Mitochondria and Reactive Oxygen Species: Physiology and Pathophysiology. Int. J. Mol. Sci. 2013, 14, 6306–6344. [Google Scholar] [CrossRef]
- Wen, X.; Tang, L.; Zhong, R.; Liu, L.; Chen, L.; Zhang, H. Role of Mitophagy in Regulating Intestinal Oxidative Damage. Antioxidants 2023, 12, 480. [Google Scholar] [CrossRef]

| KEGG Pathway | p-Value | Upregulated Genes |
|---|---|---|
| Mitophagy—animal | 1.45 × 10−4 | mapk10, bnip3l, bnip3, ubc, atg9a, ulk1, rab7a |
| Autophagy—animal | 9.89 × 10−4 | mapk10, bnip3, atg9a, ulk1, raf1, rab7a, atg4d, atg2b |
| Spinocerebellar ataxia | 1.07 × 10−3 | mapk10, por, psmd11, psmd2, psmd3, atp2a2, ulk1, atg2b |
| Protein processing in ER | 2.99 × 10−3 | mapk10, hsp90ab1, hspa1l, canx, cul1, plaa, cryaa |
| Alzheimer’s disease | 1.05 × 10−2 | mapk10, gsk3b, por, psmd11, cdk5, psmd2, psmd3, atp2a2, ulk1, raf1, atg2b |
| Antigen processing | 1.12 × 10−2 | hsp90ab1, hspa1l, hspa4, canx, rfxap |
| ErbB signaling pathway | 1.50 × 10−2 | mapk10, map2k4, gsk3b, myc, raf1 |
| Pathways of neurodegeneration | 1.70 × 10−2 | mapk10, gsk3b, por, psmd11, cdk5, psmd2, ubc, psmd3, atp2a2, ulk1, raf1, atg2b |
| Legionellosis | 2.58 × 10−2 | hspa1l, rab1b, bnip3, eef1a2 |
| mTOR signaling pathway | 3.05 × 10−2 | gsk3b, cab39, ulk1, raf1, lpin1, wdr24 |
| KEGG Pathway | p-Value | Downregulated Genes |
|---|---|---|
| Pancreatic secretion | 1.10 × 10−10 | prss1, cela2a, cpb1, ctrb2, ctrb1, amy1c, atp2a1, cel, prss3, prss2 |
| Protein digestion and absorption | 2.35 × 10−6 | prss1, cela2a, cpb1, ctrb2, ctrb1, prss3, prss2 |
| Adrenergic signaling in cardiomyocytes | 2.59 × 10−3 | cacnb1, tpm3, atp2a1, scn1b, myh7 |
| cGMP-PKG signaling pathway | 3.82 × 10−3 | atp2a1, vdac1, raf1, prkg1, myh7 |
| Influenza A | 4.15 × 10−3 | prss1, vdac1, raf1, prss3, prss2 |
| Cardiac muscle contraction | 4.57 × 10−3 | cacnb1, tpm3, atp2a1, myh7 |
| Hypertrophic cardiomyopathy | 5.03 × 10−3 | cacnb1, tpm3, atp2a1, myh7 |
| Dilated cardiomyopathy | 6.02 × 10−3 | cacnb1, tpm3, atp2a1, myh7 |
| Diabetic cardiomyopathy | 4.37 × 10−2 | atp5f1b, atp2a1, vdac1, sdha |
| Chemical carcinogenesis—ROS | 5.51 × 10−2 | atp5f1b, vdac1, sdha, raf1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molina, A.; Dettleff, P.; Valenzuela-Muñoz, V.; Gallardo-Escarate, C.; Valdés, J.A. High-Temperature Stress Induces Autophagy in Rainbow Trout Skeletal Muscle. Fishes 2023, 8, 303. https://doi.org/10.3390/fishes8060303
Molina A, Dettleff P, Valenzuela-Muñoz V, Gallardo-Escarate C, Valdés JA. High-Temperature Stress Induces Autophagy in Rainbow Trout Skeletal Muscle. Fishes. 2023; 8(6):303. https://doi.org/10.3390/fishes8060303
Chicago/Turabian StyleMolina, Alfredo, Phillip Dettleff, Valentina Valenzuela-Muñoz, Cristian Gallardo-Escarate, and Juan Antonio Valdés. 2023. "High-Temperature Stress Induces Autophagy in Rainbow Trout Skeletal Muscle" Fishes 8, no. 6: 303. https://doi.org/10.3390/fishes8060303
APA StyleMolina, A., Dettleff, P., Valenzuela-Muñoz, V., Gallardo-Escarate, C., & Valdés, J. A. (2023). High-Temperature Stress Induces Autophagy in Rainbow Trout Skeletal Muscle. Fishes, 8(6), 303. https://doi.org/10.3390/fishes8060303