Does Size Matter? Small and Large Larvae of Pikeperch (Sander lucioperca) in a Comparative Gene Expression Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Size Sorting
2.2. RNA Isolation
2.3. Design of Primer and Fluidigm Multiplex Real-Time PCR
2.4. Statistical Analyses
3. Results
3.1. Morphometry
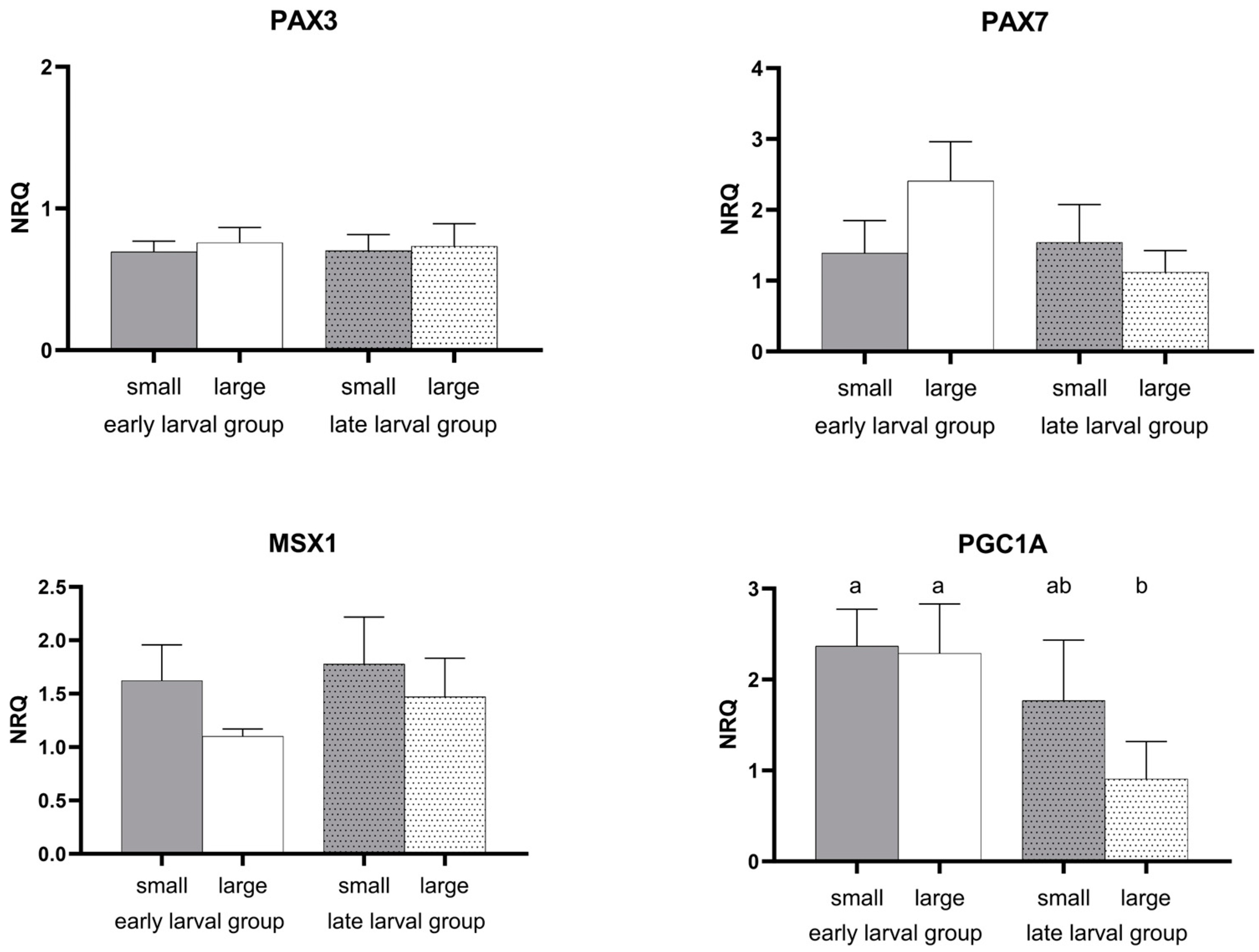
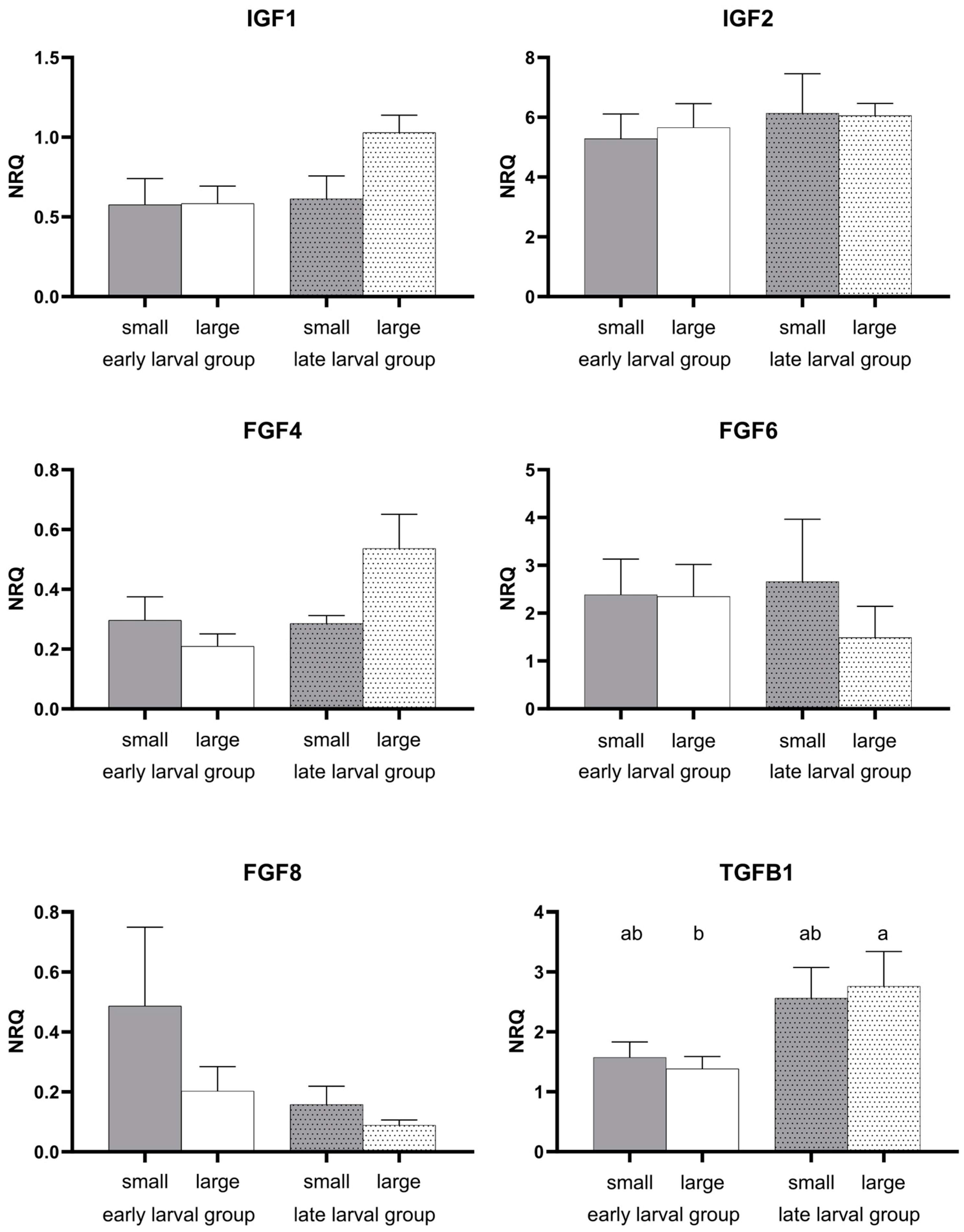
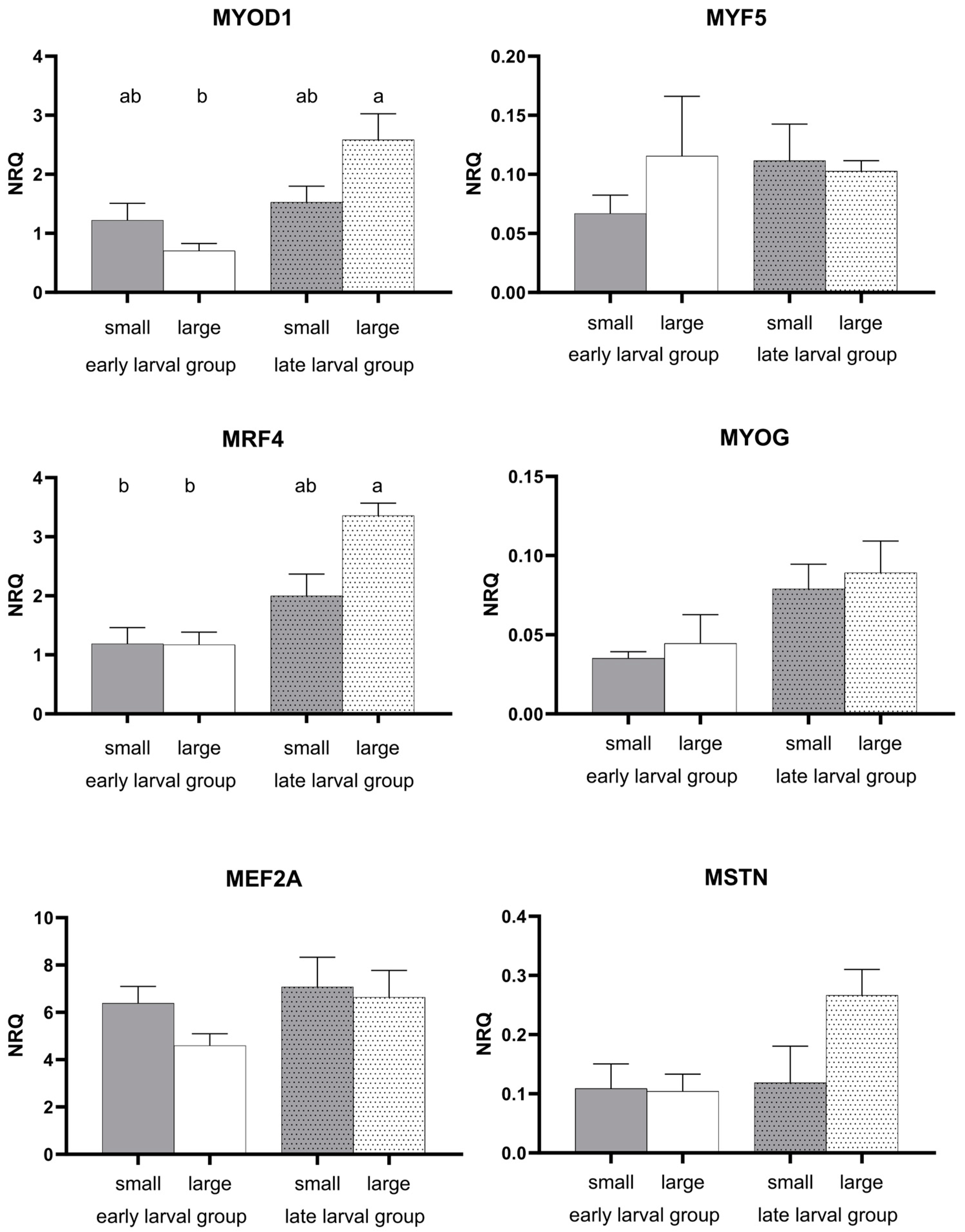
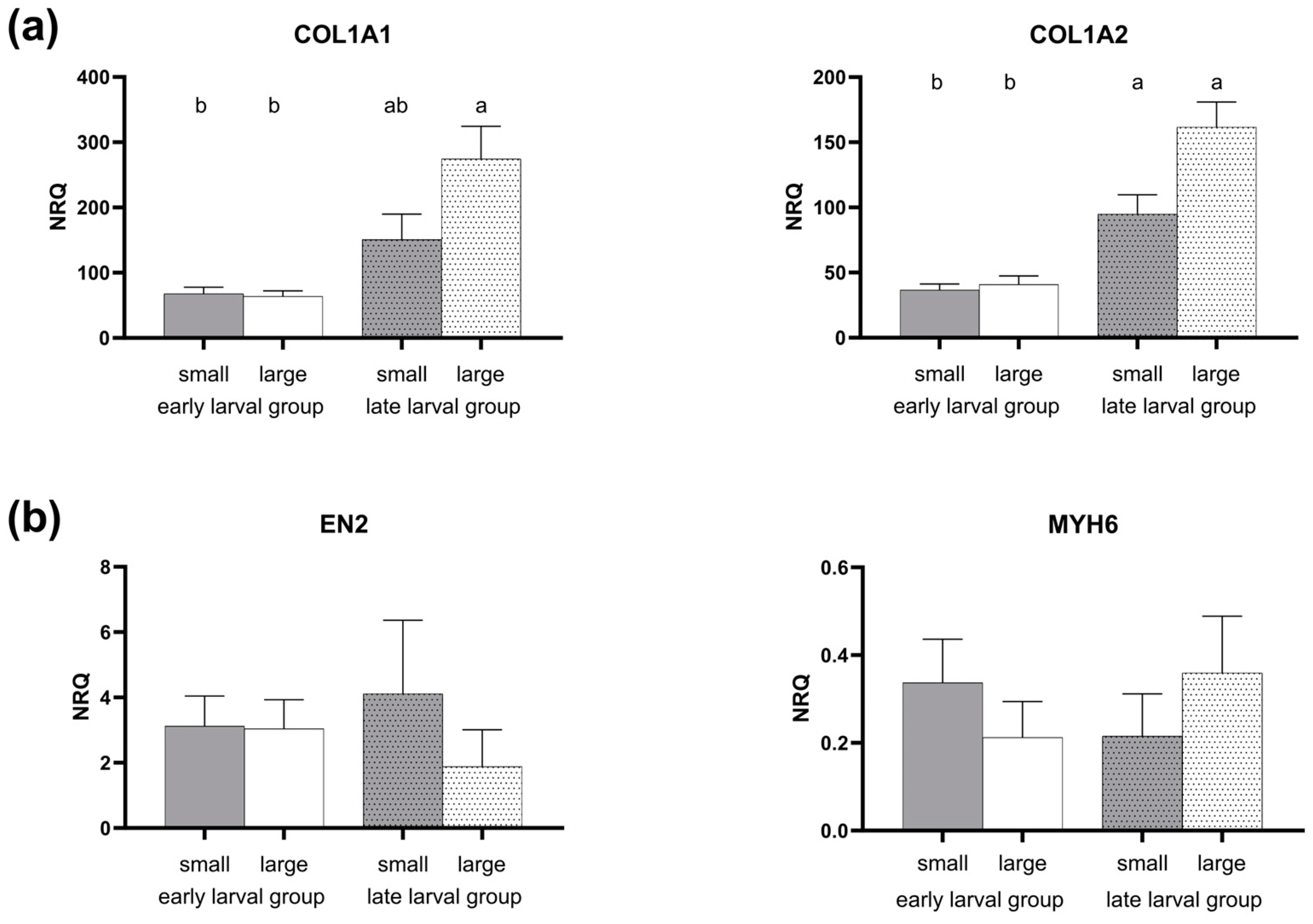
3.2. Gene Expression Pattern
4. Discussion
4.1. Morphometric Distinctions of Size Classes and Growth
4.2. Regulatory Genes Connected to Stem Cell Activation
4.3. Genes of Growth and General Development
4.4. Myogenic Development and Structural Marker Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Policar, T.; Schaefer, F.J.; Panana, E.; Meyer, S.; Teerlinck, S.; Toner, D.; Zarski, D. Recent progress in European percid fish culture production technology-tackling bottlenecks. Aquac. Int. 2019, 27, 1151–1174. [Google Scholar]
- Colchen, T.; Gisbert, E.; Krauss, D.; Ledoré, Y.; Pasquet, A.; Fontaine, P. Improving pikeperch larviculture by combining environmental, feeding and populational factors. Aquac. Rep. 2020, 17, 100337. [Google Scholar] [CrossRef]
- Colchen, T.; Fontaine, P.; Ledoré, Y.; Teletchea, F.; Pasquet, A. Intra-cohort cannibalism in early life stages of pikeperch. Aquac. Res. 2019, 50, 915–924. [Google Scholar] [CrossRef]
- Steenfeldt, S.; Lund, I.; Höglund, E. Is batch variability in hatching time related to size heterogeneity and cannibalism in pikeperch (Sander lucioperca)? Aquac. Res. 2011, 42, 727–732. [Google Scholar] [CrossRef]
- Szczepkowski, M.; Zakęś, Z.; Szczepkowska, B.; Piotrowska, I. Effect of size sorting on the survival, growth and cannibalism in pikeperch (Sander lucioperca L.) larvae during intensive culture in RAS. Czech. J. Anim. Sci. 2011, 56, 483–489. [Google Scholar] [CrossRef]
- Colchen, T.; Dias, A.; Gisbert, E.; Teletchea, F.; Fontaine, P.; Pasquet, A. The onset of piscivory in a freshwater fish species: Analysis of behavioural and physiological traits. J. Fish Biol. 2020, 96, 1463–1474. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.S.; Agostinho, A.A.; Winemiller, K.O. Revisiting cannibalism in fishes. Rev. Fish Biol. Fish. 2017, 27, 499–513. [Google Scholar]
- Szkudlarek, M.; Zakęś, Z. Effect of stocking density on survival and growth performance of pikeperch, Sander lucioperca (L.), larvae under controlled conditions. Aquac. Int. 2007, 15, 67–81. [Google Scholar]
- Franz, G.P.; Lewerentz, L.; Grunow, B. Observations of growth changes during the embryonic-larval-transition of pikeperch (Sander lucioperca) under near-natural conditions. J. Fish Biol. 2021, 99, 425–436. [Google Scholar] [CrossRef]
- Ott, A.; Löffler, J.; Ahnelt, H.; Keckeis, H. Early development of the postcranial skeleton of the pikeperch Sander lucioperca (Teleostei: Percidae) relating to developmental stages and growth. J. Morphol. 2012, 273, 894–908. [Google Scholar] [CrossRef]
- Penaz, M. A general framework of fish ontogeny: A review of the ongoing debate. Folia Zool. 2001, 50, 241–256. [Google Scholar]
- El Kertaoui, N.; Lund, I.; Assogba, H.; Dominguez, D.; Izquierdo, M.S.; Baekelandt, S.; Cornet, V.; Mandiki, S.N.M.; Montero, D.; Kestemont, P. Key nutritional factors and interactions during larval development of pikeperch (Sander lucioperca). Sci. Rep. 2019, 9, 7074. [Google Scholar] [CrossRef] [PubMed]
- Imentai, A.; Gilannejad, N.; Martínez-Rodríguez, G.; López, F.J.M.; Martínez, F.P.; Pěnka, T.; Dzyuba, V.; Dadras, H.; Policar, T. Effects of First Feeding Regime on Gene Expression and Enzyme Activity in Pikeperch (Sander lucioperca) Larvae. Front. Mar. Sci. 2022, 9, 864536. [Google Scholar] [CrossRef]
- Franz, G.P.; Tönißen, K.; Rebl, A.; Lutze, P.; Grunow, B. The expression of myogenic gene markers during the embryo-larval-transition in Pikeperch (Sander lucioperca). Aquac. Res. 2022, 53, 14. [Google Scholar] [CrossRef]
- Lavajoo, F.; Falahatkar, B.; Perelló-Amorós, M.; Moshayedi, F.; Efatpanah, I.; Gutiérrez, J. The pattern of gene expression (IGF family, muscle growth regulatory factors and osteogenesis related genes) involved in growth of skeletal muscle in pikeperch (Sander lucioperca) during ontogenesis. Preprint 2023. [Google Scholar] [CrossRef]
- Schäfer, N.; Kaya, Y.; Rebl, H.; Stüeken, M.; Rebl, A.; Nguinkal, J.A.; Franz, G.P.; Brunner, R.M.; Goldammer, T.; Grunow, B.; et al. Insights into early ontogenesis: Characterization of stress and development key genes of pikeperch (Sander lucioperca) in vivo and in vitro. Fish Physiol. Biochem. 2021, 47, 515–532. [Google Scholar] [CrossRef]
- Rebl, A.; Rebl, H.; Verleih, M.; Haupt, S.; Köbis, J.M.; Goldammer, T.; Seyfert, H.-M. At Least Two Genes Encode Many Variants of Irak3 in Rainbow Trout, but Neither the Full-Length Factor Nor Its Variants Interfere Directly With the TLR-Mediated Stimulation of Inflammation. Front. Immunol. 2019, 10, 2246. [Google Scholar] [CrossRef]
- Swirplies, F.; Wuertz, S.; Baßmann, B.; Orban, A.; Schäfer, N.; Brunner, R.M.; Hadlich, F.; Goldammer, T.; Rebl, A. Identification of molecular stress indicators in pikeperch Sander lucioperca correlating with rising water temperatures. Aquaculture 2019, 501, 260–271. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef]
- Löffler, J.; Ott, A.; Ahnelt, H.; Keckeis, H. Early development of the skull of Sander lucioperca (L.) (Teleostei: Percidae) relating to growth and mortality. J. Fish Biol. 2008, 72, 233–258. [Google Scholar] [CrossRef]
- Mehner, T.; Hüulsmann, S.; Worischka, S.; Plewa, M.; Benndorf, J. Is the midsummer decline of Daphnia really induced by age-0 fish predation? Comparison of fish consumption and M Daphnia mortality and life history parameters in a biomanipulated reservoir. J. Plankton Res. 1998, 20, 1797–1811. [Google Scholar] [CrossRef]
- Dörner, H.; Hülsmann, S.; Hölker, F.; Skov, C.; Wagner, A. Size-dependent predator–prey relationships between pikeperch and their prey fish. Ecol. Freshw. Fish 2007, 16, 307–314. [Google Scholar] [CrossRef]
- Vehanen, T.; Hyvärinen, P.; Huusko, A. Food consumption and prey orientation of piscivorous brown trout (Salmo trutta) and pikeperch (Stizostedion lucioperca) in a large regulated lake. J. Appl. Ichthyol. 1998, 14, 15–22. [Google Scholar] [CrossRef]
- Baras, E.; Jobling, M. Dynamics of intracohort cannibalism in cultured fish. Aquac. Res. 2002, 33, 461–479. [Google Scholar] [CrossRef]
- Xu, Z.; Li, C.; Ling, Q.; Gaughan, S.; Wang, G.; Han, X. Early development and the point of no return in pikeperch (Sander lucioperca L.) larvae. Chin. J. Oceanol. Limnol. 2017, 35, 1493–1500. [Google Scholar] [CrossRef]
- Güralp, H.; Pocherniaieva, K.; Blecha, M.; Policar, T.; Psenicka, M.; Saito, T. Development, and effect of water temperature on development rate, of pikeperch Sander lucioperca embryos. Theriogenology 2017, 104, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Hatta, K.; Schilling, T.F.; BreMiller, R.A.; Kimmel, C.B. Specification of Jaw Muscle Identity in Zebrafish: Correlation with engrailed-Homeoprotein Expression. Science 1990, 250, 802–805. [Google Scholar] [CrossRef]
- Knight, R.D.; Mebus, K.; Roehl, H.H. Mandibular arch muscle identity is regulated by a conserved molecular process during vertebrate development. J. Exp. Zool. Part B Mol. Dev. Evol. 2008, 310B, 355–369. [Google Scholar] [CrossRef]
- Vecino, E.; Ekström, P. Expression of the homeobox engrailed gene during the embryonic development of the nervous system of the trout (Salmo fario L.). Neurosci. Lett. 1991, 129, 311–314. [Google Scholar] [CrossRef]
- Scholpp, S.; Brand, M. Morpholino-induced knockdown of zebrafish engrailed genes eng2 and eng3 reveals redundant and unique functions in midbrain--hindbrain boundary development. Genesis 2001, 30, 129–133. [Google Scholar] [CrossRef]
- Degenhardt, K.; Sassoon, D.A. A role for Engrailed-2 in determination of skeletal muscle physiologic properties. Dev. Biol. 2001, 231, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Relaix, F.; Rocancourt, D.; Mansouri, A.; Buckingham, M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 2005, 435, 948–953. [Google Scholar] [CrossRef] [PubMed]
- Hammond, C.L.; Hinits, Y.; Osborn, D.P.S.; Minchin, J.E.N.; Tettamanti, G.; Hughes, S.M. Signals and myogenic regulatory factors restrict pax3 and pax7 expression to dermomyotome-like tissue in zebrafish. Dev. Biol. 2007, 302, 504–521. [Google Scholar] [CrossRef] [PubMed]
- Akolkar, D.B.; Asaduzzaman, M.; Kinoshita, S.; Asakawa, S.; Watabe, S. Characterization of Pax3 and Pax7 genes and their expression patterns during different development and growth stages of Japanese pufferfish Takifugu rubripes. Gene 2016, 575, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Bendall, A.J.; Ding, J.; Hu, G.; Shen, M.M.; Abate-Shen, C. Msx1 antagonizes the myogenic activity of Pax3 in migrating limb muscle precursors. Development 1999, 126, 4965–4976. [Google Scholar] [CrossRef]
- Rossi, G.; Messina, G. Comparative myogenesis in teleosts and mammals. Cell Mol. Life Sci. 2014, 71, 3081–3099. [Google Scholar] [CrossRef] [PubMed]
- Franz, A.C.; Faass, O.; Kollner, B.; Shved, N.; Link, K.; Casanova, A.; Wenger, M.; D’Cotta, H.; Baroiller, J.F.; Ullrich, O.; et al. Endocrine and Local IGF-I in the Bony Fish Immune System. Biology 2016, 5, 9. [Google Scholar] [CrossRef]
- Puigserver, P. Tissue-specific regulation of metabolic pathways through the transcriptional coactivator PGC1-alpha. Int. J. Obes. 2005, 29 (Suppl. S1), S5–S9. [Google Scholar] [CrossRef]
- Yu, E.; Foote, K.; Bennett, M. Mitochondrial function in thoracic aortic aneurysms. Cardiovasc. Res. 2018, 114, 1696–1698. [Google Scholar] [CrossRef]
- Nam, H.; Kundu, A.; Karki, S.; Brinkley, G.J.; Chandrashekar, D.S.; Kirkman, R.L.; Liu, J.; Liberti, M.V.; Locasale, J.W.; Mitchell, T.; et al. The TGF-β/HDAC7 axis suppresses TCA cycle metabolism in renal cancer. JCI Insight 2021, 6, e148438. [Google Scholar] [CrossRef]
- Darias, M.J.; Zambonino-Infante, J.L.; Hugot, K.; Cahu, C.L.; Mazurais, D. Gene Expression Patterns During the Larval Development of European Sea Bass (Dicentrarchus Labrax) by Microarray Analysis. Mar. Biotechnol. 2008, 10, 416–428. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bobe, J.; Labbé, C. Egg and sperm quality in fish. Gen. Comp. Endocrinol. 2010, 165, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, C.V.; Chapman, R.W.; Reading, B.J.; Anderson, P.E. Transcriptomics of mRNA and egg quality in farmed fish: Some recent developments and future directions. Gen. Comp. Endocrinol. 2015, 221, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Schnapp, E.; Pistocchi, A.S.; Karampetsou, E.; Foglia, E.; Lamia, C.L.; Cotelli, F.; Cossu, G. Induced early expression of mrf4 but not myog rescues myogenesis in the myod/myf5 double-morphant zebrafish embryo. J. Cell Sci. 2009, 122, 481–488. [Google Scholar] [CrossRef]
- Gelse, K.; Pöschl, E.; Aigner, T. Collagens—Structure, function, and biosynthesis. Adv. Drug Deliv. Rev. 2003, 55, 1531–1546. [Google Scholar] [CrossRef]
- Rescan, P.Y. Development of myofibres and associated connective tissues in fish axial muscle: Recent insights and future perspectives. Differentiation 2019, 106, 35–41. [Google Scholar] [CrossRef]
- Sefton, E.M.; Kardon, G. Chapter Five—Connecting muscle development, birth defects, and evolution: An essential role for muscle connective tissue. In Current Topics in Developmental Biology; Wellik, D.M., Ed.; Academic Press: Cambridge, MA, USA, 2019; Volume 132, pp. 137–176. [Google Scholar]



| Gene Symbol | Developmental Group p Value | Size Class p Value | Effect Interaction (Group × Size) p Value |
|---|---|---|---|
| COL1A1 | 0.000 | 0.196 | 0.100 |
| COL1A2 | 0.000 | 0.065 | 0.130 |
| EN2 | 0.195 | 0.445 | 0.423 |
| FGF4 | 0.032 | 0.652 | 0.143 |
| FGF6 | 0.274 | 0.691 | 0.594 |
| FGF8 | 0.178 | 0.353 | 0.887 |
| IGF1 | 0.064 | 0.201 | 0.237 |
| IGF2 | 0.454 | 0.684 | 0.915 |
| MEF2A | 0.197 | 0.133 | 0.432 |
| MRF4 | 0.001 | 0.161 | 0.353 |
| MSTN | 0.295 | 0.135 | 0.105 |
| MSX1 | 0.544 | 0.254 | 0.837 |
| MYF5 | 0.193 | 0.376 | 0.386 |
| MYH6 | 0.939 | 0.799 | 0.362 |
| MYOD1 | 0.000 | 0.977 | 0.018 |
| MYOG | 0.054 | 0.914 | 0.830 |
| PAX3 | 0.813 | 0.901 | 0.838 |
| PAX7 | 0.328 | 0.324 | 0.101 |
| PGC1A | 0.005 | 0.199 | 0.280 |
| TGFB1 | 0.004 | 0.888 | 0.603 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tönißen, K.; Franz, G.P.; Rebl, A.; Lutze, P.; Grunow, B. Does Size Matter? Small and Large Larvae of Pikeperch (Sander lucioperca) in a Comparative Gene Expression Analysis. Fishes 2024, 9, 33. https://doi.org/10.3390/fishes9010033
Tönißen K, Franz GP, Rebl A, Lutze P, Grunow B. Does Size Matter? Small and Large Larvae of Pikeperch (Sander lucioperca) in a Comparative Gene Expression Analysis. Fishes. 2024; 9(1):33. https://doi.org/10.3390/fishes9010033
Chicago/Turabian StyleTönißen, Katrin, George Philipp Franz, Alexander Rebl, Philipp Lutze, and Bianka Grunow. 2024. "Does Size Matter? Small and Large Larvae of Pikeperch (Sander lucioperca) in a Comparative Gene Expression Analysis" Fishes 9, no. 1: 33. https://doi.org/10.3390/fishes9010033
APA StyleTönißen, K., Franz, G. P., Rebl, A., Lutze, P., & Grunow, B. (2024). Does Size Matter? Small and Large Larvae of Pikeperch (Sander lucioperca) in a Comparative Gene Expression Analysis. Fishes, 9(1), 33. https://doi.org/10.3390/fishes9010033







