Rapid Monitoring of Aquatic Organism Biodiversity Based on Environmental DNA in a Medium-Sized Reservoir
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection Sites
2.2. eDNA Workflows
2.3. DNA Extraction and PCR Assay
2.4. Bioinformatics Analysis
2.5. Statistical Analysis
2.5.1. Species Composition
2.5.2. Community Diversity Analysis
2.5.3. Environmental Analysis
3. Results
3.1. Biodiversity Status Assessed by eDNA
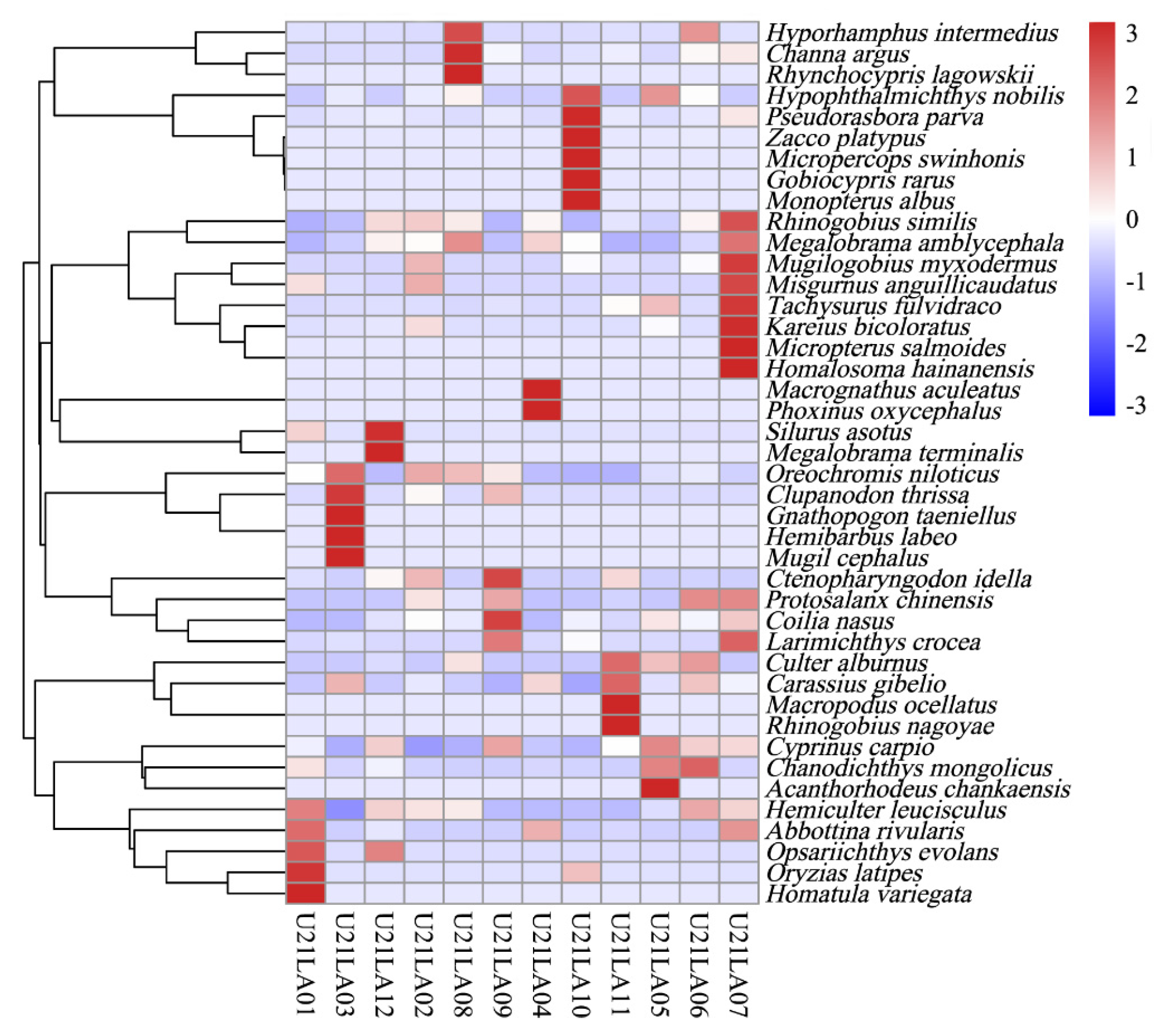
3.1.1. Fish Composition
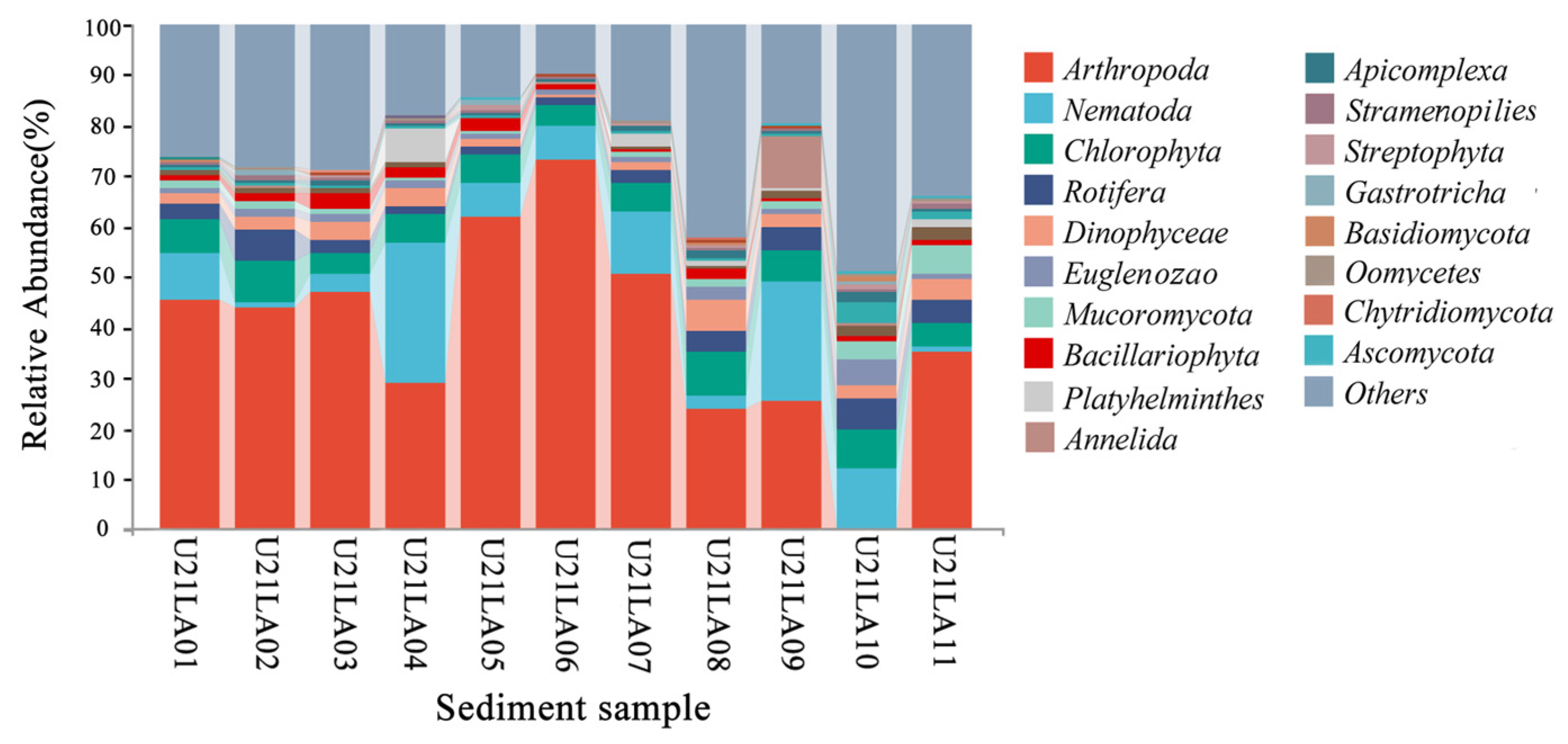
3.1.2. Benthic Species Composition
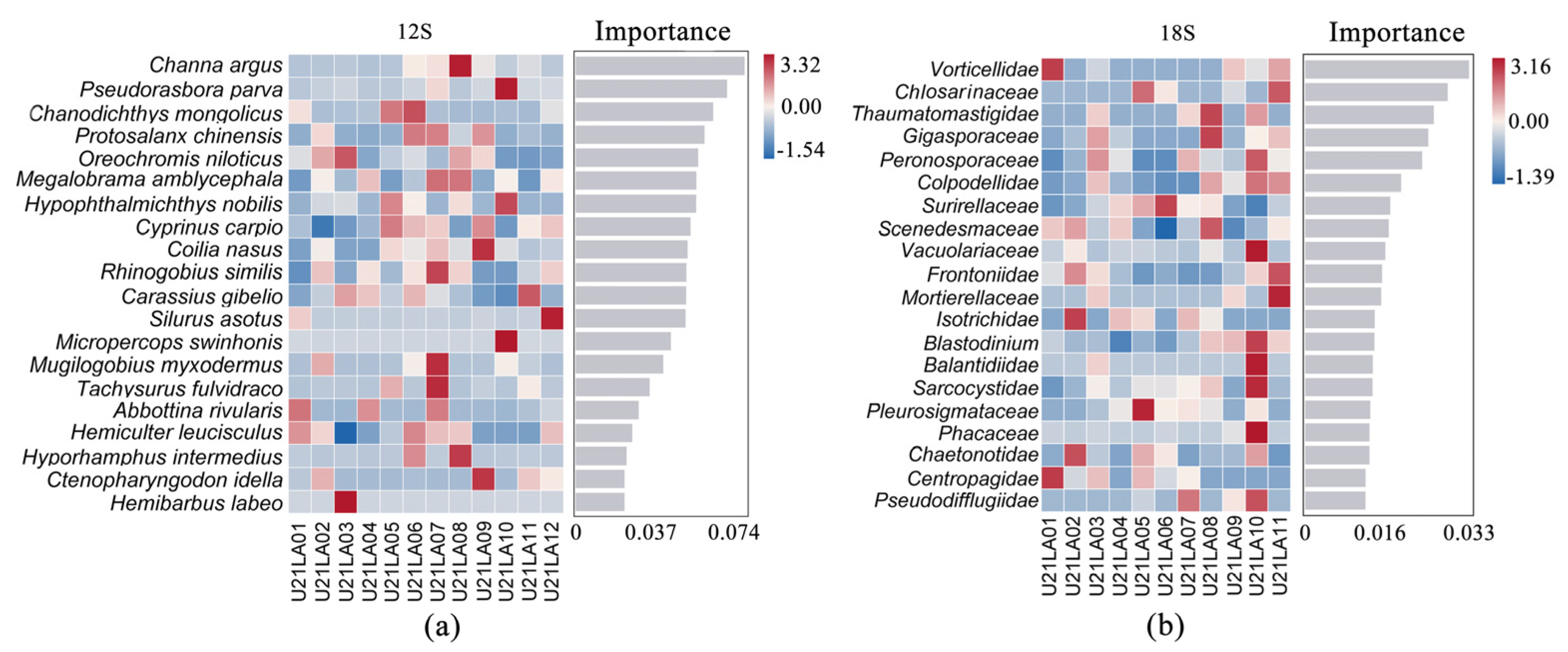
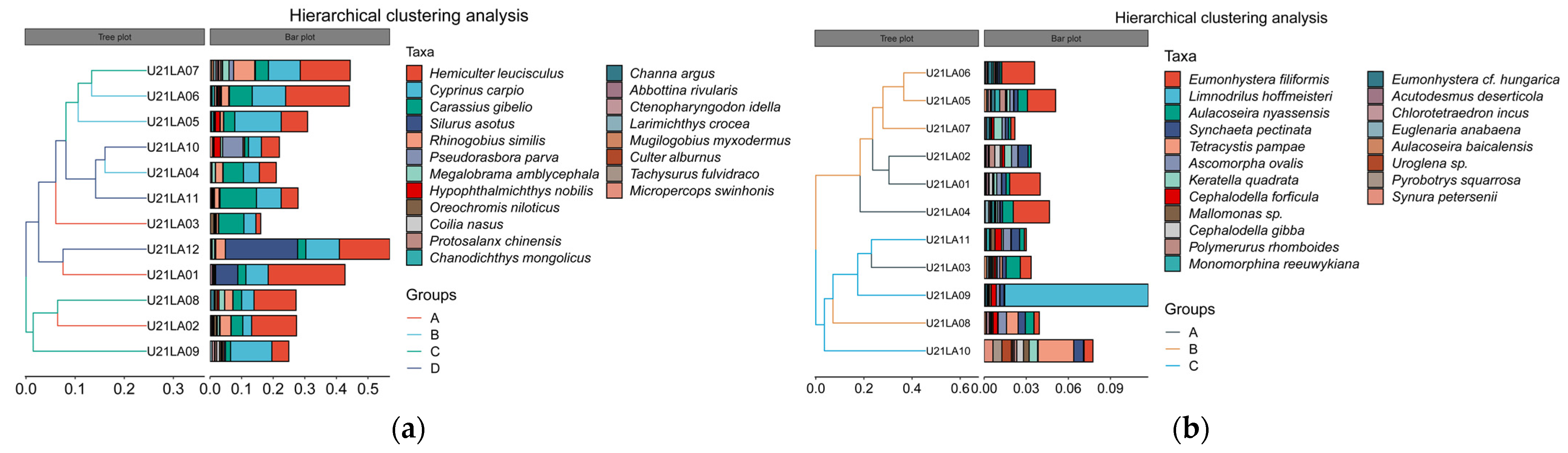
3.1.3. Biomarker Identification
3.2. Diversity Analysis
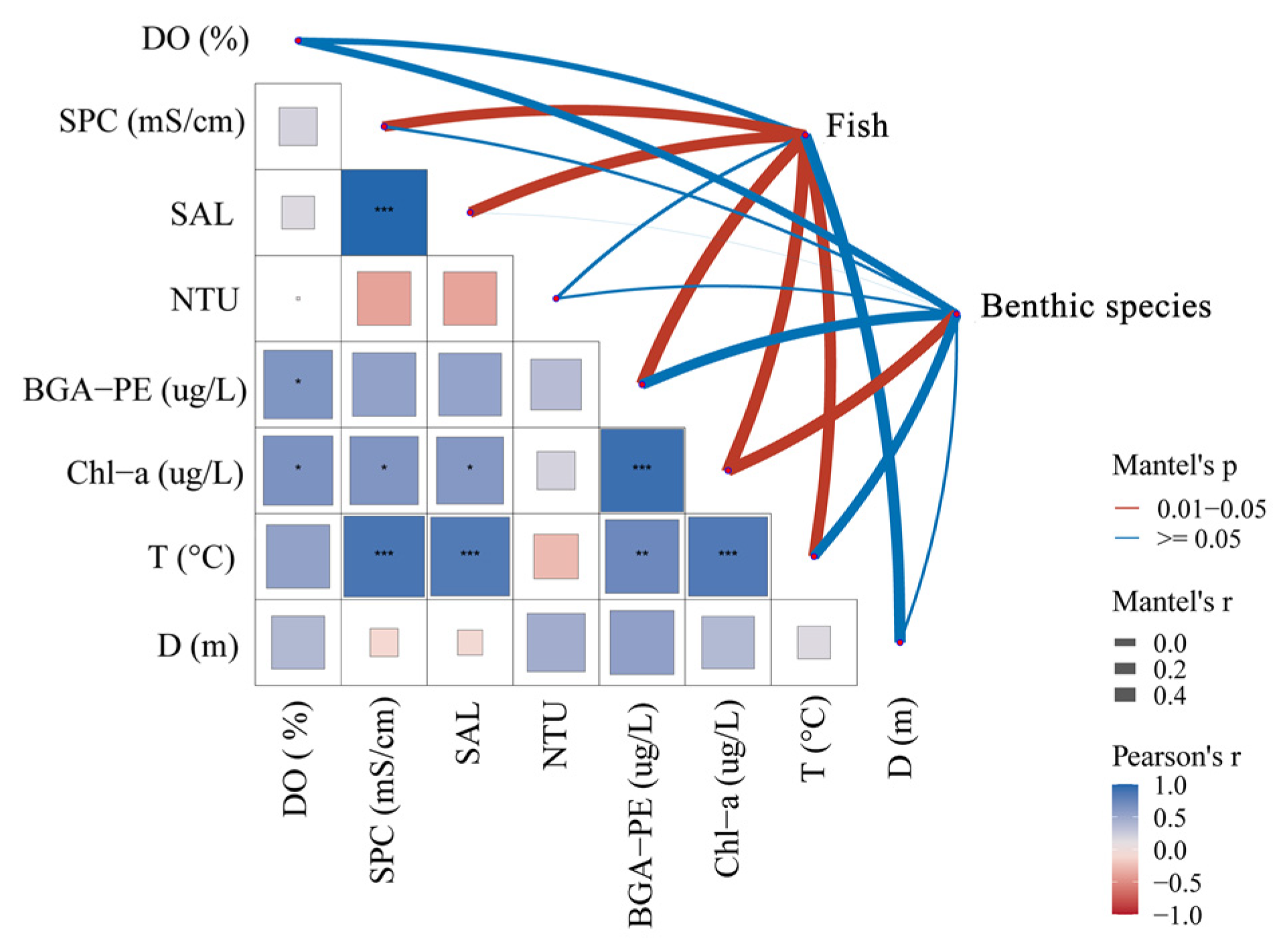
3.3. Environmental Factor Influence
4. Discussion
4.1. eDNA Assessment for Surveying Biodiversity in Shuimen Pond
4.2. Population Specific Primer Selection
4.3. Dominant Species Assemblage and Diversity in Shuimen Pond
4.4. Relationship between Species Community and Environmental Factors
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Desrosiers, M.; Usseglio-Polatera, P.; Archaimbault, V.; Larras, F.; Méthot, G.; Pinel-Alloul, B. Assessing anthropogenic pressure in the St. Lawrence River using traits of benthic macroinvertebrates. Sci. Total Environ. 2019, 649, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Liu, H.; Wu, J.; Gao, X. Spatial distribution, ecological risk assessment and source analysis of heavy metals pollution in urban lake sediments of Huaihe river basin. Int. J. Environ. Res. Public Health 2022, 19, 14653. [Google Scholar] [CrossRef] [PubMed]
- Osathanunkul, M. An eDNA detection of captive-bred Mekong Giant Catfish in the Chao Phraya River basin for further environmental impacts assessment. Aquaculture 2022, 564, 737328. [Google Scholar] [CrossRef]
- Mwaijengo, G.N.; Vanschoenwinkel, B.; Dube, T.; Njau, K.N.; Brendonck, L. Seasonal variation in benthic macroinvertebrate assemblages and water quality in an afrotropical river catchment, northeastern Tanzania. Limnologica 2020, 82, 125780. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, K.; Gao, J.; Wang, M.; Dong, J.; Xie, Y.W.; Giesy, J.P.; Jin, X.W.; Wang, B.X. Environmental DNA of preservative ethanol performed better than water samples in detecting macroinvertebrate diversity using metabarcoding. Divers. Distrib. 2021, 27, 1989–2002. [Google Scholar] [CrossRef]
- Minamoto, T.; Hayami, K.; Sakata, M.K.; Imamura, A. Real-time polymerase chain reaction assays for environmental DNA detection of three salmonid fish in Hokkaido, Japan: Application to winter surveys. Ecol. Res. 2019, 34, 237–242. [Google Scholar] [CrossRef]
- Takahashi, M.; Saccò, M.; Kestel, J.H.; Nester, G.; Campbell, M.A.; van der Heyde, M.; Heydenrych, M.J.; Juszkiewicz, D.J.; Nevill, P.; Dawkins, K.L.; et al. Aquatic environmental DNA: A review of the macro-organismal biomonitoring revolution. Sci. Total Environ. 2023, 873, 162322. [Google Scholar] [CrossRef]
- Maurer, D. The dark side of taxonomic sufficiency. Mar. Pollut. Bull. 2000, 40, 98–101. [Google Scholar] [CrossRef]
- Bush, A.; Monk, W.A.; Compson, Z.G.; Peters, D.L.; Porter, T.M.; Shokralla, S.; Wright, M.T.G.; Hajibabaei, M.; Baird, D.J. DNA metabarcoding reveals metacommunity dynamics in a threatened boreal wetland wilderness. Proc. Natl. Acad. Sci. USA 2020, 117, 8539–8545. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Møller, P.R.; Sigsgaard, E.E.; Knudsen, S.W.; Jørgensen, O.A.; Willerslev, E. Environmental DNA from seawater samples correlate with trawl catches of subarctic, deepwater fishes. PLoS ONE 2016, 11, e0165252. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yoshizawa, S.; Iwasaki, W.; Xian, W.W. Seasonal fish assemblage structure using environmental DNA in the Yangtze Estuary and its adjacent waters. Front. Mar. Sci. 2019, 6, 515. [Google Scholar] [CrossRef]
- Boivin-Delisle, D.; Laporte, M.; Burton, F.; Dion, R.; Normandeau, E.; Bernatchez, L. Using environmental DNA for biomonitoring of freshwater fish communities: Comparison with established gillnet surveys in a boreal hydroelectric impoundment. Environ. DNA 2021, 3, 105–120. [Google Scholar] [CrossRef]
- Li, H.; Yang, F.; Zhang, R.; Liu, S.G.; Yang, Z.J.; Lin, L.S.; Ye, S.Z. Environmental DNA metabarcoding of fish communities in a small hydropower dam reservoir: A comparison between the eDNA approach and established fishing methods. J. Freshw. Ecol. 2022, 37, 337–358. [Google Scholar] [CrossRef]
- Carraro, L.; Hartikainen, H.; Jokela, J.; Bertuzzo, E.; Rinaldo, A. Estimating species distribution and abundance in river networks using environmental DNA. Proc. Natl. Acad. Sci. USA 2018, 115, 11724–11729. [Google Scholar] [CrossRef] [PubMed]
- Malashenkov, D.V.; Dashkova, V.; Zhakupova, K.; Vorobjev, I.A.; Barteneva, N.S. Comparative analysis of freshwater phytoplankton communities in two lakes of Burabay National Park using morphological and molecular approaches. Sci. Rep. 2021, 11, 16130. [Google Scholar] [CrossRef] [PubMed]
- Doi, H.; Inui, R.; Akamatsu, Y.; Kanno, K.; Yamanaka, H.; Takahara, T.; Minamoto, T. Environmental DNA analysis for estimating the abundance and biomass of stream fish. Freshw. Biol. 2017, 62, 30–39. [Google Scholar] [CrossRef]
- Cilleros, K.; Valentini, A.; Allard, L.; Dejean, T.; Etienne, R.; Grenouillet, G.; Iribar, A.; Taberlet, P.; Vigouroux, R.; Brosse, S. Unlocking biodiversity and conservation studies in high-diversity environments using environmental DNA (eDNA): A test with Guianese freshwater fishes. Mol. Ecol. Resour. 2019, 19, 27–46. [Google Scholar] [CrossRef] [PubMed]
- Harrison, J.B.; Sunday, J.M.; Rogers, S.M. Predicting the fate of eDNA in the environment and implications for studying biodiversity. Proc. R. Soc. B 2019, 286, 20191409. [Google Scholar] [CrossRef] [PubMed]
- Díaz, C.; Wege, F.F.; Tang, C.Q.; Crampton-Platt, A.; Rüdel, H.; Eilebrecht, E.; Koschorreck, J. Aquatic suspended particulate matter as source of eDNA for fish metabarcoding. Sci. Rep. 2020, 10, 14352. [Google Scholar] [CrossRef]
- Jarzyna, M.A.; Jetz, W. Detecting the multiple facets of biodiversity. Trends Ecol. Evol. 2016, 31, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Osathanunkul, M.; Minamoto, T. A molecular survey based on eDNA to assess the presence of a clown featherback (Chitala ornata) in a confined environment. PeerJ 2020, 8, e10338. [Google Scholar] [CrossRef] [PubMed]
- Jiao, R.J.; Huang, Y.B.; Zhou, H.Y.; Liu, J.; Wang, S. Bird diversity and its seasonal dynamic in Shuimentang national water resources scenic area, Anhui province, China. J. Jinggangshan Univ. 2018, 39, 99–105. (In Chinese) [Google Scholar]
- Lor, Y.; Schreier, T.M.; Waller, D.L.; Merkes, C.M. Using environmental DNA (eDNA) to detect the endangered Spectaclecase mussel (Margaritifera monodonta). Freshw. Sci. 2020, 39, 837–847. [Google Scholar] [CrossRef]
- Jia, H.; Zhang, H.; Xian, W.W. Fish Diversity Monitored by Environmental DNA in the Yangtze River mainstream. Fishes 2022, 7, 1. [Google Scholar] [CrossRef]
- Yamamoto, S.; Masuda, R.; Sato, Y.; Sado, T.; Araki, H.; Kondoh, M.; Minamoto, T.; Miya, M. Environmental DNA metabarcoding reveals local fish communities in a species-rich coastal sea. Sci. Rep. 2017, 7, 40368. [Google Scholar] [CrossRef]
- Robert, S.H. The southern boundary of the Palaearctic realm in China and adjacent countries. Acta Zool. Sin. 2001, 47, 121–131. [Google Scholar]
- Miya, M.; Sato, Y.; Fukunaga, T.; Sado, T.; Poulsen, J.Y.; Sato, K.; Minamoto, T.; Yamamoto, S.; Yamanaka, H.; Araki, H.; et al. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2015, 2, 150088. [Google Scholar] [CrossRef] [PubMed]
- Amaral-Zettler, L.A.; McCliment, E.A.; Ducklow, H.W.; Huse, S.M. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS ONE 2009, 4, e6372. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable, and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. Dada2: High-resolution sample inference from illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinf. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, N.; Liu, Y.X.; Zhang, X.N.; Hu, B.; Qin, Y.; Xu, H.R.; Wang, H.; Guo, X.X.; Qian, J.M.; et al. Root microbiota shift in rice correlates with resident time in the field and developmental stage. Sci. China Life Sci. 2018, 61, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Liaw, A.; Wiener, M. Classification and regression by random forest. R News 2002, 2, 18–22. [Google Scholar]
- Whittaker, R.H. Vegetation of the Siskiyou mountains, Oregon and California. Ecol. Monogr. 1960, 30, 279–338. [Google Scholar] [CrossRef]
- Whittaker, R.H. Evolution and measurement of species diversity. Taxon 1972, 21, 213–251. [Google Scholar] [CrossRef]
- Jia, H.; Ji, D.P.; Zhang, L.B.; Zhang, T.; Xian, W.W.; Zhang, H. Application of environmental DNA technology in marine ranching-case study of Bailong Pearl Bay Demonstration area in Beibu Gulf. Ecol. Indic. 2023, 154, 110906. [Google Scholar] [CrossRef]
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Sunagawa, S.; Coelho, L.P.; Chaffron, S.; Kultima, J.R.; Labadie, K.; Salazar, G.; Djahanschiri, B.; Zeller, G.; Mende, D.R.; Alberti, A.; et al. Structure and function of the global ocean microbiome. Science 2015, 348, 1261359. [Google Scholar] [CrossRef] [PubMed]
- Reiss, M.; Martin, P.; Gerecke, R.; von Fumetti, S. Limno-ecological characteristics and distribution patterns of spring habitats and invertebrates from the Lowlands to the Alps. Environ. Earth Sci. 2016, 75, 1033. [Google Scholar] [CrossRef]
- Laporte, M.; Berger, C.S.; García-Machado, E.; Côté, G.; Morissette, O.; Bernatchez, L. Cage transplant experiment shows weak transport effect on relative abundance of fish community composition as revealed by eDNA metabarcoding. Ecol. Indic. 2022, 137, 108785. [Google Scholar] [CrossRef]
- Keeley, N.B.; Forrest, B.M.; Crawford, C.; Macleod, C.K. Exploiting salmon farm benthic enrichment gradients to evaluate the regional performance of biotic indices and environmental indicators. Ecol. Indic. 2012, 23, 453–466. [Google Scholar] [CrossRef]
- Ji, F.F.; Han, D.Y.; Yan, L.; Yan, S.H.; Zha, J.M.; Shen, J.Z. Assessment of benthic invertebrate diversity and river ecological status along an urbanized gradient using environmental DNA metabarcoding and a traditional survey method. Sci. Total Environ. 2022, 806, 150587. [Google Scholar] [CrossRef] [PubMed]
- Pochon, X.; Wood, S.A.; Keeley, N.B.; Lejzerowicz, F.; Esling, P.; Drew, J.; Pawlowski, J. Accurate assessment of the impact of salmon farming on benthic sediment enrichment using foraminiferal metabarcoding. Mar. Pollut. Bull. 2015, 100, 370–382. [Google Scholar] [CrossRef]
- He, X.P.; Gilmore, S.R.; Sutherland, T.F.; Hajibabaei, M.; Miller, K.M.; Westfall, K.M.; Pawlowski, J.; Abbott, C.L. Biotic signals associated with benthic impacts of salmon farms from eDNA metabarcoding of sediments. Mol. Ecol. 2020, 30, 3158–3174. [Google Scholar] [CrossRef] [PubMed]
- Schubert, B.; Heiniger, P.; Keller, M.; Claus, E.; Ricking, M. Monitoring of contaminants in suspended particulate matter as an alternative to sediments. TrAC Trends Anal. Chem. 2012, 36, 58–70. [Google Scholar] [CrossRef]
- Roussel, J.M.; Paillisson, J.M.; Treguier, A.; Petit, E. The downside of eDNA as a survey tool in water bodies. J. Appl. Ecol. 2015, 52, 823–826. [Google Scholar] [CrossRef]
- Yao, W.Q. Systematic Synopsis of Fishes in Anhui; Anhui University Press: Hefei, China, 2020. [Google Scholar]
- Yao, M.; Zhang, S.; Lu, Q.; Chen, X.Y.; Zhang, S.Y.; Kong, Y.Q.; Zhao, J.D. Fishing for fish environmental DNA: Ecological applications, methodological considerations, surveying designs, and ways forward. Mol. Ecol. 2022, 31, 5132–5164. [Google Scholar] [CrossRef] [PubMed]
- Deiner, K.; Bik, H.M.; Machler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; de Vere, N.; et al. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef] [PubMed]
- Bylemans, J.; Gleeson, D.M.; Hardy, C.M.; Furlan, E. Toward an ecoregion scale evaluation of eDNA metabarcoding primers: A case study for the freshwater fish biodiversity of the Murray-Darling Basin (Australia). Ecol. Evol. 2018, 8, 8697–8712. [Google Scholar] [CrossRef] [PubMed]
- Goutte, A.; Molbert, N.; Guérin, S.; Richoux, R.; Rocher, V. Monitoring freshwater fish communities in large rivers using environmental DNA metabarcoding and a long-term electrofishing survey. J. Fish Biol. 2020, 97, 444–452. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, H.B. Combining multiple markers in environmental DNA metabarcoding to assess deep-sea benthic biodiversity. Front. Mar. Sci. 2021, 8, 9. [Google Scholar] [CrossRef]
- Leray, M.; Yang, J.Y.; Meyer, C.P.; Mills, S.C.; Agudelo, N.; Ranwez, V.; Boehm, J.T.; Machida, R.J. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: Application for characterizing coral reef fish gut contents. Front. Zool. 2013, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.L.; Zhao, G.F.; Yang, J.H.; Wang, Z.H.; Xu, Y.P.; Zhang, X.W.; Wang, Z.J. eDNA metabarcoding revealed differential structures of aquatic communities in a dynamic freshwater ecosystem shaped by habitat heterogeneity. Environ. Res. 2021, 201, 111602. [Google Scholar] [CrossRef] [PubMed]
- Barnes, M.A.; Turner, C.R.; Jerde, C.L.; Renshaw, M.A.; Chadderton, W.L.; Lodge, D.M. Environmental conditions influence eDNA persistence in aquatic systems. Environ. Sci. Technol. 2014, 48, 1819–1827. [Google Scholar] [CrossRef]
- Carroll, M.L.; Cochrane, S.; Fieler, R.; Velvin, R.; White, P. Organic enrichment of sediments from salmon farming in Norway: Environmental factors, management practices and monitoring techniques. Aquaculture 2003, 226, 165–180. [Google Scholar] [CrossRef]
- Shu, L.; Chen, S.J.; Li, P.; Peng, Z.G. Environmental DNA metabarcoding reflects fish DNA dynamics in lentic ecosystems: A case study of freshwater ponds. Fishes 2022, 7, 257. [Google Scholar] [CrossRef]
- Sun, X.; Wang, K.; Zhang, G.; Ren, H.; Yu, H. Spatial–temporal patterns of fish trophic guilds in a freshwater river wetland ecosystem of northeastern China. Ecol. Evo. 2024, 14, e11711. [Google Scholar] [CrossRef]
- Volkoff, H. The effects of environmental changes on the endocrine regulation of feeding in fishes. Phil. Trans. R. Soc. B 2024, 379, 20220503. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Wang, Y.; Yoshizawa, S.; Iwasaki, W.; Li, Y.Q.; Xian, W.W.; Zhang, H. Seasonal variation and assessment of fish resources in the Yangtze Estuary based on environmental DNA. Water 2020, 12, 2874. [Google Scholar] [CrossRef]
- Kaenel, B.R.; Matthaei, C.D.; Uehlinger, U. Disturbance by aquatic plant management in streams: Effects on benthic invertebrates. Regul. Ric. Res. Manag. 1998, 14, 341–356. [Google Scholar] [CrossRef]
- Tokeshi, M. Resource utilization, overlap and temporal community dynamics: A null model analysis of an epiphytic chironomid community. J. Anim. Ecol. 1986, 55, 491–506. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, H.; Xia, M.; Zhang, T.; Zhang, H. Rapid Monitoring of Aquatic Organism Biodiversity Based on Environmental DNA in a Medium-Sized Reservoir. Fishes 2024, 9, 396. https://doi.org/10.3390/fishes9100396
Jia H, Xia M, Zhang T, Zhang H. Rapid Monitoring of Aquatic Organism Biodiversity Based on Environmental DNA in a Medium-Sized Reservoir. Fishes. 2024; 9(10):396. https://doi.org/10.3390/fishes9100396
Chicago/Turabian StyleJia, Hui, Mingxia Xia, Tao Zhang, and Hui Zhang. 2024. "Rapid Monitoring of Aquatic Organism Biodiversity Based on Environmental DNA in a Medium-Sized Reservoir" Fishes 9, no. 10: 396. https://doi.org/10.3390/fishes9100396
APA StyleJia, H., Xia, M., Zhang, T., & Zhang, H. (2024). Rapid Monitoring of Aquatic Organism Biodiversity Based on Environmental DNA in a Medium-Sized Reservoir. Fishes, 9(10), 396. https://doi.org/10.3390/fishes9100396






