Comparative Metabolomic Analysis Reveals the Impact of the Photoperiod on the Hepatopancreas of Chinese Grass Shrimp (Palaemonetes sinensis)
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Design
2.3. Sample Collection
2.4. Enzyme Activity Assay
2.5. Hepatopancreas Metabolomic Analysis
2.6. Analysis of Differential Metabolites
2.7. Statistical Analysis
3. Results
3.1. Survival and Growth Performance
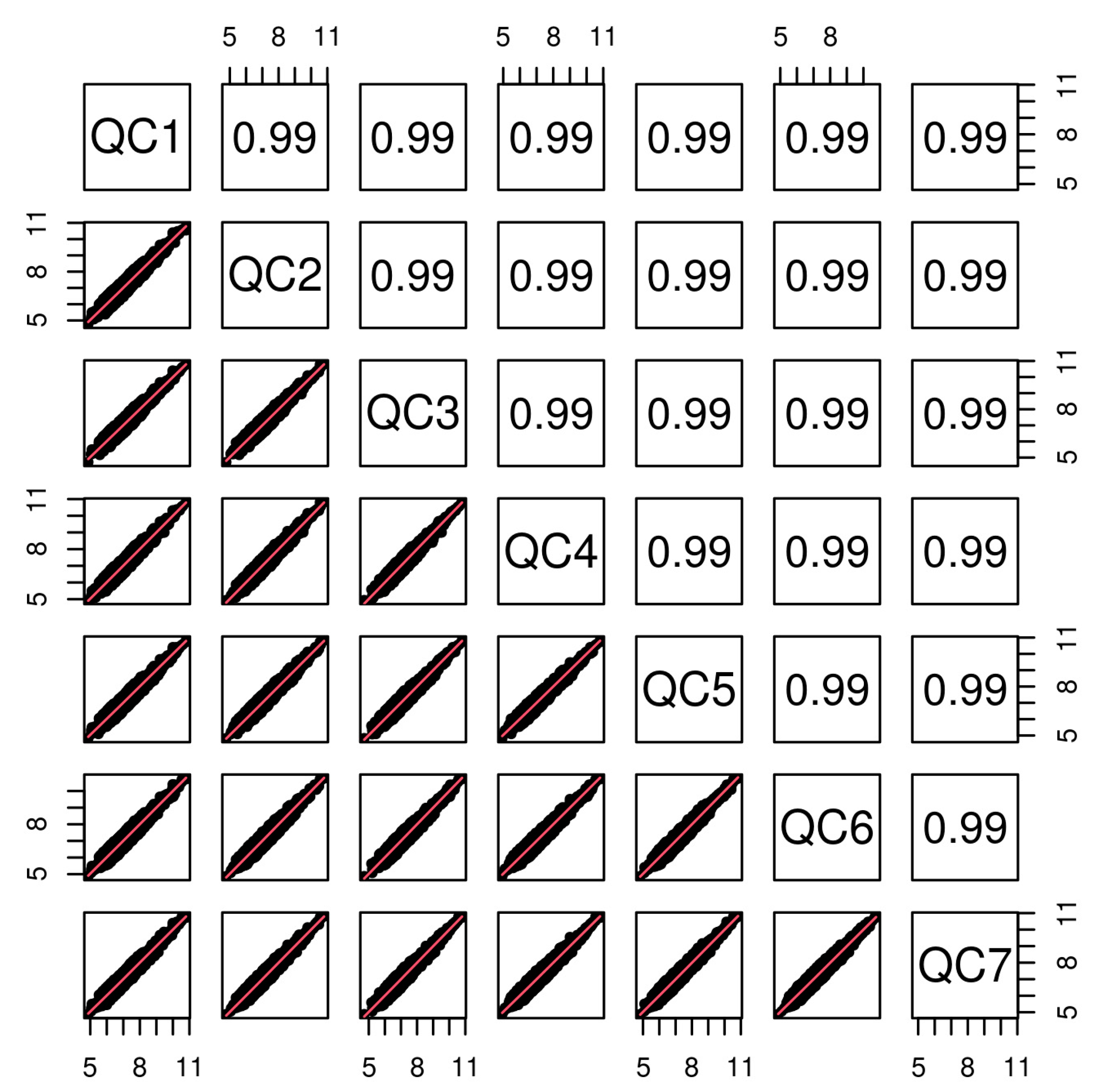
3.2. Quality Control and Data Checking
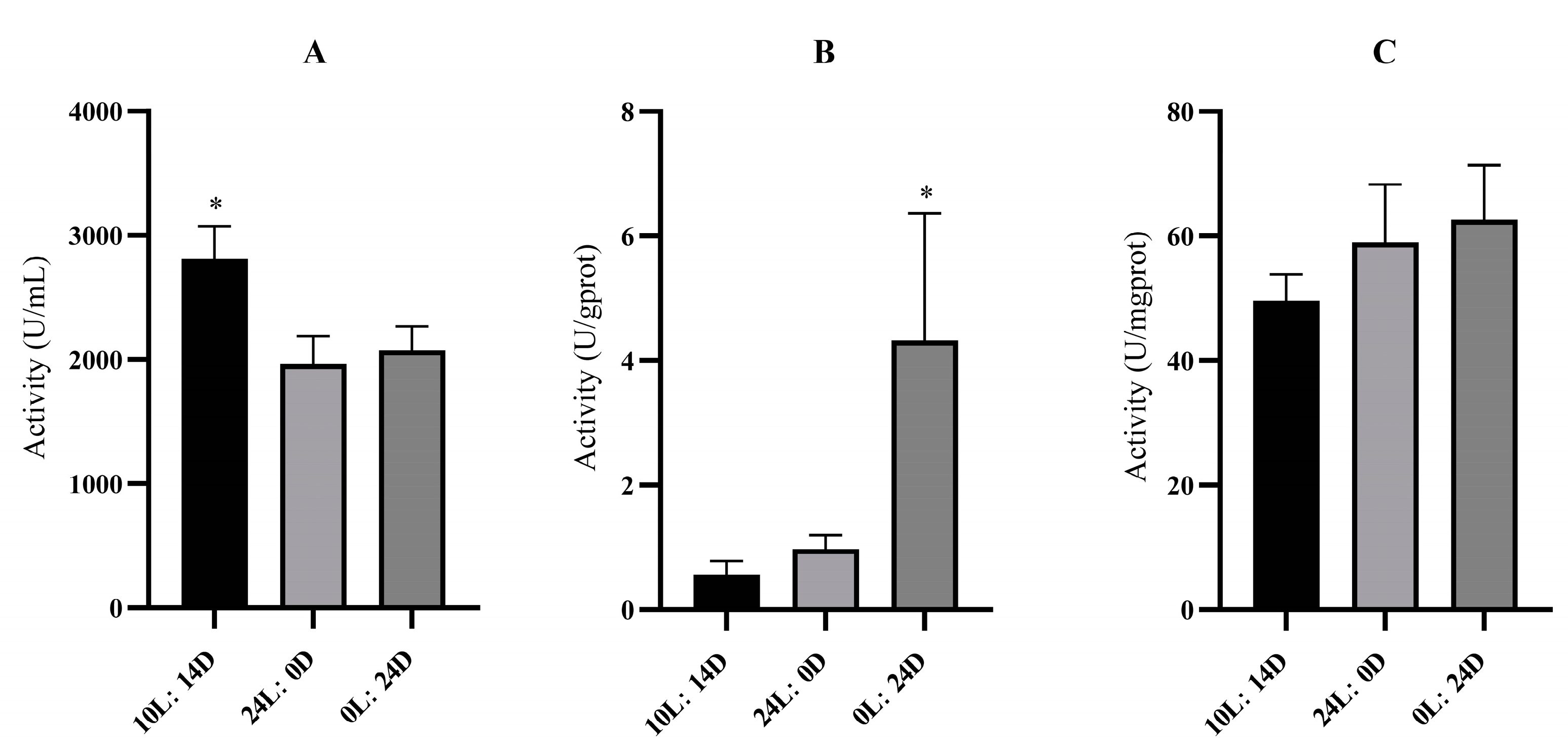
3.3. Enzyme Activity
3.4. Metabolite Identification and Comparison
3.5. Identification of Metabolites with Significant Differences
3.6. Comparison of Hepatopancreatic Metabolites
3.7. KEGG Pathway Analysis of Key Metabolic Pathways
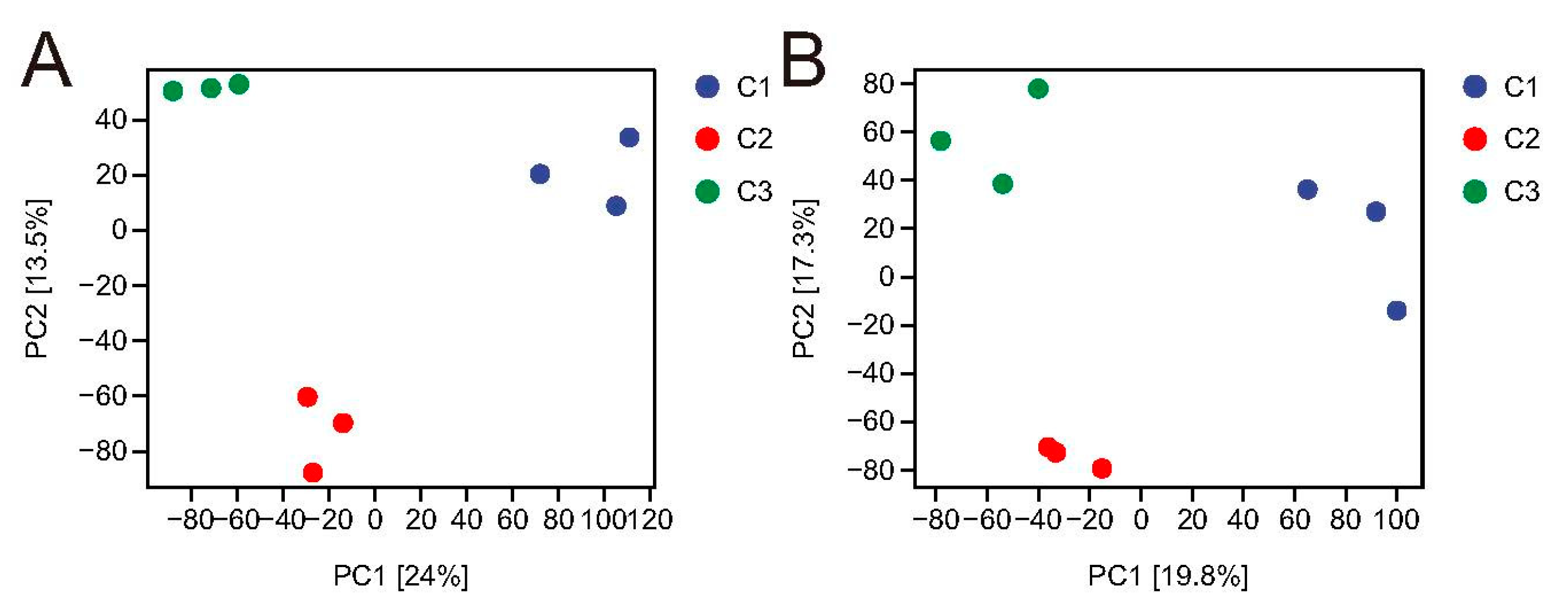
3.8. Cluster Analysis
4. Discussion
4.1. Effects on Immune Function
4.2. Effects on the Digestive System
4.3. Influence on Metabolism and Energy Metabolism
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Frimmel, F.H. Impact of light on the properties of aquatic natural organic matter. Environ. Int. 1998, 24, 559–571. [Google Scholar] [CrossRef]
- Han, Z.; Li, X.; Xu, W.; She, Q.; Liang, S.; Li, X.; Li, Y. Melatonin concentrations in Chinese mitten crabs (Eriocheir sinesis) are affected by artificial photoperiods. Biol. Rhythm Res. 2020, 51, 362–372. [Google Scholar] [CrossRef]
- Zhang, B.; Yu, C.; Xu, Y.; Huang, Z.; Cai, Y.; Li, Y. Hepatopancreas immune response during different photoperiods in the Chinese mitten crab, Eriocheir sinensis. Fish Shellfish Immunol. 2023, 132, 108482. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Chai, Y.; Xu, Y.; Huang, Z.; Hu, X.; Li, Y. Impact analysis of photoperiodic disorder on the eyestalk of Chinese mitten crab (Eriocheir sinensis) through high-throughput sequencing technology. Life 2024, 14, 209. [Google Scholar] [CrossRef]
- Nie, X.; Huang, C.; Wei, J.; Wang, Y.; Hong, K.; Mu, X.; Liu, C.; Chu, Z.; Zhu, X.; Yu, L. Effects of photoperiod on survival, growth, physiological, and biochemical indices of redclaw crayfish (Cherax quadricarinatus) juveniles. Animals 2024, 14, 411. [Google Scholar] [CrossRef]
- Chen, S.; Liu, J.; Shi, C.; Migaud, H.; Ye, Y.; Song, C.; Mu, C.; Ren, Z.; Wang, C. Effect of photoperiod on growth, survival, and lipid metabolism of mud crab Scylla paramamosain juveniles. Aquac. Res. 2023, 567, e739279. [Google Scholar] [CrossRef]
- Gao, X.; Pang, G.; Luo, X.; You, W.; Ke, C. Effects of light cycle on circadian feeding activity and digestive physiology in Haliotis discus hannai. Aquac. Res. 2021, 539, e736642. [Google Scholar] [CrossRef]
- Imai, T.; Oonuki, T. Records of Chinese grass shrimp, Palaemonetes sinensis (Sollaud, 1911) from western Japan and simple differentiation method with native freshwater shrimp, Palaemon paucidens De Haan, 1844 using eye size and carapace color pattern. BioInvasions Rec. 2014, 3, 163–168. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhu, X.; Jiang, Y.; Li, Z.; Li, X.; Xu, W.; Wei, H.; Li, Y.; Li, X. Genetic diversity and variation of seven Chinese grass shrimp (Palaemonetes sinensis) populations based on the mitochondrial COI gene. BMC Ecol. Evol. 2021, 21, 167. [Google Scholar] [CrossRef]
- Li, Y.; Liang, S.; She, Q.; Han, Z.; Li, Y.; Li, X. Influence of temperature and size on menthol anaesthesia in Chinese grass shrimp Palaemonetes sinensis (Sollaud, 1911). Aquac. Res. 2018, 49, 2091–2098. [Google Scholar] [CrossRef]
- Li, Y.; She, Q.; Han, Z.; Sun, N.; Liu, X.; Li, X. Anaesthetic effects of eugenol on grass shrimp (Palaemonetes sinensis) of different sizes at different concentrations and temperatures. Sci. Rep. 2018, 8, 11007. [Google Scholar] [CrossRef] [PubMed]
- Aoki, Y.; Sugimoto, Y.; Imai, T.; Tani, S.; Tettey, P.A.; Khalfan, A.-W.M.; Saito, H. Appearance of exotic shrimp Palaemon sinensis (Sollaud, 1911) and other freshwater shrimps before and after the 2018 extreme flood in western Japan. BioInvasions Rec. 2024, 13, 183–194. [Google Scholar] [CrossRef]
- Han, Z.; Li, X.; Li, X.; Xu, W.; Li, Y. Circadian rhythms of melatonin in haemolymph and optic lobes of Chinese mitten crab (Eriocheir sinensis) and Chinese grass shrimp (Palaemonetes sinensis). Biol. Rhythm Res. 2019, 50, 400–407. [Google Scholar] [CrossRef]
- Yingdong, L.; Huiling, X.; Xiaodong, L. Transcriptome analysis of the Chinese grass shrimp Palaemonetes sinensis (Sollaud 1911) and its predicted feeding habit. J. Oceanol. Limnol. 2018, 36, 1778–1787. [Google Scholar]
- Changyue, Y.; Weibin, X.; Xin, L.; Jiaxin, J.; Xinmiao, Z.; Simiao, W.; Zhiyuan, Z.; Yanyu, W.; Qijun, C.; Yingdong, L. Comparative transcriptome analysis of Chinese grass shrimp (Palaemonetes sinensis) hepatopancreas under ectoparasitic isopod (Tachaea chinensis) infection. Fish Shellfish Immunol. 2021, 117, 211–219. [Google Scholar]
- Mark, R.V. Metabolomics of aquatic organisms: The new ‘omics’ on the block. Mar. Ecol. Prog. Ser. 2007, 332, 301–306. [Google Scholar]
- Hillyer, K.E.; Beale, D.J.; Shima, J.S. Artificial light at night interacts with predatory threat to alter reef fish metabolite profiles. Sci. Total Environ. 2021, 769, e144482. [Google Scholar] [CrossRef]
- Yu, C.; Zhang, B.; Hu, N.; Li, L.; Wang, S.; Zhang, Z.; Wei, T.; Zhao, Y.; We, I.H.; Hu, Q.; et al. Comparative metabolomic analysis of Chinese mitten crab (Eriocheir sinensis) during the daily cycle. Aquac. Res. 2022, 53, 2947–2958. [Google Scholar] [CrossRef]
- Zhou, X.; Li, Q.; Zhu, J.; Xie, S.; Wang, M.; Yang, T. Effects of hypoxic stress on tissue structure and gut bacterial community of Exopalaemon carinicauda. Prog. Fish. Sci. 2024, 45, 167–177. [Google Scholar]
- Wang, L.; Xue, Y.; Jiang, G.; Shi, L.; Huang, X. The Effects of Temperature and Feed on the Growth, Development, and Reproductive Performances of Neocaridina denticulata; Shanghai Ocean University: Shanghai, China, 2024; pp. 1–22. [Google Scholar]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; He, W.-Y.; Wang, W.-N.; Yang, C.-W.; Wang, L.; Xin, Y.; Wu, J.; Cai, D.-x.; Liu, Y.; Wang, A.-L. Molecular cloning and characterization of an ATP-binding cassette (ABC) transmembrane transporter from the white shrimp Litopenaeus vannamei. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2009, 150, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.-S.; Chen, X.-L.; Wang, A.-J.; Liu, Q.-Y.; Peng, M.; Yang, C.-L.; Yin, C.-C.; Zhu, W.-L.; Zeng, D.-G.; Zhang, B.; et al. Genome-wide analysis of ATP-binding cassette (ABC) transporter in Penaeus vannamei and identification of two ABC genes involved in immune defense against Vibrio parahaemolyticus by affecting NF-κB signaling pathway. Int. J. Biol. Macromol. 2024, 262, 129984. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.-K.; Kwon, B. Immune regulation through tryptophan metabolism. Exp. Mol. Med. 2023, 55, 1371–1379. [Google Scholar] [CrossRef] [PubMed]
- Arumugham, V.B.; Baldari, C.T. cAMP: A multifaceted modulator of immune synapse assembly and T cell activation. J. Leukoc. Biol. 2017, 101, 1301–1316. [Google Scholar] [CrossRef]
- Cameron, E.G.; Kapiloff, M.S. Intracellular compartmentation of cAMP promotes neuroprotection and regeneration of CNS neurons. Neural Regener. Res. 2017, 12, 201–202. [Google Scholar] [CrossRef]
- Naderi, F.; Hernández-Pérez, J.; Chivite, M.; Soengas, J.L.; Míguez, J.M.; López-Patiño, M.A. Involvement of cortisol and sirtuin1 during the response to stress of hypothalamic circadian system and food intake-related peptides in rainbow trout, Oncorhynchus mykiss. Chronobiol. Int. 2018, 35, 1122–1141. [Google Scholar] [CrossRef]
- Ren, X.; Zhang, J.; Wang, L.; Wang, Z.; Wang, Y. Diel variation in cortisol, glucose, lactic acid and antioxidant system of black sea bass Centropristis striata under natural photoperiod. Chronobiol. Int. 2020, 37, 176–188. [Google Scholar] [CrossRef]
- Espinosa-Chaurand, D.; Vega-Villasante, F.; Carrillo-Farnés, O.; Nolasco-Soria, H. Effect of circadian rhythm, photoperiod, and molt cycle on digestive enzymatic activity of Macrobrachium tenellum juveniles. Aquac. Res. 2017, 479, 225–232. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, R.; Fu, Z.; Ma, Z.; Bai, Z. The photoperiod significantly influences the growth rate, digestive efficiency, immune response, and antioxidant activities in the juvenile scalloped spiny lobster (Panulirus homarus). J. Mar. Sci. Eng. 2024, 12, e389. [Google Scholar] [CrossRef]
- Shiau, S.-Y.; Chin, Y.-H. Dietary biotin requirement for maximum growth of juvenile grass shrimp, Penaeus monodon. J. Nutr. 1998, 128, 2494–2497. [Google Scholar] [CrossRef] [PubMed]

| Items | 10L:14D | 24L:0D | 0L:24D |
|---|---|---|---|
| Survive rate (%) | 91.83 ± 3.13 | 93.43 ± 2.32 | 96.13 ± 0.45 |
| Weight gain rate (%) | 10.93 ± 0.02 | 6.08 ± 0.02 | 8.80 ± 0.03 |
| Regulation | Name | VIP | Fold Change | p-Value | FDR |
|---|---|---|---|---|---|
| Up | Anabasine | 1.907297316 | 3.158097785 | 1.74 × 10−5 | 0.318782 |
| Methoprene | 1.684382823 | 2.780965383 | 0.0258962 | 0.656741 | |
| Down | 3-Hydroxymethylglutaric acid | 1.905099882 | 0.190579202 | 0.0009552 | 0.403403 |
| Guanosine | 1.911404573 | 0.466353051 | 0.00236 | 0.654583 | |
| Maltol | 1.837813673 | 0.119762921 | 0.0025269 | 0.46636 | |
| Agmatine | 1.832581464 | 0.212675525 | 0.0035182 | 0.488711 | |
| Palmitoyl-L-carnitine | 1.871665251 | 0.421684127 | 0.0037924 | 0.488711 | |
| Indole | 1.809790956 | 0.862476991 | 0.0069588 | 0.535137 | |
| L-Tryptophan | 1.783906192 | 0.866652658 | 0.0086536 | 0.553457 | |
| 3-Dehydroshikimate | 1.730013181 | 0.853776833 | 0.015607 | 0.606582 | |
| 12-Keto-leukotriene B4 | 1.720753783 | 0.58867918 | 0.0208922 | 0.790759 | |
| 3-Methylindole | 1.662382191 | 0.88113951 | 0.0253116 | 0.65533 | |
| Taurocholic acid | 1.69563276 | 0.414046236 | 0.025464 | 0.656555 | |
| 4-Acetylbutyrate | 1.762004096 | 0.862065771 | 0.0287383 | 0.663664 | |
| 1,5-Naphthalenediamine | 1.636454482 | 0.896950592 | 0.0308542 | 0.670219 | |
| Cytidine | 1.705254513 | 0.424843618 | 0.0318796 | 0.672933 | |
| Uridine | 1.701526212 | 0.435161805 | 0.0343067 | 0.790759 | |
| 8-Amino-7-oxononanoate | 1.592304996 | 0.68752891 | 0.0432281 | 0.68288 | |
| Kinetin | 1.641848437 | 0.66737036 | 0.044375 | 0.68288 | |
| N-Alpha-acetyllysine | 1.58166688 | 0.876164025 | 0.0481752 | 0.686258 |
| Regulation | Name | VIP | Fold Change | p-Value | FDR |
|---|---|---|---|---|---|
| Up | 12,13-DHOME | 1.848892946 | 7.944503725 | 0.0025492 | 0.435487 |
| Vanillylmandelic acid | 1.698582067 | 2.287584995 | 0.0146389 | 0.572589 | |
| 1-Hexadecanol | 1.608151478 | 2.041461624 | 0.0173203 | 0.304102 | |
| Deoxyuridine | 1.587256606 | 1.272344109 | 0.0219119 | 0.322259 | |
| PC(18_3(6Z,9Z,12Z)_18_3(6Z,9Z,12Z)) | 1.624879576 | 9.744630256 | 0.0231141 | 0.329669 | |
| Glutathionylspermidine | 1.475731058 | 4.137433822 | 0.0314225 | 0.350268 | |
| Porphobilinogen | 1.466742525 | 2.877259275 | 0.0369976 | 0.364777 | |
| Gamma-Linolenic acid | 1.65502262 | 1.365807338 | 0.0378202 | 0.612128 | |
| Alpha-dimorphecolic acid | 1.618926287 | 2.597285846 | 0.040452 | 0.614582 | |
| 7,8-Diaminononanoate | 1.507837379 | 1.273066329 | 0.0419005 | 0.374433 | |
| 8,11,14-Eicosatrienoic acid | 1.513521616 | 2.351772099 | 0.0429527 | 0.378504 | |
| Down | (R)-4-Hydroxymandelate | 1.71739743 | 0.776230178 | 0.0001679 | 0.163141 |
| N-Methyltryptamine | 1.834687656 | 0.302541618 | 0.0018306 | 0.398592 | |
| Guanosine 3′-phosphate | 1.661328012 | 0.55848625 | 0.0029291 | 0.22165 | |
| Guanosine | 1.820377684 | 0.368505666 | 0.0039691 | 0.483225 | |
| Indole | 1.640341233 | 0.848668536 | 0.0064791 | 0.246949 | |
| Taurine | 1.592918474 | 0.715815838 | 0.0106643 | 0.27098 | |
| CMP | 1.563437429 | 0.571401423 | 0.0115314 | 0.276959 | |
| 4-Acetylbutyrate | 1.588548367 | 0.842573158 | 0.0121322 | 0.281471 | |
| 3-Dehydroshikimate | 1.560734708 | 0.860089133 | 0.0125116 | 0.284814 | |
| 1,5-Naphthalenediamine | 1.598978464 | 0.869860663 | 0.0132279 | 0.287593 | |
| 3-Methylindole | 1.593774137 | 0.865101666 | 0.0145031 | 0.291362 | |
| 4-Guanidinobutanoic acid | 1.651596962 | 0.160116061 | 0.0150216 | 0.292051 | |
| 8-Amino-7-oxononanoate | 1.553996072 | 0.59167106 | 0.0151476 | 0.292051 | |
| Cytidine | 1.659082161 | 0.251674321 | 0.0181636 | 0.309742 | |
| L-Tryptophan | 1.526508918 | 0.886507002 | 0.0186632 | 0.311918 | |
| Guanidoacetic acid | 1.520163573 | 0.829851625 | 0.0199308 | 0.315627 | |
| Palmitoyl-L-carnitine | 1.569867597 | 0.310984095 | 0.0305007 | 0.348488 | |
| Uridine | 1.665952542 | 0.408655048 | 0.0342888 | 0.607128 | |
| Glycitein | 1.576053112 | 0.317608942 | 0.034728 | 0.359354 | |
| Cortexolone | 1.521167154 | 0.533997038 | 0.0353217 | 0.360789 | |
| Propionylcarnitine | 1.532435504 | 0.612049756 | 0.0389239 | 0.367213 | |
| Pyrrolidonecarboxylic acid | 1.58033653 | 0.255350103 | 0.0446104 | 0.384618 | |
| D-Galactose | 1.68348797 | 0.455843304 | 0.045287 | 0.619777 | |
| epsilon-(gamma-L-Glutamyl)-L-lysine | 1.418573297 | 0.621361739 | 0.0462443 | 0.388083 | |
| Cyclic AMP | 1.430922335 | 0.625015239 | 0.046507 | 0.389122 | |
| Kinetin | 1.560722322 | 0.783293537 | 0.0472655 | 0.390247 | |
| Creatinine | 1.404212767 | 0.71705939 | 0.0473558 | 0.390353 | |
| Saccharopine | 1.484150092 | 0.839835089 | 0.048923 | 0.393707 |
| Pathway | Total | Hits | p-Value | FDR | Compounds |
|---|---|---|---|---|---|
| Phenylalanine, tyrosine and tryptophan biosynthesis | 35 | 3 | 0.000138 | 0.00344 | L-Tryptophan; Indole; 3-Dehydroshikimate |
| Tryptophan metabolism | 83 | 3 | 0.001774 | 0.022176 | L-Tryptophan; Indole; 3-Methylindole |
| ABC transporters | 138 | 3 | 0.007504 | 0.053808 | Uridine; Guanosine; Cytidine |
| Protein digestion and absorption | 47 | 2 | 0.008609 | 0.053808 | L-Tryptophan; Indole |
| Pyrimidine metabolism | 64 | 2 | 0.015593 | 0.077965 | Uridine; Cytidine |
| Axon regeneration | 7 | 1 | 0.021192 | 0.088298 | L-Tryptophan |
| Cholesterol metabolism | 10 | 1 | 0.030146 | 0.107665 | Taurocholic acid |
| Pathway | Total | Hits | p-Value | FDR | Compounds |
|---|---|---|---|---|---|
| Linoleic acid metabolism | 28 | 5 | 1.51 × 10−6 | 0.00014 | PC(18_3(6Z,9Z,12Z)_18_3(6Z,9Z,12Z)); 8,11,14-Eicosatrienoic acid; Gamma-Linolenic acid; Alpha-dimorphecolic; 12,13-DHOME |
| Pyrimidine metabolism | 64 | 4 | 0.001217 | 0.037738 | CMP; Uridine; Cytidine; Deoxyuridine |
| ABC transporters | 138 | 5 | 0.003307 | 0.045784 | Taurine; Uridine; Guanosine; Cytidine; Deoxyuridine |
| Retrograde endocannabinoid signaling | 19 | 2 | 0.008641 | 0.089285 | Cyclic AMP; PC(18_3(6Z,9Z,12Z)_18_3(6Z,9Z,12Z)) |
| Biotin metabolism | 29 | 2 | 0.019579 | 0.121392 | 8-Amino-7-oxononanoate; 7,8-Diaminononanoate |
| PPAR signaling pathway | 5 | 1 | 0.037033 | 0.136271 | Alpha-dimorphecolic acid |
| Axon regeneration | 7 | 2 | 0.001123 | 0.037738 | L-Tryptophan; Cyclic AMP |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 35 | 3 | 0.002144 | 0.045784 | L-Tryptophan; Indole; 3-Dehydroshikimate |
| Tryptophan metabolism | 83 | 4 | 0.003186 | 0.045784 | L-Tryptophan; Indole; N-Methyltryptamine; 3-Methylindole |
| Cortisol synthesis and secretion | 12 | 2 | 0.003446 | 0.045784 | Cyclic AMP; Cortexolone |
| Hedgehog signaling pathway | 1 | 1 | 0.007515 | 0.087365 | Cyclic AMP |
| Arginine and proline metabolism | 69 | 3 | 0.014438 | 0.107135 | Guanidoacetic acid; Creatinine; 4-Guanidinobutanoic acid |
| Longevity regulating pathway—multiple species | 2 | 1 | 0.014976 | 0.107135 | Cyclic AMP |
| Circadian rhythm | 2 | 1 | 0.014976 | 0.107135 | Cyclic AMP |
| Vasopressin-regulated water reabsorption | 2 | 1 | 0.014976 | 0.107135 | Cyclic AMP |
| Mineral absorption | 29 | 2 | 0.019579 | 0.121392 | L-Tryptophan; D-Galactose |
| Oocyte meiosis | 4 | 1 | 0.029734 | 0.136271 | Cyclic AMP |
| Leukocyte transendothelial migration | 4 | 1 | 0.029734 | 0.136271 | Cyclic AMP |
| Insulin signaling pathway | 4 | 1 | 0.029734 | 0.136271 | Cyclic AMP |
| Progesterone-mediated oocyte maturation | 4 | 1 | 0.029734 | 0.136271 | Cyclic AMP |
| Growth hormone synthesis, secretion and action | 4 | 1 | 0.029734 | 0.136271 | Cyclic AMP |
| MAPK signaling pathway | 5 | 1 | 0.037033 | 0.136271 | Cyclic AMP |
| Rap1 signaling pathway | 5 | 1 | 0.037033 | 0.136271 | Cyclic AMP |
| Chemokine signaling pathway | 5 | 1 | 0.037033 | 0.136271 | Cyclic AMP |
| Purine metabolism | 101 | 3 | 0.039073 | 0.136271 | Guanosine; Cyclic AMP; Guanosine 3′-phosphate |
| Serotonergic synapse | 42 | 2 | 0.03909 | 0.136271 | L-Tryptophan; Cyclic AMP |
| GnRH signaling pathway | 6 | 1 | 0.044278 | 0.136271 | Cyclic AMP |
| Melanogenesis | 6 | 1 | 0.044278 | 0.136271 | Cyclic AMP |
| Relaxin signaling pathway | 6 | 1 | 0.044278 | 0.136271 | Cyclic AMP |
| Protein digestion and absorption | 47 | 2 | 0.047956 | 0.136271 | L-Tryptophan; Indole |
| Glycine, serine and threonine metabolism | 48 | 2 | 0.04981 | 0.136271 | L-Tryptophan; Guanidoacetic acid |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qu, D.; Fu, C.; Han, M.; Li, Y. Comparative Metabolomic Analysis Reveals the Impact of the Photoperiod on the Hepatopancreas of Chinese Grass Shrimp (Palaemonetes sinensis). Fishes 2024, 9, 444. https://doi.org/10.3390/fishes9110444
Qu D, Fu C, Han M, Li Y. Comparative Metabolomic Analysis Reveals the Impact of the Photoperiod on the Hepatopancreas of Chinese Grass Shrimp (Palaemonetes sinensis). Fishes. 2024; 9(11):444. https://doi.org/10.3390/fishes9110444
Chicago/Turabian StyleQu, Duojia, Chunyan Fu, Muyu Han, and Yingdong Li. 2024. "Comparative Metabolomic Analysis Reveals the Impact of the Photoperiod on the Hepatopancreas of Chinese Grass Shrimp (Palaemonetes sinensis)" Fishes 9, no. 11: 444. https://doi.org/10.3390/fishes9110444
APA StyleQu, D., Fu, C., Han, M., & Li, Y. (2024). Comparative Metabolomic Analysis Reveals the Impact of the Photoperiod on the Hepatopancreas of Chinese Grass Shrimp (Palaemonetes sinensis). Fishes, 9(11), 444. https://doi.org/10.3390/fishes9110444








