The Effect of Copper–Cadmium Co-Exposure and Hormone Remediation on the Ovarian Transcriptome of Nile Tilapia (Oreochromis niloticus)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Animals and Sample Collection
2.3. Determination of Cu and Cd Bioaccumulation
2.4. Histomorphological Analysis
2.5. Biochemical Analysis
2.6. Total RNA Isolation, Library Construction, and Sequencing
2.7. Bioinformatics Analysis
2.8. qRT-PCR Analysis of Selected Genes
2.9. Statistical Analysis
3. Results
3.1. Metal Bioaccumulation in the Ovaries of Nile tilapia
3.2. Effects on Growth and Ovarian Development of Nile tilapia
3.3. Effects on Histopathological Changes in the Ovaries of Nile Tilapia
3.4. Effects of Hormone Levels on Nile Tilapia
3.5. Quality Analysis of Transcriptome Sequencing and Identification of Differential Genes in Nile Tilapia Ovaries
3.6. The Validated Results of qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Béné, C.; Arthur, R.; Norbury, H.; Allison, E.H.; Beveridge, M.; Bush, S.; Campling, L.; Leschen, W.; Little, D.; Squires, D.; et al. Contribution of Fisheries and Aquaculture to Food Security and Poverty Reduction: Assessing the Current Evidence. World Dev. 2016, 79, 177–196. [Google Scholar] [CrossRef]
- Emenike, E.C.; Iwuozor, K.O.; Anidiobi, S.U. Heavy Metal Pollution in Aquaculture: Sources, Impacts and Mitigation Techniques. Biol. Trace Elem. Res. 2022, 200, 4476–4492. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.J.; Shimmield, T.M.; Black, K.D. Copper, zinc and cadmium in marine cage fish farm sediments: An extensive survey. Environ. Pollut. 2007, 145, 84–95. [Google Scholar] [CrossRef]
- Adhikari, S.; Ghosh, L.; Rai, S.P.; Ayyappan, S. Metal concentrations in water, sediment, and fish from sewage-fed aquaculture ponds of Kolkata, India. Environ. Monit. Assess. 2009, 159, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Perea-Juárez, R.N.; Frías-Espericueta, M.G.; Páez-Osuna, F.; Voltolina, D. Copper, zinc, cadmium and lead inputs and outputs in the maternity section of a commercial shrimp hatchery. Lat. Am. J. Aquat. Res. 2016, 44, 595–601. [Google Scholar] [CrossRef]
- Salami, I.R.S.; Rahmawati, S.; Indri, R.; Sutarto, H.; Jaya, P.M. Accumulation of Heavy Metals in Freshwater Fish in Cage Aquaculture at Cirata Reservoir, West Java, Indonesia. In Environmental Challenges in the Pacific Basin; Carpenter, D.O., Ed.; Wiley-Blackwell: Malden, MA, USA, 2008; Volume 1140, pp. 290–296. [Google Scholar]
- Squadrone, S.; Brizio, P.; Stella, C.; Prearo, M.; Pastorino, P.; Serracca, L.; Ercolini, C.; Abete, M.C. Presence of trace metals in aquaculture marine ecosystems of the northwestern Mediterranean Sea (Italy). Environ. Pollut. 2016, 215, 77–83. [Google Scholar] [CrossRef]
- Dixit, R.; Wasiullah; Malaviya, D.; Pandiyan, K.; Singh, U.B.; Sahu, A.; Shukla, R.; Singh, B.P.; Rai, J.P.; Sharma, P.K.; et al. Bioremediation of Heavy Metals from Soil and Aquatic Environment: An Overview of Principles and Criteria of Fundamental Processes. Sustainability 2015, 7, 2189–2212. [Google Scholar] [CrossRef]
- Chen, C.W.; Kao, C.M.; Chen, C.F.; Dong, C.D. Distribution and accumulation of heavy metals in the sediments of Kaohsiung Harbor, Taiwan. Chemosphere 2007, 66, 1431–1440. [Google Scholar] [CrossRef]
- Tchounwou, P.B.; Yedjou, C.G.; Patlolla, A.K.; Sutton, D.J. Heavy metal toxicity and the environment. Mol. Clin. Environ. Toxicol. 2012, 3, 133–164. [Google Scholar] [CrossRef]
- Malhotra, N.; Ger, T.R.; Uapipatanakul, B.; Huang, J.C.; Chen, K.H.C.; Hsiao, C.D. Review of Copper and Copper Nanoparticle Toxicity in Fish. Nanomaterials 2020, 10, 28. [Google Scholar] [CrossRef]
- Thompson, J.; Bannigan, J. Cadmium: Toxic effects on the reproductive system and the embryo. Reprod. Toxicol. 2008, 25, 304–315. [Google Scholar] [CrossRef]
- Liu, Y.A.; Chen, Q.Q.; Li, Y.Q.; Bi, L.L.; Jin, L.B.; Peng, R.Y. Toxic Effects of Cadmium on Fish. Toxics 2022, 10, 19. [Google Scholar] [CrossRef]
- Jezierska, B.; Witeska, M. The metal uptake and accumulation in fish living in polluted waters. In Proceedings of the NATO Advanced Research Workshop on Viable Methods of Soil and Water Pollution Monitoring, Protection and Remediation, Cracow, Poland, 27 June–1 July 2006; pp. 107–114. [Google Scholar]
- Milla, S.; Depiereux, S.; Kestemont, P. The effects of estrogenic and androgenic endocrine disruptors on the immune system of fish: A review. Ecotoxicology 2011, 20, 305–319. [Google Scholar] [CrossRef] [PubMed]
- Schreck, C.B.; Tort, L. The concept of stress in fish. Fish Physiol. 2016, 35, 1–34. [Google Scholar]
- Atli, G.; Canli, M. Response of antioxidant system of freshwater fish Oreochromis niloticus to acute and chronic metal (Cd, Cu, Cr, Zn, Fe) exposures. Ecotox. Environ. Saf. 2010, 73, 1884–1889. [Google Scholar] [CrossRef] [PubMed]
- Atli, G.; Canli, M. Responses of metallothionein and reduced glutathione in a freshwater fish Oreochromis niloticus following metal exposures. Environ. Toxicol. Pharmacol. 2008, 25, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Sato, T.; Xing, B.S.; Tao, S. Health risks of heavy metals to the general public in Tianjin, China via consumption of vegetables and fish. Sci. Total Environ. 2005, 350, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.X.; Lampi, M.A.; Huang, X.D.; Gerhardt, K.; Dixon, D.G.; Greenberg, B.M. Assessment of Mixture Toxicity of Copper, Cadmium, and Phenanthrenequinone to the Marine Bacterium Vibrio fischeri. Environ. Toxicol. 2009, 24, 166–177. [Google Scholar] [CrossRef] [PubMed]
- Firat, Ö.; Kargin, F. Individual and Combined Effects of Heavy Metals on Serum Biochemistry of Nile Tilapia Oreochromis niloticus. Arch. Environ. Contam. Toxicol. 2010, 58, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.B.; Wang, N.B.; Zhang, M.; Dai, H.X.; Dawood, M.; Zhang, G.P.; Wu, F.B. Comparative study of alleviating effects of GSH, Se and Zn under combined contamination of cadmium and chromium in rice (Oryza sativa). Biometals 2013, 26, 297–308. [Google Scholar] [CrossRef]
- Yang, X.X.; Song, Y.F.; Kai, J.R.; Cao, X.F. Enzymatic biomarkers of earthworms Eisenia fetida in response to individual and combined cadmium and pyrene. Ecotoxicol. Environ. Saf. 2012, 86, 162–167. [Google Scholar] [CrossRef]
- Jeppe, K.J.; Carew, M.E.; Long, S.M.; Lee, S.F.; Pettigrove, V.; Hoffmann, A.A. Genes involved in cysteine metabolism of Chironomus tepperi are regulated differently by copper and by cadmium. Comp. Biochem. Physiol. C-Toxicol. Pharmacol. 2014, 162, 1–6. [Google Scholar] [CrossRef]
- Gebeyew, K.; Jiang, C.Y.; Gao, Q.H.; Zhang, L.P.; Zhu, H.H.; Tian, Y.S.; Wang, Q.; Wei, Y.Q.; Tan, Z.L.; Han, X.F. Cadmium Accumulation in the Goat Liver and Kidney Is Partially Promoted by the Upregulation of Metal Transporter Genes. Animals 2022, 12, 15. [Google Scholar] [CrossRef]
- Heo, D.H.; Baek, I.J.; Kang, H.J.; Kim, J.H.; Chang, M.; Jeong, M.Y.; Kim, T.H.; Choi, I.D.; Yun, C.W. Cadmium regulates copper homoeostasis by inhibiting the activity of Mac1, a transcriptional activator of the copper regulon, in Saccharomyces cerevisiae. Biochem. J. 2010, 431, 257–265. [Google Scholar] [CrossRef]
- El Asely, A.M.; Reda, R.M.; Salah, A.S.; Mahmoud, M.A.; Dawood, M.A.O. Overall performances of Nile tilapia (Oreochromis niloticus) associated with using vegetable oil sources under suboptimal temperature. Aquac. Nutr. 2020, 26, 1154–1163. [Google Scholar] [CrossRef]
- Almeida, J.A.; Diniz, Y.S.; Marques, S.F.G.; Faine, L.A.; Ribas, B.O.; Burneiko, R.C.; Novelli, E.L.B. The use of the oxidative stress responses as biomarkers in Nile tilapia (Oreochromis niloticus) exposed to in vivo cadmium contamination. Environ. Int. 2002, 27, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.M.; Lin, H.C.; Yang, W.L. The effects of maternal Cd on the metallothionein expression in tilapia (Oreochromis mossambicus) embryos and larvae. Aquat. Toxicol. 2008, 87, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Parra, L.; Lloret, G.; Lloret, J.; Rodilla, M. Physical Sensors for Precision Aquaculture: A Review. IEEE Sens. J. 2018, 18, 3915–3923. [Google Scholar] [CrossRef]
- Wu, Y.J.; Huang, T.; Yan, X.; Xiao, J.; Ma, Z.R.; Luo, L.M.; Chen, L.T.; Cao, J.L.; Tang, Z.Y.; Wei, X.X.; et al. Effects of four hormones on the mitigation of ovarian damage in tilapia (Oreochromis niloticus) after copper and cadmium exposure. Aquat. Toxicol. 2023, 258, 8. [Google Scholar] [CrossRef] [PubMed]
- LaFrentz, B.R.; Shoemaker, C.A.; Klesius, P.H. Immunoproteomic analysis of the antibody response obtained in Nile tilapia following vaccination with a Streptococcus iniae vaccine. Vet. Microbiol. 2011, 152, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Landmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357-U121. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 38. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.Z.; Cai, T.; Olyarchuk, J.G.; Wei, L.P. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Kozera, B.; Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef]
- Wang, J.J.; Xiao, J.; Zhang, J.Y.; Chen, H.X.; Li, D.; Li, L.Q.; Cao, J.L.; Xie, L.T.; Luo, Y.J. Effects of dietary Cu and Zn on the accumulation, oxidative stress and the expressions of immune-related genes in the livers of Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 2020, 100, 198–207. [Google Scholar] [CrossRef]
- Younis, N.A.; Laban, S.E.; Al-Mokaddem, A.K.; Attia, M.M. Immunological status and histopathological appraisal of farmed Oreochromis niloticus exposed to parasitic infections and heavy metal toxicity. Aquac. Int. 2020, 28, 2247–2262. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Capriello, T.; Grimaldi, M.C.; Cofone, R.; D’Aniello, S.; Ferrandino, I. Effects of aluminium and cadmium on hatching and swimming ability in developing zebrafish. Chemosphere 2019, 222, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Driessnack, M.K.; Matthews, A.L.; Raine, J.C.; Niyogi, S. Interactive effects of chronic waterborne copper and cadmium exposure on tissue-specific metal accumulation and reproduction in fathead minnow (Pimephales promelas). Comp. Biochem. Physiol. C-Toxicol. Pharmacol. 2016, 179, 165–173. [Google Scholar] [CrossRef]
- Tilton, S.C.; Foran, C.M.; Benson, W.H. Effects of cadmium on the reproductive axis of Japanese medaka (Oryzias latipes). Comp. Biochem. Physiol. C-Toxicol. Pharmacol. 2003, 136, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Suvi, R.; Giovanna, M.; Katja, A. Experimental copper exposure, but not heat stress, leads to elevated intraovarian thyroid hormone levels in three-spined sticklebacks (Gasterosteus aculeatus). Ecotoxicology 2020, 29, 1431–1440. [Google Scholar] [CrossRef] [PubMed]
- Genchi, G.; Sinicropi, M.S.; Lauria, G.; Carocci, A.; Catalano, A. The Effects of Cadmium Toxicity. Int. J. Environ. Res. Public Health 2020, 17, 24. [Google Scholar] [CrossRef]
- Akan, J.C.; Mohmoud, S.; Yikala, B.S.; Ogugbuaja, V.O. Bioaccumulation of some heavy metals in fish samples from River Benue in Vinikilang, Adamawa State, Nigeria. Am. J. Anal. Chem. 2012, 3, 24468. [Google Scholar] [CrossRef]
- Pelgrom, S.; Lock, R.A.C.; Balm, P.H.M.; Bonga, S.E.W. Integrated physiological response of tilapia, Oreochromis mossambicus, to sublethal copper exposure. Aquat. Toxicol. 1995, 32, 303–320. [Google Scholar] [CrossRef]
- Bettini, S.; Ciani, F.; Franceschini, V. Recovery of the olfactory receptor neurons in the African Tilapia mariae following exposure to low copper level. Aquat. Toxicol. 2006, 76, 321–328. [Google Scholar] [CrossRef]
- Mowat, F.S.; Bundy, K.J. A mathematical algorithm to identify toxicity and prioritize pollutants in field sediments. Chemosphere 2002, 49, 499–513. [Google Scholar] [CrossRef]
- West, G. Methods of assessing ovarian development in fishes: A review. Aust. J. Mar. Freshw. Res. 1990, 41, 199–222. [Google Scholar] [CrossRef]
- Nagahama, Y. Endocrine regulation of gametogenesis in fish. Int. J. Dev. Biol. 1994, 38, 217–229. [Google Scholar]
- Chen, J.; Hu, W.; Zhu, Z.Y. Progress in studies of fish reproductive development regulation. Chin. Sci. Bull. 2013, 58, 7–16. [Google Scholar] [CrossRef]
- Bock, S.L.; Chow, M.I.; Forsgren, K.L.; Lema, S.C. Widespread alterations to hypothalamic-pituitary-gonadal (HPG) axis signaling underlie high temperature reproductive inhibition in the eurythermal sheepshead minnow (Cyprinodon ariegatus). Mol. Cell. Endocrinol. 2021, 537, 16. [Google Scholar] [CrossRef] [PubMed]
- Kavanaugh, S.I.; Nozaki, M.; Sower, S.A. Origins of gonadotropin-releasing hormone (GnRH) in vertebrates: Identification of a novel GnRH in a basal vertebrate, the sea lamprey. Endocrinology 2008, 149, 3860–3869. [Google Scholar] [CrossRef] [PubMed]
- Tyler, C.R.; vanderEerden, B.; Jobling, S.; Panter, G.; Sumpter, J.P. Measurement of vitellogenin, a biomarker for exposure to oestrogenic chemicals, in a wide variety of cyprinid fish. J. Comp. Physiol. B-Biochem. Syst. Environ. Physiol. 1996, 166, 418–426. [Google Scholar] [CrossRef]
- Zheng, X.C.; Liu, W.B.; Liu, J.D.; Zhang, C.Y.; Zhang, L.; Gao, F.; Zhang, D.D.; Chi, C. Dietary Supplementation With Icariin Affects Estrogen Synthesis, Vitellogenesis, and Oocyte Development in the Chinese Mitten Crab, Eriocheir sinensis. Front. Mar. Sci. 2020, 7, 13. [Google Scholar] [CrossRef]
- Babich, H.; Shopsis, C.; Borenfreund, E. In vitro cytotoxicity testing of aquatic pollutants (cadmium, copper, zinc, nickel) using established fish cell lines. Ecotoxicol. Environ. Saf. 1986, 11, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Funk, A.E.; Day, F.A.; Brady, F.O. Displacement of zinc and copper from copper-induced metallothionein by cadmium and by mercury: In vivo and ex vivo studies. Comp. Biochem. Physiol. Part C Comp. Pharmacol. 1987, 86, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kamunde, C.; MacPhail, R. Metal-metal interactions of dietary cadmium, copper and zinc in rainbow trout, Oncorhynchus mykiss. Ecotoxicol. Environ. Saf. 2011, 74, 658–667. [Google Scholar] [CrossRef] [PubMed]
- Tuormaa, T.E. Chromium, selenium and copper and other trace minerals in health and reproduction. J. Orthomol. Med. 2000, 15, 145–156. [Google Scholar]
- Minghetti, M.; Leaver, M.J.; Taggart, J.B.; Casadei, E.; Auslander, M.; Tom, M.; George, S.G. Copper induces Cu-ATPase ATP7A mRNA in a fish cell line, SAF1. Comp. Biochem. Physiol. C-Toxicol. Pharmacol. 2011, 154, 93–99. [Google Scholar] [CrossRef]
- Puig, S.; Thiele, D.J. Molecular mechanisms of copper uptake and distribution. Curr. Opin. Chem. Biol. 2002, 6, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Lutsenko, S.; Gupta, A.; Burkhead, J.L.; Zuzel, V. Cellular multitasking: The dual role of human Cu-ATPases in cofactor delivery and intracellular copper balance. Arch. Biochem. Biophys. 2008, 476, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Blaby-Haas, C.E.; Flood, J.A.; de Crécy-Lagard, V.; Zamble, D.B. YeiR: A metal-binding GTPase from Escherichia coli involved in metal homeostasis. Metallomics 2012, 4, 488–497. [Google Scholar] [CrossRef] [PubMed]
- Vetter, I.R.; Wittinghofer, A. Signal transduction—The guanine nucleotide-binding switch in three dimensions. Science 2001, 294, 1299–1304. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Maxfield, F.R. Role of membrane organization and membrane domains in endocytic lipid trafficking. Traffic 2000, 1, 203–211. [Google Scholar] [CrossRef] [PubMed]
- McNew, J.A.; Parlati, F.; Fukuda, R.; Johnston, R.J.; Paz, K.; Paumet, F.; Söllner, T.H.; Rothman, J.E. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature 2000, 407, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Parlati, F.; McNew, J.A.; Fukuda, R.; Miller, R.; Söllner, T.H.; Rothman, J.E. Topological restriction of SNARE-dependent membrane fusion. Nature 2000, 407, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Zerial, M.; McBride, H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001, 2, 107–117. [Google Scholar] [CrossRef]
- Pascale, M.C.; Franceschelli, S.; Moltedo, O.; Belleudi, F.; Torrisi, M.R.; Bucci, C.; La Fontaine, S.; Mercer, J.F.B.; Leone, A. Endosomal trafficking of the Menkes copper ATPase ATP7A is mediated by vesicles containing the Rab7 and Rab5 GTPase proteins. Exp. Cell Res. 2003, 291, 377–385. [Google Scholar] [CrossRef]
- Lemmon, S.K.; Traub, L.M. Sorting in the endosomal system in yeast and animal cells. Curr. Opin. Cell Biol. 2000, 12, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Sönnichsen, B.; De Renzis, S.; Nielsen, E.; Rietdorf, J.; Zerial, M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5 and Rab11. J. Cell Biol. 2000, 149, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Bucci, C.; Parton, R.G.; Mather, I.H.; Stunnenberg, H.; Simons, K.; Hoflack, B.; Zerial, M. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell 1992, 70, 715–728. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.S.; Lv, Z.M.; Zhu, A.Y.; Zheng, J.L.; Wu, C.W. Negative effect of chronic cadmium exposure on growth, histology, ultrastructure, antioxidant and innate immune responses in the liver of zebrafish: Preventive role of blue light emitting diodes. Ecotoxicol. Environ. Saf. 2017, 139, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Eyckmans, M.; Celis, N.; Horemans, N.; Blust, R.; De Boeck, G. Exposure to waterborne copper reveals differences in oxidative stress response in three freshwater fish species. Aquat. Toxicol. 2011, 103, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Lushchak, V.I. Environmentally induced oxidative stress in aquatic animals. Aquat. Toxicol. 2011, 101, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Cowan, K.J.; Storey, K.B. Mitogen-activated protein kinases: New signaling pathways functioning in cellular responses to environmental stress. J. Exp. Biol. 2003, 206, 1107–1115. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.; Forman, H.J. Redox signaling and the MAP kinase pathways. Biofactors 2003, 17, 287–296. [Google Scholar] [CrossRef]
- Adhikari, S.; Bhatia, M. H2S-induced pancreatic acinar cell apoptosis is mediated via JNK and p38 MAP kinase. J. Cell. Mol. Med. 2008, 12, 1374–1383. [Google Scholar] [CrossRef]
- Chan, L.-P.; Liu, C.; Chiang, F.-Y.; Wang, L.-F.; Lee, K.-W.; Chen, W.-T.; Kuo, P.-L.; Liang, C.-H. IL-8 promotes inflammatory mediators and stimulates activation of p38 MAPK/ERK-NF-κB pathway and reduction of JNK in HNSCC. Oncotarget 2017, 8, 56375. [Google Scholar] [CrossRef]
- Son, Y.; Kim, S.; Chung, H.T.; Pae, H.O. Reactive Oxygen Species in the Activation of MAP Kinases. In Hydrogen Peroxide and Cell Signaling, Pt C; Cadenas, E., Packer, L., Eds.; Elsevier Academic Press Inc.: San Diego, CA, USA, 2013; Volume 528, pp. 27–48. [Google Scholar]
- Araya, M.; Pizarro, F.; Olivares, M.; Arredondo, M.; González, M.; Méndez, M. Understanding copper homeostasis in humans and copper effects on health. Biol. Res. 2006, 39, 183–187. [Google Scholar] [CrossRef]
- Rae, T.D.; Schmidt, P.J.; Pufahl, R.A.; Culotta, V.C.; O’Halloran, T.V. Undetectable intracellular free copper: The requirement of a copper chaperone for superoxide dismutase. Science 1999, 284, 805–808. [Google Scholar] [CrossRef]
- Zheng, J.L.; Yuan, S.S.; Wu, C.W.; Lv, Z.M. Acute exposure to waterborne cadmium induced oxidative stress and immunotoxicity in the brain, ovary and liver of zebrafish (Danio rerio). Aquat. Toxicol. 2016, 180, 36–44. [Google Scholar] [CrossRef]
- Tümer, Z.; Moller, L.B. Menkes disease. Eur. J. Hum. Genet. 2010, 18, 511–518. [Google Scholar] [CrossRef]
- Liaw, K.Y.; Lee, P.H.; Wu, F.C.; Tsai, J.S.; LinShiau, S.Y. Zinc, copper, and superoxide dismutase in hepatocellular carcinoma. Am. J. Gastroenterol. 1997, 92, 2260–2263. [Google Scholar]
- Xiao, J.; Zhou, Y.; Luo, Y.J.; Zhong, H.; Huang, Y.L.; Zhang, Y.; Luo, Z.J.; Ling, Z.B.; Zhang, M.; Gan, X. Suppression effect of LHRH-A and hCG on Piwi expression in testis of Nile tilapia Oreochromis niloticus. Gen. Comp. Endocrinol. 2013, 189, 43–50. [Google Scholar] [CrossRef]
- Ahmadnezhad, M.; Oryan, S.; Sahafi, H.H.; Khara, H. Effect of Synthetic Luteinizing Hormone—Releasing Hormone (LHRH-A2) Plus Pimozide and Chlorpromazine on Ovarian Development and Levels of Gonad Steroid Hormones in Female Kutum Rutilus frisii kutum. Turk. J. Fish. Quat. Sci. 2013, 13, 95–100. [Google Scholar] [CrossRef]
- Matsuyama, M.; Takeuchi, H.; Kashiwagi, M.; Hirose, K.; Kagawa, H. Induced gonadal development and spawning of immature red sea bream Pagrus major with LHRH-a administration in different ways during winter season. Fish. Sci. 1995, 61, 472–477. [Google Scholar] [CrossRef]
- Legler, D.F.; Thelen, M. New insights in chemokine signaling. FResearch 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. mTOR Signaling in Growth Control and Disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Z.; Liu, Z.Q.; Kang, T.; Li, M.H.; Wang, D.S.; Cheng, C.H.K. Igf3: A novel player in fish reproduction. Biol. Reprod. 2021, 104, 1194–1204. [Google Scholar] [CrossRef]
- Elakkanai, P.; Francis, T.; Ahilan, B.; Jawahar, P.; Padmavathy, P.; Jayakumar, N.; Subburaj, A. Role of GnRH, HCG and Kisspeptin on reproduction of fishes. Indian J. Sci. Technol. 2015, 8, 65166. [Google Scholar] [CrossRef]
- Strehl, C.; Ehlers, L.; Gaber, T.; Buttgere, F. Glucocorticoids-All-Rounders Tackling the Versatile Players of the Immune System. Front. Immunol. 2019, 10, 20. [Google Scholar] [CrossRef]
- Busada, J.T.; Cidlowski, J.A. Mechanisms of Glucocorticoid Action During Development. In Nuclear Receptors in Development and Disease; Forrest, D., Tsai, S., Eds.; Elsevier Academic Press Inc.: San Diego, CA, USA, 2017; Volume 125, pp. 147–170. [Google Scholar]
- Whirledge, S.; Cidlowski, J.A. Glucocorticoids and Reproduction: Traffic Control on the Road to Reproduction. Trends Endocrinol. Metab. 2017, 28, 399–415. [Google Scholar] [CrossRef] [PubMed]
- Hashida, S.; Kawai-Yamada, M. Inter-Organelle NAD Metabolism Underpinning Light Responsive NADP Dynamics in Plants. Front. Plant Sci. 2019, 10, 7. [Google Scholar] [CrossRef] [PubMed]
- Pollard, C.L.; Gibb, Z.; Clulow, J.; Ruiz, A.; Sheridan, A.; Bahrami, M.; Swegen, A.; Grupen, C.G. Supplemental Nicotinic Acid Elevates NAD+Precursors in the Follicular Fluid of Mares. Animals 2022, 12, 12. [Google Scholar] [CrossRef] [PubMed]
- Leroy, J.; Vanholder, T.; Mateusen, B.; Christophe, A.; Opsomer, G.; de Kruif, A.; Genicot, G.; Van Soom, A. Non-esterified fatty acids in follicular fluid of dairy cows and their effect on developmental capacity of bovine oocytes in vitro. Reproduction 2005, 130, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Bowater, R.; Doherty, A.J. Making ends meet: Repairing breaks in bacterial DNA by non-homologous end-joining. PLoS Genet. 2006, 2, 93–99. [Google Scholar] [CrossRef] [PubMed]



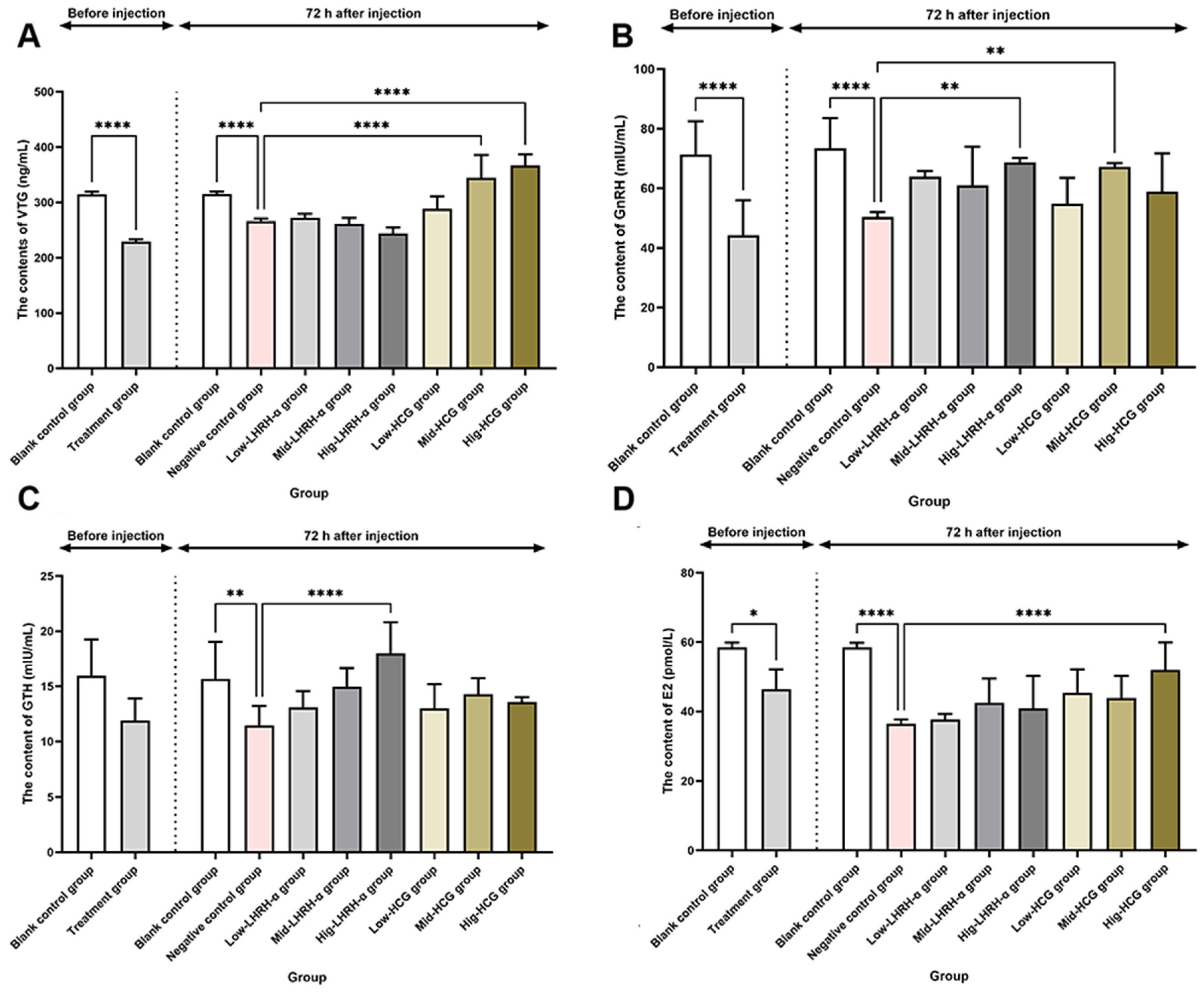
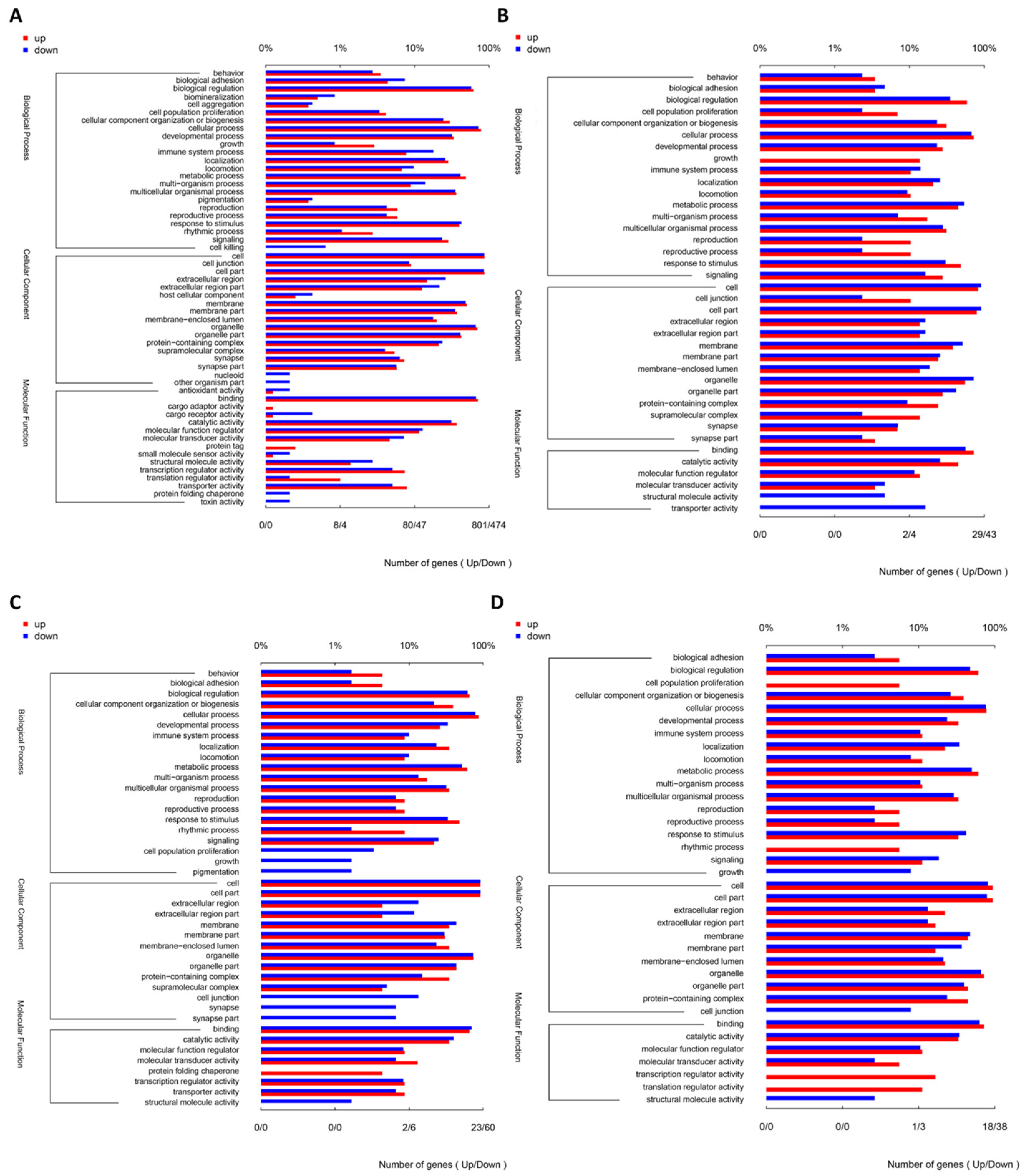
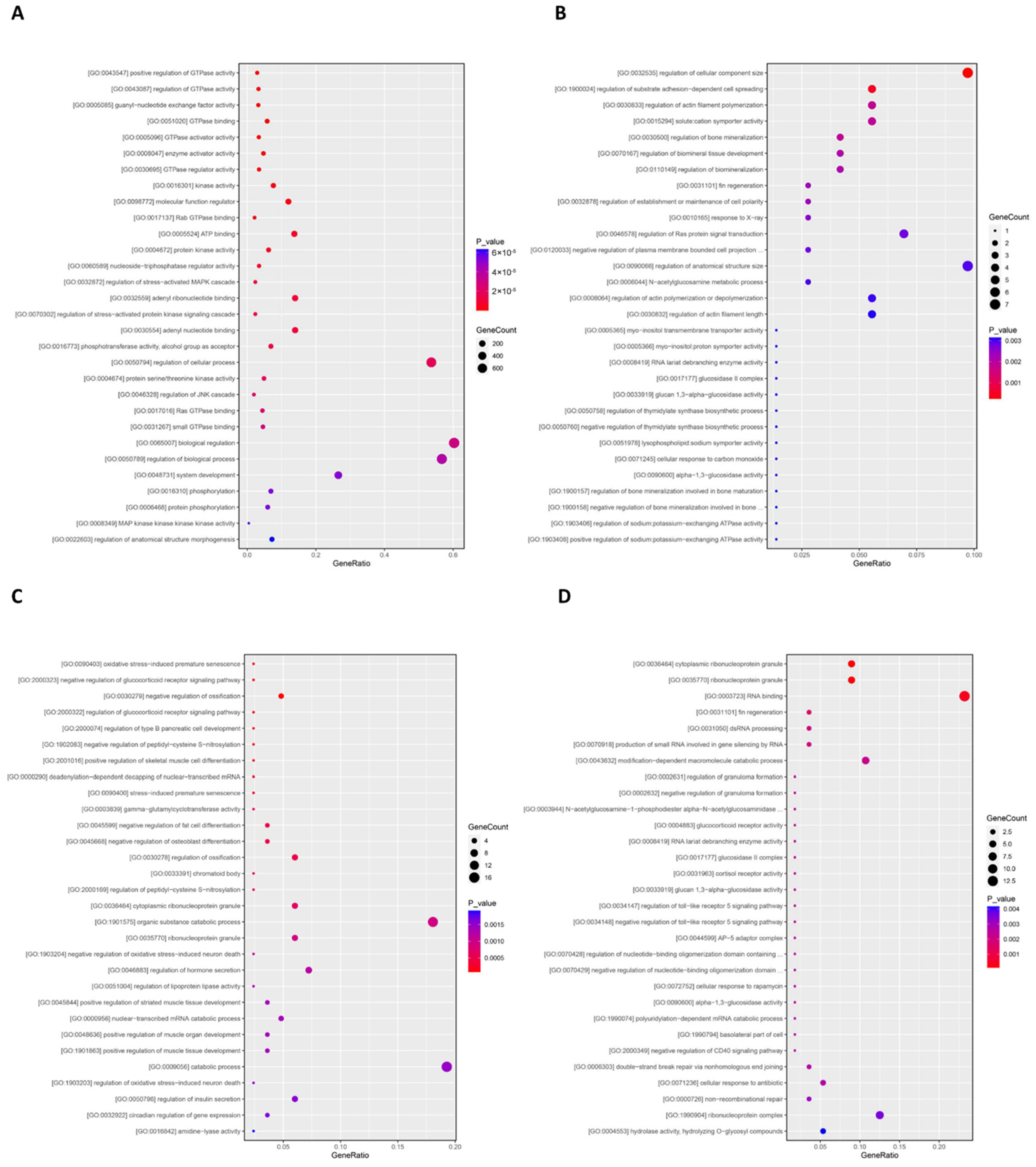

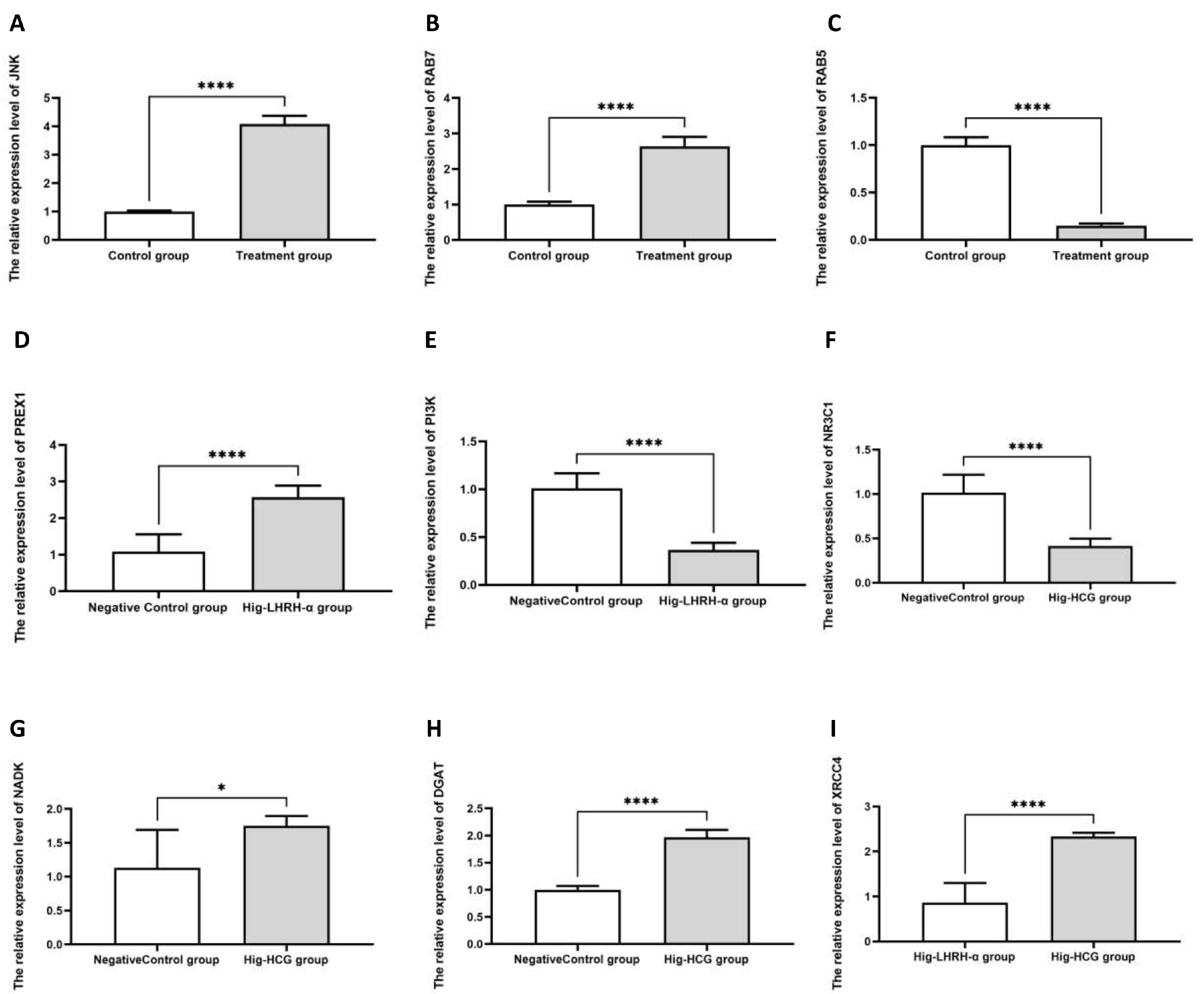
| Group | First Experimental Phase | Second Experimental Phase | |||
|---|---|---|---|---|---|
| Culture Water | Number of Organisms | Intraperitoneal Injection | Number of Organisms | ||
| Blank control group | Aerated pure water | 96 (in 2 tanks; 48 per tank) | Nothing | 48 | |
| Treatment group | Negative control group | Water containing 300 μg/L Cu2+ and 100 μg/L Cd2+ | 384 (in 8 tanks; 48 per tank) | PBS (200 μL) | 48 |
| Low-LHRH-α group | 5 μg/g LHRH-α (in 200 μL PBS) | 48 | |||
| Mid-LHRH-α group | 10 μg/g LHRH-α (in 200 μL PBS) | 48 | |||
| Hig-LHRH-α group | 20 μg/g LHRH-α (in 200 μL PBS) | 48 | |||
| Low-HCG group | 0.5 IU/g HCG (in 200 μL PBS) | 48 | |||
| Mid-HCG group | 1.0 IU/g HCG (in 200 μL PBS) | 48 | |||
| Hig-HCG group | 1.5 IU/g HCG (in 200 μL PBS) | 48 | |||
| Name | GenBank Accession Number | Sequence (5′–3′) |
|---|---|---|
| JNK | NC_031984.2 | F-GCACAGCCAAGTGGAGGTAT |
| R-TGAGTGCTGCAAGGGCATTA | ||
| RAB5 | NC_031985.2 | F-CCCTACCGCAAAGTGGAGTT |
| R-GAGCCCTAAACTGGGCTTGA | ||
| RAB7A | NC_031970.2 | F-TTTTGAAACCAGCGCCAAGG |
| R-AGAGTCTGTGTGCCGCTCTA | ||
| PREX1 | NC_031984.2 | F-TGCAATCAAGTCACGTGTAAAA |
| R-TACTCGCTGTCCCCTGATGA | ||
| PI3K | NC_031982.2 | F-ACTTGTAGCCTTTCCCCGTG |
| R-GCACTCGGGGGAGAATCAAA | ||
| NR3C1 | NC_031966.2 | F-TGTGCGGAAATCTACGGAGT |
| R-GCTGGATTGATTTGGACGGC | ||
| NADK | NC_031984.2 | F-GTGCTGCATGCTGATAAGGC |
| R-TTGGCAAGCACGCAAATACC | ||
| DGAT | NC_031985.2 | F-GATGCCTTTTCTATGGCCTGC |
| R-GCTTGTTTCGCAGACTGGTG | ||
| XRCC4 | NC_031977.2 | F-CAGCCCTGGTAACCACTCAG |
| R-ACTCATGCGAATTGTTGCCG | ||
| BETA-ACTIN | NC_031969.2 | F-ACTAGCGGAAGTTCACGAGC |
| R-GCAAAGCTGCTCAAAACCGA |
| Compare | All | Up | Down |
|---|---|---|---|
| Blank control group vs. Treatment group | 1792 | 977 | 815 |
| Negative control group vs. Hig-LHRH-α group | 99 | 36 | 63 |
| Negative control group vs. Hig-HCG group | 121 | 31 | 90 |
| Hig-LHRH-α group vs. Hig-HCG group | 75 | 28 | 47 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Chen, L.; Yan, X.; Xiao, J.; Ma, Z.; Tang, Z.; Guo, Z.; Li, L.; Tong, G.; Tan, H.; et al. The Effect of Copper–Cadmium Co-Exposure and Hormone Remediation on the Ovarian Transcriptome of Nile Tilapia (Oreochromis niloticus). Fishes 2024, 9, 67. https://doi.org/10.3390/fishes9020067
Wu Y, Chen L, Yan X, Xiao J, Ma Z, Tang Z, Guo Z, Li L, Tong G, Tan H, et al. The Effect of Copper–Cadmium Co-Exposure and Hormone Remediation on the Ovarian Transcriptome of Nile Tilapia (Oreochromis niloticus). Fishes. 2024; 9(2):67. https://doi.org/10.3390/fishes9020067
Chicago/Turabian StyleWu, Yijie, Liting Chen, Xin Yan, Jun Xiao, Zhirui Ma, Zhanyang Tang, Zhongbao Guo, Liping Li, Guixiang Tong, Honglian Tan, and et al. 2024. "The Effect of Copper–Cadmium Co-Exposure and Hormone Remediation on the Ovarian Transcriptome of Nile Tilapia (Oreochromis niloticus)" Fishes 9, no. 2: 67. https://doi.org/10.3390/fishes9020067
APA StyleWu, Y., Chen, L., Yan, X., Xiao, J., Ma, Z., Tang, Z., Guo, Z., Li, L., Tong, G., Tan, H., Chen, F., Wei, X., Huang, T., & Luo, Y. (2024). The Effect of Copper–Cadmium Co-Exposure and Hormone Remediation on the Ovarian Transcriptome of Nile Tilapia (Oreochromis niloticus). Fishes, 9(2), 67. https://doi.org/10.3390/fishes9020067





