Exploring Seasonal Variations in Fish Communities: A Study of the Yellow River Estuary and Its Adjacent Waters Using eDNA and Trawl Surveys
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Sites
2.2. Sample Collection and DNA Extraction
2.3. Library Construction and Sequencing
2.4. Bioinformatics Analysis
3. Results
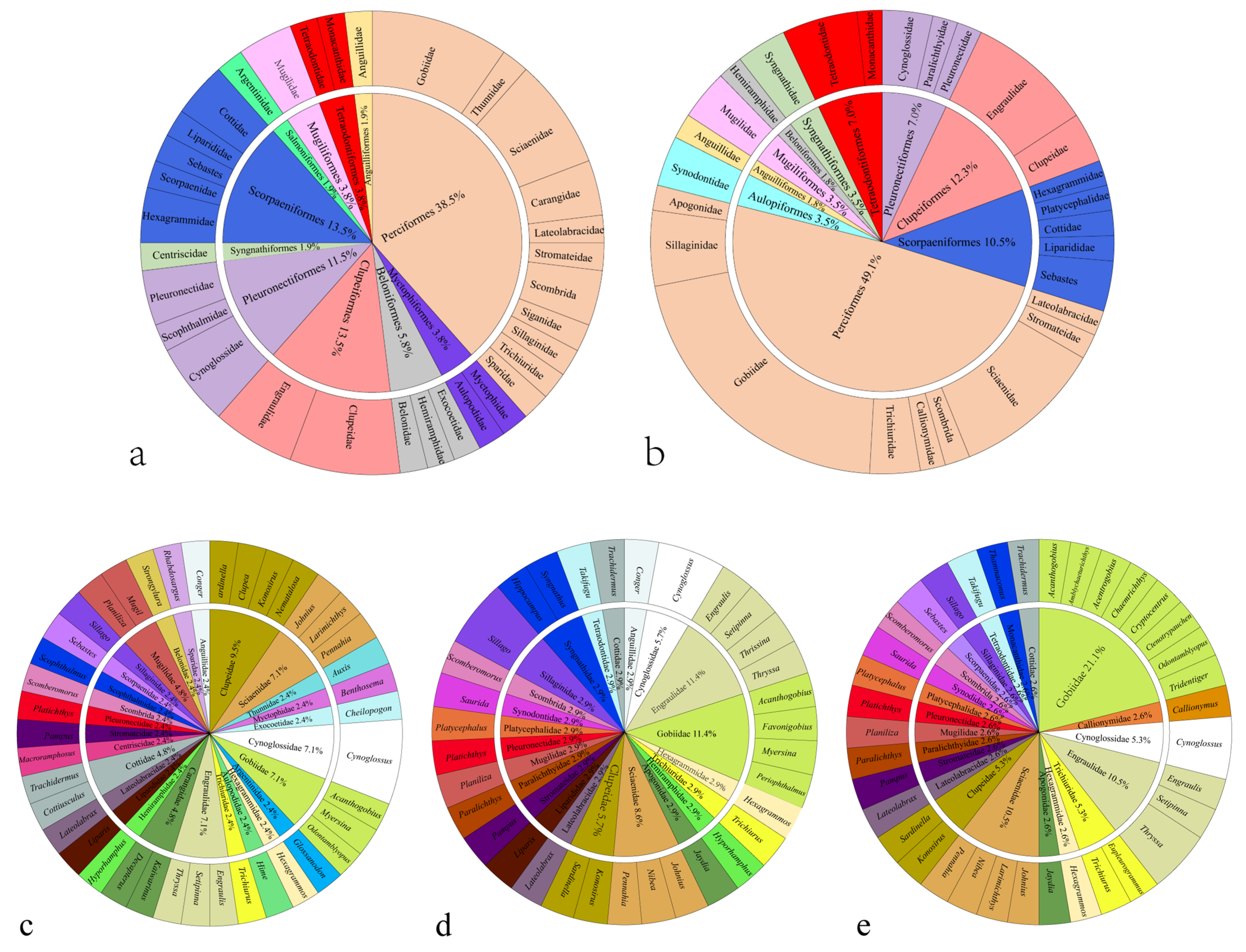
3.1. Survey Results and Taxonomic Composition
3.2. Diversity of Fish Communities
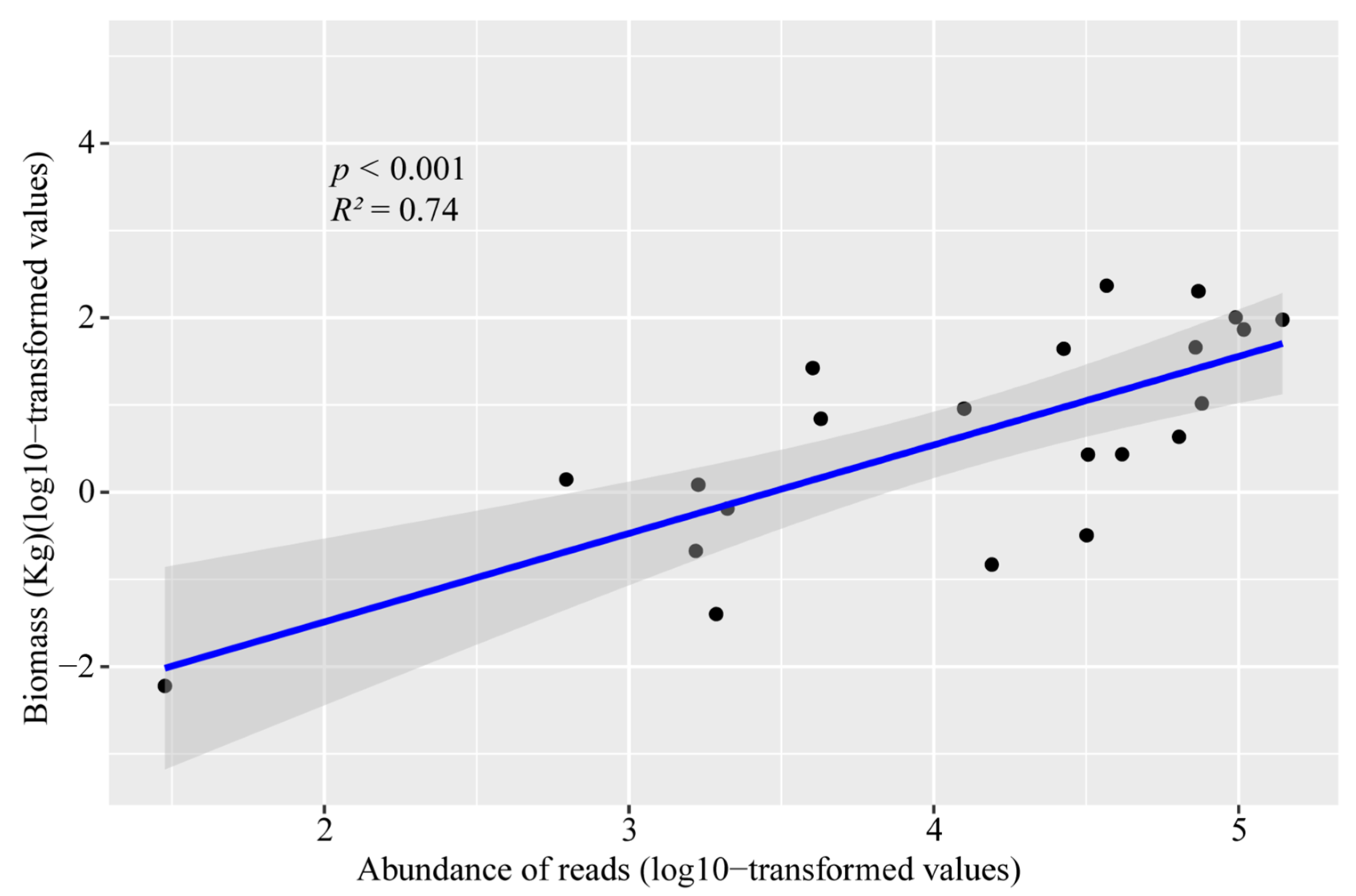
3.3. Relationship between Fish Community and Environmental Factors
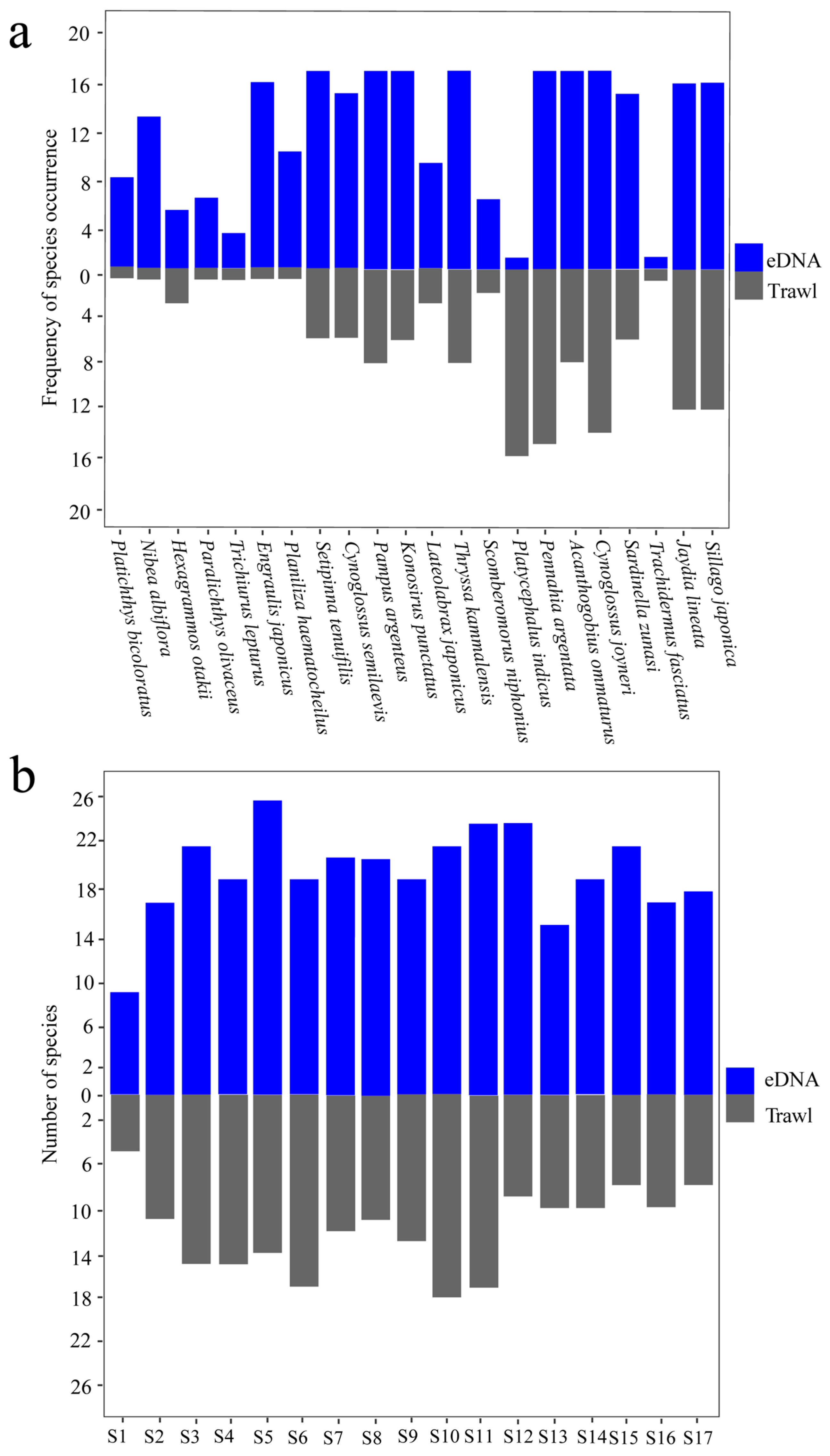
3.4. Comparison with Fish Trawling
4. Discussion
4.1. Fish Diversity in Yellow River Estuary
| Year | Number of Investigations | Species Number | Dominant Species | Method |
|---|---|---|---|---|
| 1959 [34] | 4 | 53 | Trichiurus lepturus Larimichthys polyactis Cynoglossus semilaevis Platycephalus indicus | Trawl |
| 1983 [34] | 4 | 45 | Setipinna tenuifilis Engraulis japonicus Nibea albiflora Larimichthys polyactis | Trawl |
| 1993 [42] | 4 | 35 | Engraulis japonicus Thryssa kammalensis Setipinna tenuifilis Konosirus punctatus | Trawl |
| 1999 [34] | 4 | 30 | Thryssa kammalensis Engraulis japonicus Setipinna tenuifilis Konosirus punctatus | Trawl |
| 2012 [32] | 3 | 39 | Chaemrichthys stigmatias Konosirus punctatus Sardinella zunasi Engraulis japonicus | Trawl |
| 2014 [33] | 7 | 51 | - | Trawl |
| 2019 [43] | 3 | 46 | Chaemrichthys stigmatias Cynoglossus joyneri | Trawl |
| 2020 [31] | 3 | 54 | Cynoglossus joyneri Chaemrichthys stigmatias Thryssa kammalensis Callionymus beniteguri | Trawl |
| 2022–2023 | 2 | 55 | Sardinella zunasi Hexagrammos otakii Engraulis japonicus Sebastes spp. | eDNA |
| 2023 | 1 | 38 | Chaemrichthys stigmatias Pampus argenteus Cynoglossus joyneri Thryssa kammalensis | Trawl |
4.2. Relationships between Fish Community and Environmental Factors
4.3. Comparison of eDNA with Trawl Survey
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yeoh, D.E.; Valesini, F.J.; Hallett, C.S.; Abdo, D.A.; Williams, J. Diel shifts in the structure and function of nearshore estuarine fish communities. J. Fish Biol. 2017, 90, 1214–1243. [Google Scholar] [CrossRef]
- Qian, M.; Wang, Z.; Zhou, Q.; Wang, J.; Shao, Y.; Qiao, Q.; Fan, J.; Yan, Z. Environmental DNA unveiling the fish community structure and diversity features in the Yangtze River basin. Environ. Res. 2023, 239, 117198. [Google Scholar] [CrossRef] [PubMed]
- Van Niekerk, L.; Taljaard, S.; Lamberth, S.J.; Adams, J.B.; Weerts, S.P.; MacKay, C.F. Disaggregation and assessment of estuarine pressures at the country-level to better inform management and resource protection–the South African experience. Afr. J. Aquat. Sci. 2022, 47, 127–148. [Google Scholar] [CrossRef]
- Worm, B.; Barbier, E.B.; Beaumont, N.; Duffy, J.E.; Folke, C.; Halpern, B.S.; Jackson, J.B.C.; Lotze, H.K.; Micheli, F.; Palumbi, S.R.; et al. Impacts of Biodiversity Loss on Ocean Ecosystem Services. Science 2006, 314, 787–790. [Google Scholar] [CrossRef] [PubMed]
- Cinner, J.E.; Zamborain-Mason, J.; Gurney, G.G.; Graham, N.A.J.; MacNeil, M.A.; Hoey, A.S.; Mora, C.; Villéger, S.; Maire, E.; McClanahan, T.R.; et al. Meeting fisheries, ecosystem function, and biodiversity goals in a human-dominated world. Science 2020, 368, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Seitzinger, S.P.; Mayorga, E.; Bouwman, A.F.; Kroeze, C.; Beusen, A.H.W.; Billen, G.; Van Drecht, G.; Dumont, E.; Fekete, B.M.; Garnier, J.; et al. Global river nutrient export: A scenario analysis of past and future trends. Glob. Biogeochem. Cycles 2010, 24, 2621–2628. [Google Scholar] [CrossRef]
- Lyu, D.; Qian, T.; Li, F.; Sun, S.; Wang, W.; Shan, X. Monitoring and assessing the species diversity and abundance of marine teleost around the Yellow River estuary in June using environmental DNA. Front. Mar. Sci. 2023, 10, 1123831. [Google Scholar] [CrossRef]
- Wang, H.; Yang, Z.; Saito, Y.; Liu, J.P.; Sun, X.; Wang, Y. Stepwise decreases of the Huanghe (Yellow River) sediment load (1950-2005): Impacts of climate change and human activities. Gobal Planet. Chang. 2007, 57, 331–354. [Google Scholar] [CrossRef]
- Jin, X.; Shan, X.; Li, X.; Wang, J.; Cui, Y.; Zuo, T. Long-term changes in the fishery ecosystem structure of Laizhou Bay, China. Sci. China Earth Sci. 2013, 56, 366–374. [Google Scholar] [CrossRef]
- Fraija-Fernández, N.; Bouquieaux, M.C.; Rey, A.; Mendibil, I.; Cotano, U.; Irigoien, X.; Rodríguez-Ezpeleta, N. Marine water environmental DNA metabarcoding provides a comprehensive fish diversity assessment and reveals spatial patterns in a large oceanic area. Ecol. Evol. 2020, 10, 7560–7584. [Google Scholar] [CrossRef]
- Sakuma, K.M.; Field, J.C.; Mantua, N.J.; Ralston, S.; Marinovic, B.B.; Carrion, C.N. Anomalous epipelagic micronekton assemblage patterns in the neritic waters of the California Current in spring 2015 during a period of extreme ocean conditions. CalCOFI Rep. 2016, 57, 163–183. [Google Scholar]
- Thomsen, P.F.; Willerslev, E. Environmental DNA—An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Doi, H.; Fukaya, K.; Oka, S.I.; Sato, K.; Kondoh, M.; Miya, M. Evaluation of detection probabilities at the water-filtering and initial PCR steps in environmental DNA metabarcoding using a multispecies site occupancy model. Sci. Rep. 2019, 9, 3581. [Google Scholar] [CrossRef] [PubMed]
- Doi, H.; Inui, R.; Akamatsu, Y.; Kanno, K.; Yamanaka, H.; Takahara, T.; Minamoto, T. Environmental DNA analysis for estimating the abundance and biomass of stream fish. Freshw. Biol. 2017, 62, 30–39. [Google Scholar] [CrossRef]
- Wang, S.; Yan, Z.; Hänfling, B.; Zheng, X.; Wang, P.; Fan, J.; Li, J. Methodology of fish eDNA and its applications in ecology and environment. Sci. Total Environ. 2021, 755, 142622. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhao, J.; Yao, M. A comprehensive and comparative evaluation of primers for metabarcoding eDNA from fish. Methods Ecol. Evol. 2020, 11, 1609–1625. [Google Scholar] [CrossRef]
- Ogram, A.; Sayler, G.S.; Barkay, T. The extraction and purification of microbial dna from sediments. J. Microbiol. Methods 1987, 7, 57–66. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Miaud, C.; Pompanon, F.; Taberlet, P. Species detection using environmental DNA from water samples. Biol. Lett. 2008, 4, 423–425. [Google Scholar] [CrossRef] [PubMed]
- Sigsgaard, E.E.; Carl, H.; Moller, P.R.; Thomsen, P.F. Monitoring the near-extinct European weather loach in Denmark based on environmental DNA from water samples. Biol. Conserv. 2014, 183, 46–52. [Google Scholar] [CrossRef]
- Tréguier, A.; Paillisson, J.M.; Dejean, T.; Valentini, A.; Schlaepfer, M.A.; Roussel, J.M. Environmental DNA surveillance for invertebrate species: Advantages and technical limitations to detect invasive crayfish Procambarus clarkii in freshwater ponds. J. Appl. Ecol. 2014, 51, 871–879. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, X.; Chen, Z.; Zhong, L.; Gao, T. Minary study on spatial distribution pattern of fish in Zhoushan and its adjacent waters based on environmental DNA metabarcoding. J. Fish. China 2022, 1–13. Available online: https://kns.cnki.net/kcms/detail//31.1283.S.20221226.1142.003.html (accessed on 15 May 2024). (In Chinese).
- Shu, L.; Lin, J.; Xu, Y.; Cao, T.; Feng, J.; Peng, Z. Investigating the fish diversity in Erhai lake based on environmental DNA metabarcoding. Acta Hydrobiol. Sin. 2020, 44, 1080–1086. [Google Scholar] [CrossRef]
- Miya, M.; Sato, Y.; Fukunaga, T.; Sado, T.; Poulsen, J.Y.; Sato, K.; Poulsen, J.Y.; Sato, K.; Minamoto, T.; Yamamoto, S.; et al. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2015, 2, 150088. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Berger, C.S.; Hernandez, C.; Laporte, M.; Cté, G.; Bernatchez, L. Fine: Cale environmental heterogeneity shapes fluvial fish communities as revealed by eDNA metabarcoding. Environ. DNA 2020, 2, 647–666. [Google Scholar] [CrossRef]
- Shen, A.; Ishizaka, J.; Yang, M.; Ouyang, L.; Yin, Y.; Ma, Z. Changes in community structure and photosynthetic activities of total phytoplankton species during the growth, maintenance, and dissipation phases of a Prorocentrum donghaiense bloom. Harmful Algae 2019, 82, 35–43. [Google Scholar] [CrossRef]
- Martin, G.R.; Twigg, L.E.; Robinson, D.J. Comparison of the diet of feral cats from rural and pastoral Western Australia. Wildl. Res. 1996, 23, 475–484. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. vegan: Community Ecology Package, R package version 2.6-4; 2022. Available online: https://CRAN.R-project.org/package=vegan (accessed on 15 May 2024).
- Froese, R.; Pauly, D. FishBase. World Wide Web Electronic Publication. Available online: http://www.fishbase.org (accessed on 10 May 2024).
- Sun, Y.; Wang, L.; Wang, Z.; Zhang, Y.; Zhao, G.; Kong, Q.; Xu, X.; Qiu, S. Changes of community structure of swimming organisms in the Yellow River Estuary and adjacent waters and its relationship with environmental factors. J. Yantai Univ. Nat. Sci. Eng. Ed. 2023, 36, 186–195. (In Chinese) [Google Scholar] [CrossRef]
- Zheng, L.; Lv, Z.; Li, F.; Zhang, L.; Yu, W. Fish community structure in the Yellow River estuary: Effect of water and sediment discharge regulations. J. Fish. Sci. China 2014, 21, 602–610. (In Chinese) [Google Scholar] [CrossRef]
- Wang, J.; Zhang, C.; Xue, Y.; Ren, Y.; Xu, B. Taxonomic diversity of fish community in the Yellow River Estuary. Acta Oceanol. Sin. 2018, 40, 86–95. (In Chinese) [Google Scholar] [CrossRef]
- Jin, X.; Deng, J. Variations in community structure of fishery resources and biodiversity in the Laizhou Bay, Shandong, Chinese. Biodiversity 2000, 8, 65–72. (In Chinese) [Google Scholar] [CrossRef]
- Ryder, R.A.; Kerr, S.R.; Taylor, W.W.; Larkin, P.A. Community consequence of fish stock diversity. Can. J. Fish. Aquat. Sci. 1981, 38, 1856–1866. [Google Scholar] [CrossRef]
- Lyman, J.A.; Sanchez, D.E.; Hershauer, S.N.; Sobek, C.J.; Chambers, C.L.; Zahratka, J.; Walker, F.M. Mammalian eDNA on herbaceous vegetation? Validating a qPCR assay for detection of an endangered rodent. Environ. DNA 2022, 4, 1187–1197. [Google Scholar] [CrossRef]
- Dai, S.; Bai, M.; Jia, H.; Xian, W.; Zhang, H. An assessment of seasonal differences in fish populations in Laizhou Bay using environmental DNA and conventional resource survey techniques. Fishes 2022, 7, 250. [Google Scholar] [CrossRef]
- Abidin, D.H.Z.; Nor, S.A.M.; Lavoué, S.; Rahim, M.A.; Akib, N.A.M. Assessing a megadiverse but poorly known community of fishes in a tropical mangrove estuary through environmental DNA (eDNA) metabarcoding. Sci. Rep. 2022, 12, 16346. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Tang, Q. Changes in fish species diversity and dominant species composition in the Yellow Sea. Fish. Res. 1996, 26, 337–352. [Google Scholar] [CrossRef]
- Jaureguizar, A.J.; Solari, A.; Cortés, F.; Milessi, A.C.; Militelli, M.I.; Camiolo, M.D.; García, M. Fish diversity in the Río de la Plata and adjacent waters: An overview of environmental influences on its spatial and temporal structure. J. Fish Biol. 2016, 89, 569–600. [Google Scholar] [CrossRef]
- Zhou, F.; Huang, D.; Su, J. Numerical simulation of the dual-core structure of the Bohai Sea cold bottom water in summer. Chin. Sci. Bull. 2009, 54, 1591–1599. [Google Scholar] [CrossRef]
- Jin, X. The dynamics of major fishery resources in the Bohai Sea. J. Fish. Sci. China 2000, 7, 22–26. (In Chinese) [Google Scholar] [CrossRef]
- Gao, H.; Wang, L.; Qiu, S.; Gao, K. Current Status of Fishery Resources in the Yellow River Estuary and Its Adjacent Sea Area. J. Yantai Univ. Nat. Sci. Eng. Ed. 2022, 35, 191–200. (In Chinese) [Google Scholar] [CrossRef]
- Kamrani, E.; Sharifinia, M.; Hashemi, S.H. Analyses of fish community structure changes in three subtropical estuaries from the Iranian coastal waters. Mar. Biol. 2016, 46, 561–577. [Google Scholar] [CrossRef]
- Beukhof, E.; Dencker, T.S.; Pecuchet, L.; Lindegren, M. Spatio-temporal variation in marine fish traits reveals community-wide responses to environmental change. Mar. Ecol. Prog. Ser. 2019, 610, 205–222. [Google Scholar] [CrossRef]
- Luo, Z.; Yang, C.; Wang, L.; Liu, Y.; Shan, B.; Liu, M.; Sun, D. Relationships between Fish Community Structure and Environmental Factors in the Nearshore Waters of Hainan Island, South China. Diversity 2023, 15, 901. [Google Scholar] [CrossRef]
- Wu, L.; Osugi, T.; Inagawa, T.; Okitsu, J.; Sakamoto, S.; Minamoto, T. Monitoring of multiple fish species by quantitative environmental DNA metabarcoding surveys over two summer seasons. Mol. Ecol. Resour. 2023, 24, e13875. [Google Scholar] [CrossRef]
- Carlisle, A.B.; Kochevar, R.E.; Arostegui, M.C.; Ganong, J.E.; Castleton, M.; Schratwieser, J.; Block, B.A. Influence of temperature and oxygen on the distribution of blue marlin (Makaira nigricans) in the Central Pacific. Fish. Oceanogr. 2017, 26, 34–48. [Google Scholar] [CrossRef]
- Liu, H.; Niu, J.; Liu, C.; Zhang, T.; Mina, M.; Chen, H.; Cai, L. Fish community structure and its relationship with environmental factors of main stream of Emin River, Xinjiang. Sheng Tai Xue Za Zhi 2017, 36, 3558–3563. (In Chinese) [Google Scholar] [CrossRef]
- Baptista, J.; Martinho, F.; Martins, R.; Carneiro, M.; Azevedo, M.; Vieira, A.R.; Gomes, P.; Pardal, M.A. Water temperature gradient shapes the structure and composition of nearshore marine fish communities in southern Europe. J. Sea Res. 2019, 154, 101807. [Google Scholar] [CrossRef]
- Guo, C.; Konar, B.H.; Gorman, K.B.; Walker, C.M. Environmental factors important to high-latitude nearshore estuarine fish community structure. Deep Sea Res. Part II Top. Stud. Oceanogr. 2022, 201, 105109. [Google Scholar] [CrossRef]
- Díaz, J.; Vanina, V.; Felipe, D.P.; Florencia, B.; Alexis, G.; Silvia, E.A. First DNA barcode reference library for the identification of South American freshwater fish from the lower Paraná River. PLoS ONE 2016, 11, e0157419. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef]
- Pentinsaari, M.; Ratnasingham, S.; Miller, S.E.; Hebert, P.D.N. BOLD and GenBank revisited—Do identification errors arise in the lab or in the sequence libraries? PLoS ONE 2020, 15, e0231814. [Google Scholar] [CrossRef]
- Itakura, H.; Wakiya, R.; Yamamoto, S.; Kaifu, K.; Sato, T.; Minamoto, T. Environmental DNA analysis reveals the spatial distribution, abundance, and biomass of Japanese eels at the river-basin scale. Aquat. Conserv. Mar. Freshw. Ecosyst. 2019, 29, 361–373. [Google Scholar] [CrossRef]
- Shu, L.; Ludwig, A.; Peng, Z. Environmental dna metabarcoding primers for freshwater fish detection and quantification: In silico and in tanks. Ecol. Evol. 2021, 11, 8281–8294. [Google Scholar] [CrossRef]




| Season | Survey Site | Surface eDNA | Bottom eDNA | Season | Survey Site | Surface eDNA | Trawl Survey | Environmental Factors |
|---|---|---|---|---|---|---|---|---|
| Spring | A1 | + | Summer | S1 | + | + | ||
| Spring | A2 | + | + | Summer | S2 | + | + | |
| Spring | A3 | + | + | Summer | S3 | + | + | + |
| Spring | A4 | + | + | Summer | S4 | + | + | + |
| Spring | A5 | + | + | Summer | S5 | + | + | + |
| Spring | A6 | + | Summer | S6 | + | + | + | |
| Spring | A7 | + | + | Summer | S7 | + | + | |
| Spring | A8 | + | + | Summer | S8 | + | + | |
| Spring | A9 | + | + | Summer | S9 | + | + | + |
| Spring | A10 | + | + | Summer | S10 | + | + | + |
| Spring | A11 | + | + | Summer | S11 | + | + | + |
| Spring | A12 | + | Summer | S12 | + | + | + | |
| Spring | A13 | + | Summer | S13 | + | + | ||
| Spring | A14 | + | + | Summer | S14 | + | + | |
| Spring | A15 | + | + | Summer | S15 | + | + | |
| Spring | A16 | + | Summer | S16 | + | + | ||
| Spring | A17 | + | Summer | S17 | + | + | ||
| Spring | A18 | + | ||||||
| Spring | A19 | + |
| Dominant Species | Spring/eDNA (Surface) | Spring/eDNA (Bottom) | Summer/eDNA (Surface) | Summer/Trawl (Bottom) |
|---|---|---|---|---|
| Acanthogobius ommaturus | + | + | + | |
| Chaemrichthys stigmatias | + | |||
| Cynoglossus joyneri | + | + | ||
| Engraulis japonicus | + | + | + | |
| Hexagrammos otakii | + | + | ||
| Jaydia lineata | + | |||
| Johnius grypotus | + | |||
| Konosirus punctatus | + | + | + | |
| Pampus argenteus | + | + | ||
| Pennahia argentata | + | |||
| Sardinella zunasi | + | + | + | |
| Saurida microlepis | + | |||
| Sebastes spp. | + | + | ||
| Setipinna tenuifilis | + | + | ||
| Sillago japonica | + | |||
| Thryssa kammalensis | + | + | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Li, F.; Shao, F.; Song, H.; Song, N.; Zhang, X.; Zhao, L. Exploring Seasonal Variations in Fish Communities: A Study of the Yellow River Estuary and Its Adjacent Waters Using eDNA and Trawl Surveys. Fishes 2024, 9, 192. https://doi.org/10.3390/fishes9060192
Wang X, Li F, Shao F, Song H, Song N, Zhang X, Zhao L. Exploring Seasonal Variations in Fish Communities: A Study of the Yellow River Estuary and Its Adjacent Waters Using eDNA and Trawl Surveys. Fishes. 2024; 9(6):192. https://doi.org/10.3390/fishes9060192
Chicago/Turabian StyleWang, Xiaoyang, Fan Li, Fei Shao, Hongjun Song, Na Song, Xiaomin Zhang, and Linlin Zhao. 2024. "Exploring Seasonal Variations in Fish Communities: A Study of the Yellow River Estuary and Its Adjacent Waters Using eDNA and Trawl Surveys" Fishes 9, no. 6: 192. https://doi.org/10.3390/fishes9060192
APA StyleWang, X., Li, F., Shao, F., Song, H., Song, N., Zhang, X., & Zhao, L. (2024). Exploring Seasonal Variations in Fish Communities: A Study of the Yellow River Estuary and Its Adjacent Waters Using eDNA and Trawl Surveys. Fishes, 9(6), 192. https://doi.org/10.3390/fishes9060192





