Identification and Characterization of microRNAs in Morphological Color Change of Polychromatic Midas Cichlids (Amphilophus citrinellus)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fish Farming and Sample Collection
2.2. Total RNA Extraction, Construction, and Sequencing of a Small RNA Library
2.3. Basic Analysis of Sequencing Data
2.4. Differential Expression Analysis of miRNAs
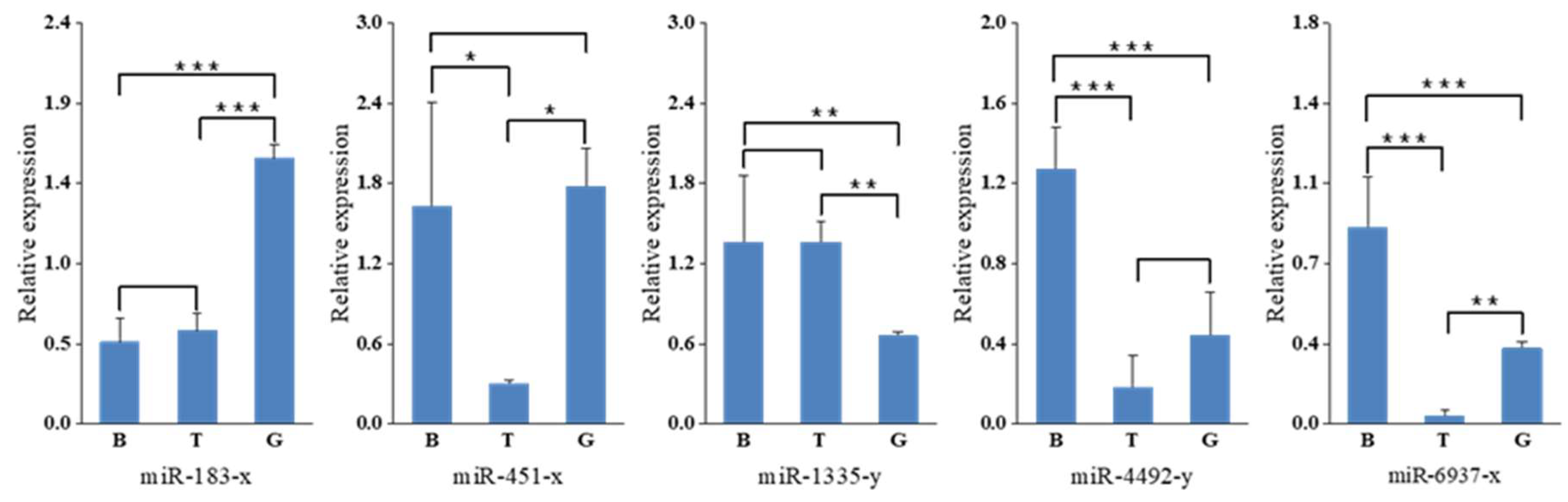
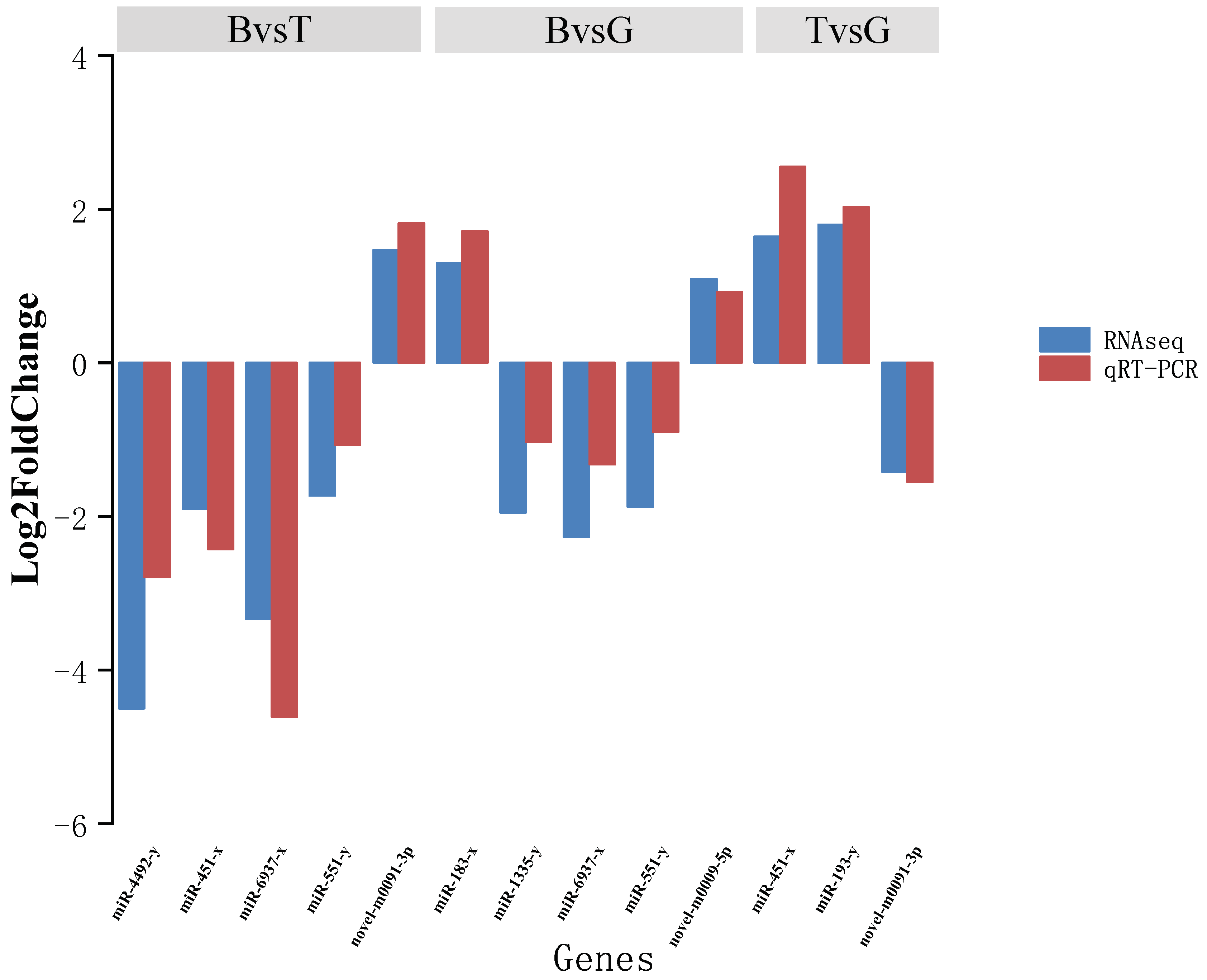
2.5. Validation of miRNA Expression by Stem-Loop qRT-PCR
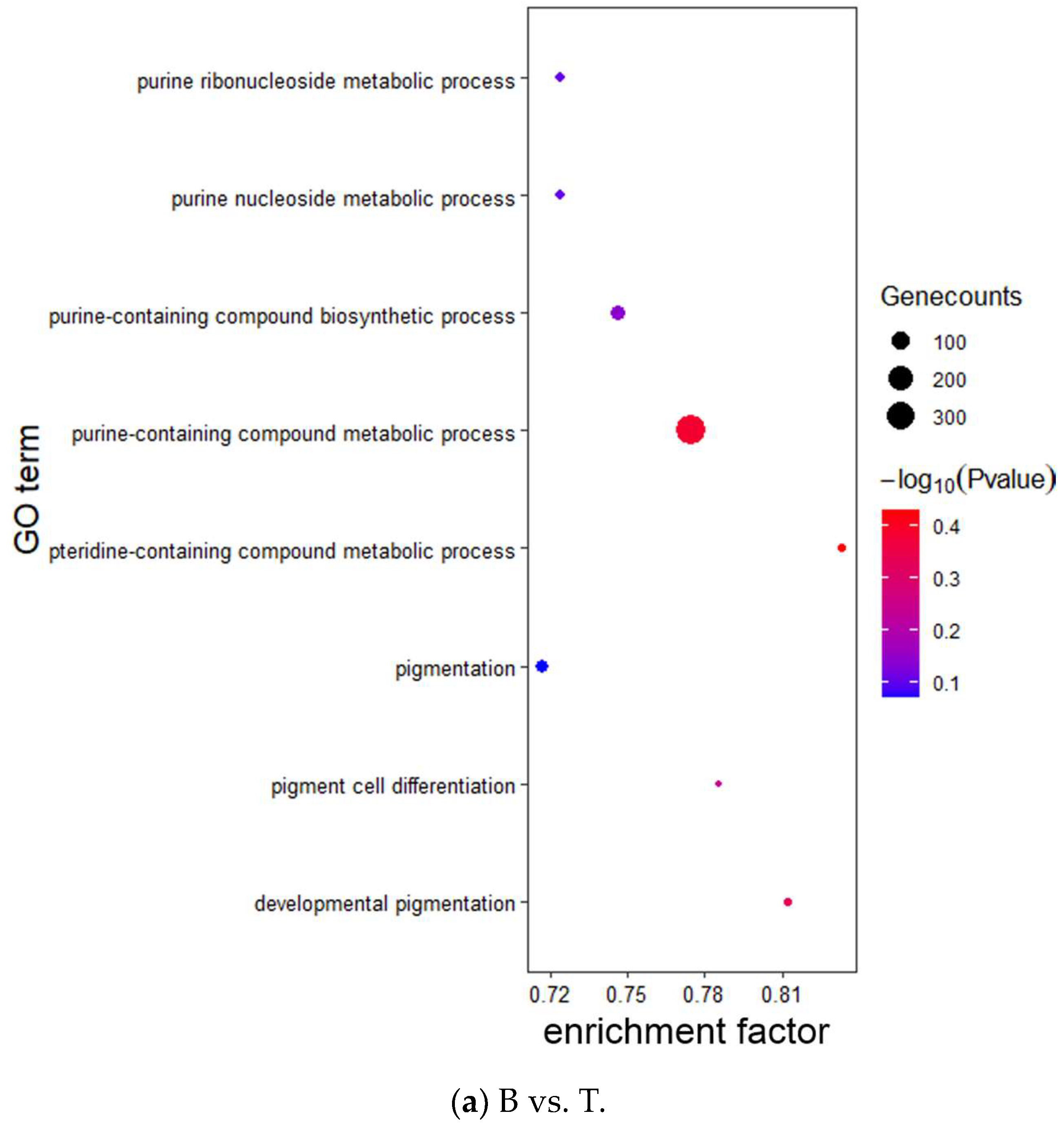
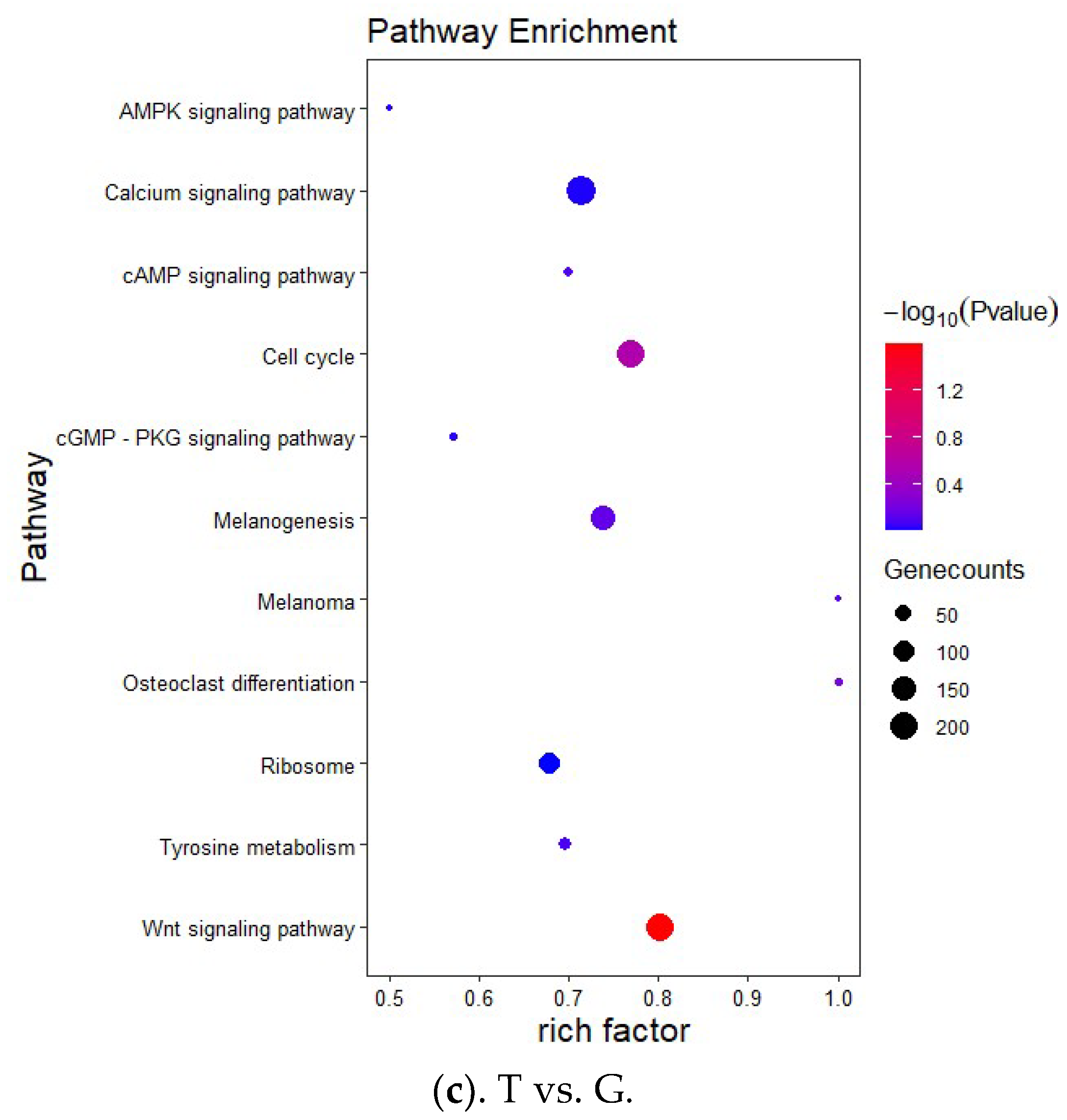
2.6. Target Gene Prediction and Functional Enrichment Analysis
3. Results
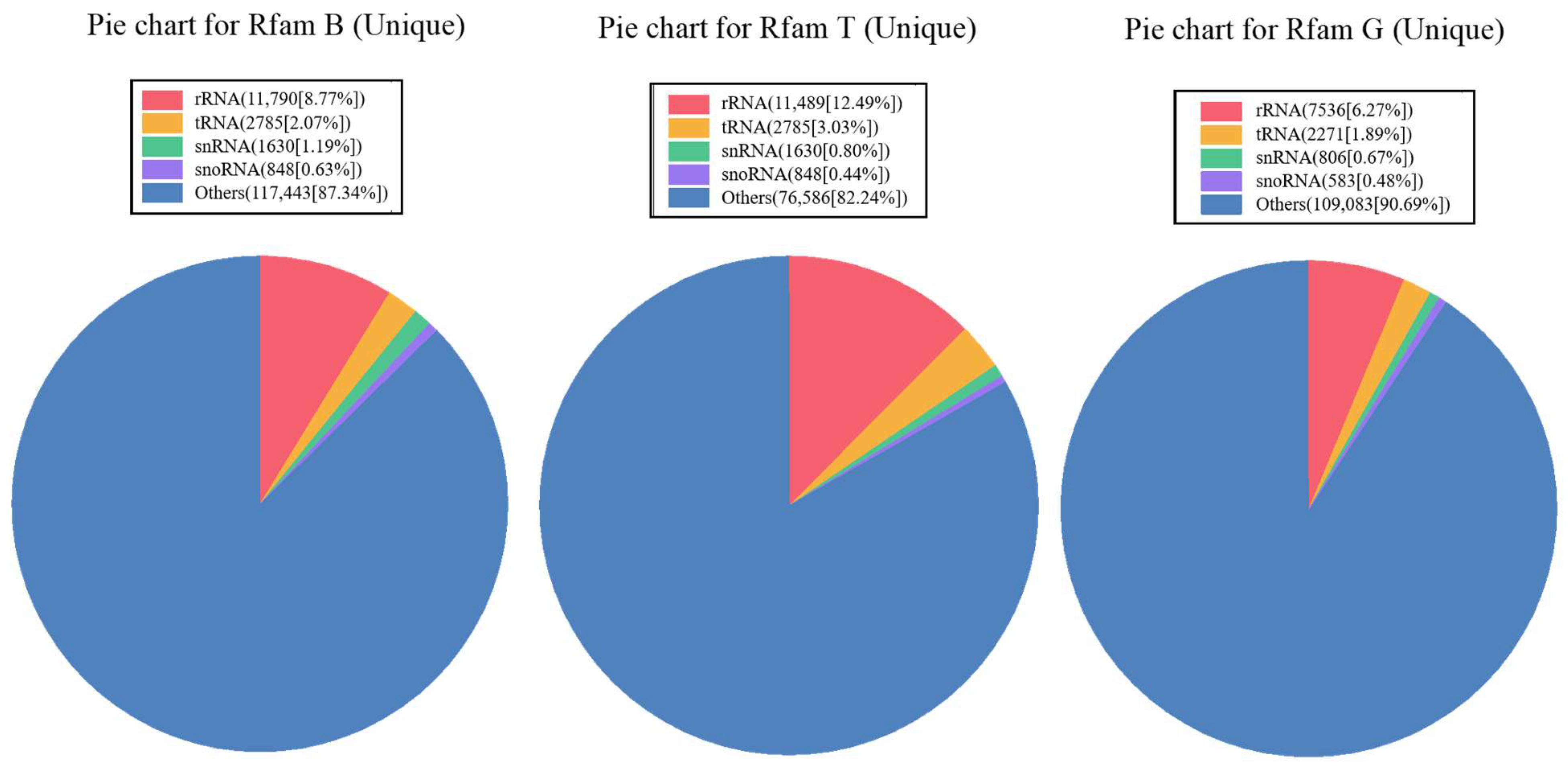
3.1. Small RNA Sequence Analysis
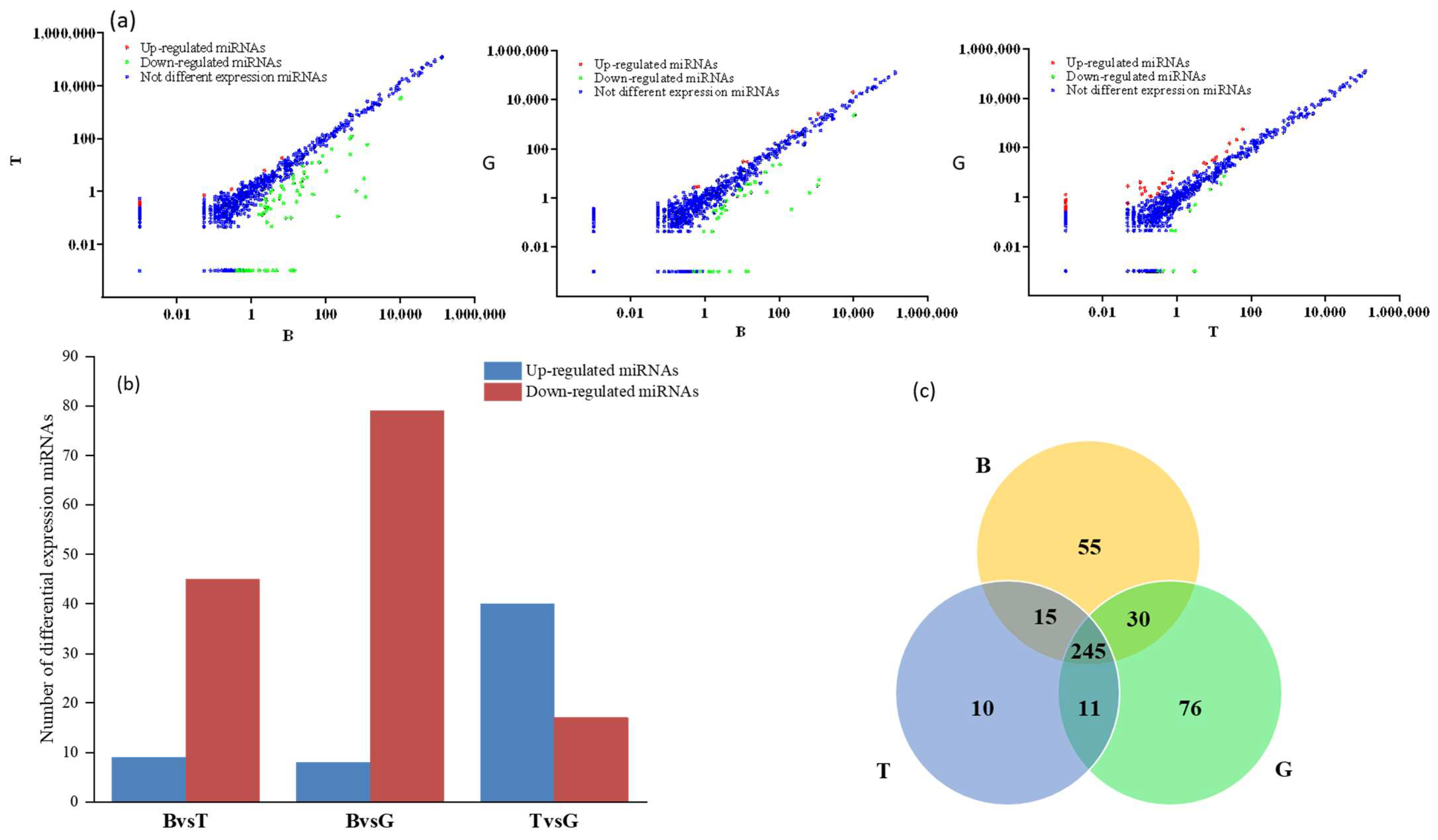
3.2. Analysis of miRNA Expression Trends and Differences
3.3. Target Prediction and Enrichment Analysis for miRNAs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, G.; Tang, L.; Huang, J.; Wang, Y.; Wang, H.; Fan, Y.; Yuan, X.; Liu, W.; Peng, L.; Liu, J.; et al. Formation of asymmetric body color in the caudal fin of common carp (Cyprinus carpio). Aquaculture 2023, 577, 739970. [Google Scholar] [CrossRef]
- Marcoli, R.; Jones, D.B.; Massault, C.; Marc, A.F.; Moran, M.; Harrison, P.J.; Cate, H.S.; Lopata, A.L.; Jerry, D.R. The skin structure in multiple color variants of barramundi (Lates calcarifer): A histological, immunohistochemical and ultrastructural overview. Aquaculture 2023, 576, 739859. [Google Scholar] [CrossRef]
- Chen, X.; Wu, G.; Song, H.; Wang, X.; Mou, X.; Liu, Y.; Liu, C.; Hu, Y. Expression analysis of mitf gene relating to body color variation in Amphilophus citrinellus. Prog. Fish. Sci. 2021, 42, 107–118. [Google Scholar]
- Ahi, E.P.; Lecaudey, L.A.; Ziegelbecker, A.; Steiner, O.; Glabonjat, R.; Goessler, W.; Hois, V.; Wagner, C.; Lass, A.; Sefc, K.M. Comparative transcriptomics reveals candidate carotenoid color genes in an East African cichlid fish. BMC Genom. 2020, 21, 54. [Google Scholar] [CrossRef]
- Jiang, Y.; Song, H.; Liu, Y.; Wei, M.; Wang, X.; Hu, Y.; Luo, J. Cloning and expression analysis of the developing sequence and tissue expression of TYR gene in Amphilophus citrinellus. J. Agric. Biotechnol. 2016, 24, 697–707. [Google Scholar]
- Andrade, P.; Pinho, C.; Pérez I De Lanuza, G.; Afonso, S.; Brejcha, J.; Rubin, C.; Wallerman, O.; Pereira, P.; Sabatino, S.J.; Bellati, A.; et al. Regulatory changes in pterin and carotenoid genes underlie balanced color polymorphisms in the wall lizard. Proc. Natl. Acad. Sci. USA 2019, 116, 5633–5642. [Google Scholar] [CrossRef] [PubMed]
- Inaba, M.; Yamanaka, H.; Kondo, S. Pigment pattern formation by contact-dependent depolarization. Science 2012, 335, 677. [Google Scholar] [CrossRef] [PubMed]
- Volkening, A.; Sandstede, B. Iridophores as a source of robustness in zebrafish stripes and variability in Danio patterns. NAT Commun. 2018, 9, 3231. [Google Scholar] [CrossRef]
- Huang, Y.; Luo, Y.; Liu, J.; Gui, S.; Wang, M.; Liu, W.; Peng, L.; Xiao, Y. A light-colored region of caudal fin: A niche of melanocyte progenitors in crucian carp (Cyprinus carpio L.). Cell Biol. Int. 2017, 41, 42–50. [Google Scholar] [CrossRef]
- Cal, L.; Suarez-Bregua, P.; Cerdá-Reverter, J.M.; Braasch, I.; Rotllant, J. Fish pigmentation and the melanocortin system. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2017, 211, 26–33. [Google Scholar] [CrossRef]
- Kautt, A.F.; Kratochwil, C.F.; Nater, A.; Machado-Schiaffino, G.; Olave, M.; Henning, F.; Torres-Dowdall, J.; Härer, A.; Hulsey, C.D.; Franchini, P.; et al. Contrasting signatures of genomic divergence during sympatric speciation. Nature 2020, 588, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Ma, B.; Guo, X.; Wang, H.; Ma, A.; Sun, Z.; Wang, Q. Comparative transcriptome analysis of the molecular mechanism underlying the golden red colour in mutant Taiwanese loach. Aquaculture 2021, 543, 736979. [Google Scholar] [CrossRef]
- Chen, Y.; Gong, Q.; Lai, J.; Song, M.; Liu, Y.; Wu, Y.; Ai, J.; Long, Z. Transcriptome analysis identifies candidate genes associated with skin color variation in Triplophysa siluroides. Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 35, 100682. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Janpoom, S.; Prasertlux, S.; Rongmung, P.; Ratdee, O.; Zhang, W.; Khamnamtong, B.; Klinbunga, S. Transcriptome comparison for identification of pigmentation-related genes in different color varieties of Siamese fighting fish Betta splendens. Comp. Biochem. Physiol. Part. D Genom. Proteom. 2022, 43, 101014. [Google Scholar] [CrossRef]
- Rani, V.; Sengar, R.S. Biogenesis and mechanisms of microRNA-mediated gene regulation. Biotechnol. Bioeng. 2022, 3, 119. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Huang, Y.; Xie, J.; Wang, G.; Yu, D.; Sun, X. Genome-wide identification and characterization of conserved and novel microRNAs in grass carp (Ctenopharyngodon idella) by deep sequencing. Comput. Biol. Chem. 2017, 68, 92–100. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Zhang, A.; Chu, P.; Li, Y.; Huang, R.; Liao, L.; Zhu, Z.; Wang, Y. Deep Illumina sequencing reveals conserved and novel microRNAs in grass carp in response to grass carp reovirus infection. BMC Genom. 2017, 18, 195. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Huang, Y.; Xie, J.; Wang, G.; Yu, D.; Sun, X.; Zhang, K.; Li, Z.; Ermeng, Y.; Tian, J.; et al. Identification and expression analysis of miRNA in hybrid snakehead by deep sequencing approach and their targets prediction. Genomics 2019, 111, 1315–1324. [Google Scholar] [CrossRef] [PubMed]
- Franchini, P.; Xiong, P.; Fruciano, C.; Meyer, A. The role of microRNAs in the repeated parallel diversification of lineages of Midas cichlid fish from Nicaragua. Genome Biol. Evol. 2016, 8, 1543–1555. [Google Scholar] [CrossRef]
- Franchini, P.; Xiong, P.; Fruciano, C.; Schneider, R.F.; Woltering, J.M.; Hulsey, C.D.; Meyer, A. MicroRNA gene regulation in extremely young and parallel adaptive radiations of crater lake cichlid fish. Mol. Biol. Evol. 2019, 36, 2498–2511. [Google Scholar] [CrossRef]
- Wu, S.; Huang, J.; Li, Y.; Zhao, L. Involvement of miR-495 in the skin pigmentation of rainbow trout (Oncorhynchus mykiss) through the regulation of mc1r. Int. J. Biol. Macromol. 2024, 254, 127638. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Song, F.; Yin, H.; Zhu, W.; Fu, J.; Dong, Z.; Xu, P. Comparative microRNAs expression profiles analysis during embryonic development of common carp, Cyprinus carpio. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 37, 100754. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zeng, D.; Xiong, G.; Zhou, X.; Wang, X. Integrated analysis of mRNA-seq and microRNA-seq depicts the potential roles of miRNA-mRNA networks in pigmentation of Chinese soft-shelled turtle (Pelodiscus sinensis). Aquac. Rep. 2021, 20, 100686. [Google Scholar] [CrossRef]
- Guo, Y.; Wu, W.; Yang, X. Coordinated microRNA/mRNA Expression Profiles Reveal Unique Skin Color Regulatory Mechanisms in Chinese Giant Salamander (Andrias davidianus). Animals 2023, 13, 1181. [Google Scholar] [CrossRef] [PubMed]
- Botchkareva, N.V. MicroRNA/mRNA regulatory networks in the control of skin development and regeneration. Cell Cycle 2012, 11, 468–474. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.; Zhu, Z.; Yang, S.; Ji, K.; Hu, S.; Liu, X.; Yao, J.; Fan, R.; Dong, C. Role of microRNA508-3p in melanogenesis by targeting microphthalmia transcription factor in melanocytes of alpaca. Animal 2017, 11, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Luo, M.; Wang, L.; Yin, H.; Zhu, W.; Fu, J. MicroRNA-206 Regulation of Skin Pigmentation in Koi Carp (Cyprinus carpio L.). Front. Genet. 2020, 11, 47. [Google Scholar] [CrossRef]
- Jiang, S.; Yu, X.; Dong, C. MiR-137 affects melanin synthesis in mouse melanocyte by repressing the expression of c-Kit and Tyrp2 in SCF/c-Kit signaling pathway. J. Agric. Chem. Soc. Jpn. 2016, 80, 2115–2121. [Google Scholar]
- Zhu, Z.; Cai, Y.; Li, Y.; Li, H.; Zhang, L.; Xu, D.; Yu, X.; Li, P.; Lv, L. miR-148a-3p inhibits alpaca melanocyte pigmentation by targeting MITF. Small Rumin. Res. 2019, 177, 44–49. [Google Scholar] [CrossRef]
- Wei, M.; Song, H.; Qi, B.; Liu, C.; Luo, J.; Hu, Y. Pigment cells development and body color variation of postembryonic development in Amphilophus citrinellus (Günther 1864). J. Shanghai Ocean. Univ. 2015, 24, 28–35. [Google Scholar]
- Forsman, A.; Polic, D.; Sunde, J.; Betzholtz, P.E.; Franzén, M. Variable colour patterns indicate multidimensional, intraspecific trait variation and ecological generalization in moths. Ecography 2020, 43, 823–833. [Google Scholar] [CrossRef]
- Henning, F.; Jones, J.C.; Franchini, P.; Meyer, A. Transcriptomics of morphological color change in polychromatic Midas cichlids. BMC Genom. 2013, 14, 171. [Google Scholar] [CrossRef] [PubMed]
- Kratochwil, C.F.; Kautt, A.F.; Nater, A.; Härer, A.; Liang, Y.; Henning, F.; Meyer, A. An intronic transposon insertion associates with a trans-species color polymorphism in Midas cichlid fishes. Nat. Commun. 2022, 13, 296. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Mu, X.; Song, H.; Liu, Y.; Yang, Y.; Liu, C. Characterization and functional analysis of pax3 in body color transition of polychromatic Midas cichlids (Amphilophus citrinellus). Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 2023, 263, 110779. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Liu, G.; Chen, Z.; Ren, Q.; Yin, H.; Luo, J.; Wang, H. Identification and characterization of microRNAs by deep-sequencing in Hyalomma anatolicum anatolicum (Acari: Ixodidae) ticks. Gene 2015, 564, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Betel, D.; Koppal, A.; Agius, P.; Sander, C.; Leslie, C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010, 11, R90. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Zhu, Y.; Yan, J.; Zhang, L.; Zhu, S.; An, L.; Meng, Q.; Zhang, Z.; Wang, X. Full-length transcriptome sequencing analysis reveals differential skin color regulation in snakeheads fish Channa argus. Aquac. Fish. 2023. [Google Scholar] [CrossRef]
- Tian, X.; Peng, N.; Ma, X.; Wu, L.; Shi, X.; Liu, H.; Song, H.; Wu, Q.; Meng, X.; Li, X. microRNA-430b targets scavenger receptor class B member 1 (scarb1) and inhibits coloration and carotenoid synthesis in koi carp (Cyprinus carpio L.). Aquaculture 2022, 546, 737334. [Google Scholar] [CrossRef]
- Gong, W.; Huang, Y.; Xie, J.; Wang, G.; Yu, D.; Zhang, K.; Li, Z.; Yu, E.; Tian, J.; Zhu, Y. Identification and comparative analysis of the miRNA expression profiles from four tissues of Micropterus salmoides using deep sequencing. Genomics 2018, 110, 414–422. [Google Scholar] [CrossRef]
- Zhang, R. miR-199a-5p Reduces Chondrocyte Hypertrophy and Attenuates Osteoarthritis Progression via the Indian Hedgehog Signal Pathway. J. Clin. Med. 2023, 12, 1313. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Zhang, Z.; Qin, X.; Gao, Y.; Zhao, P.; Liu, J.; Zeng, W. Gambogic Acid Inhibits Melanoma through Regulation of miR-199a-3p/ZEB1 Signalling. Basic Clin. Pharmacol. 2018, 123, 692–703. [Google Scholar] [CrossRef] [PubMed]
- Stein, C.S.; McLendon, J.M.; Witmer, N.H.; Boudreau, R.L. Modulation of miR-181 influences dopaminergic neuronal degeneration in a mouse model of Parkinson’s disease. Mol. Ther. Nucleic Acids 2022, 28, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Deng, Y.N.; Wang, X.; Xia, Z.; He, Y.; Zhang, P.; Syed, S.E.; Li, Q.; Liang, S. miR-26a enhances colorectal cancer cell growth by targeting RREB1 deacetylation to activate AKT-mediated glycolysis. Cancer Lett. 2021, 521, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; O’Carroll, D.; Pasolli, H.A.; Zhang, Z.; Dietrich, F.S.; Tarakhovsky, A.; Fuchs, E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat. Genet. 2006, 38, 356–362. [Google Scholar] [CrossRef]
- Zhu, Z.; He, J.; Jia, X.; Jiang, J.; Bai, R.; Yu, X.; Lv, L.; Fan, R.; He, X.; Geng, J.; et al. MicroRNA-25 functions in regulation of pigmentation by targeting the transcription factor MITF in alpaca (Lama pacos) skin melanocytes. Domest. Anim. Endocrinol. 2010, 38, 200–209. [Google Scholar] [CrossRef]
- Zhang, H.; Fu, Y.; Su, Y.; Shi, Z.; Zhang, J. Identification and expression of HDAC4 targeted by miR-1 and miR-133a during early development in Paralichthys olivaceus. Comp. Biochem. Physiol. Part. B Biochem. Mol. Biol. 2015, 179, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ceinos, R.M.; Guillot, R.; Kelsh, R.N.; Cerdá-Reverter, J.M.; Rotllant, J. Pigment patterns in adult fish result from superimposition of two largely independent pigmentation mechanisms. Pigment Cell Melanoma Res. 2015, 28, 196–209. [Google Scholar] [CrossRef]
- Xu, F.; Zhang, H.; Su, Y.; Kong, J.; Yu, H.; Qian, B. Up-regulation of microRNA-183-3p is a potent prognostic marker for lung adenocarcinoma of female non-smokers. Clin. Transl. Oncol. 2014, 16, 980–985. [Google Scholar] [CrossRef]
- Zhang, C.; Gu, H.; Liu, D.; Tong, F.; Wei, H.; Zhou, D.; Fang, J.; Dai, X.; Tian, H. The circ_FAM53B-miR-183-5p-CCDC6 axis modulates the malignant behaviors of papillary thyroid carcinoma cells. Mol. Cell. Biochem. 2022, 477, 2627–2641. [Google Scholar] [CrossRef]
- Kaken, H.; Wang, S.; Zhao, W. P53 Regulates Osteogenic Differentiation Through miR-153-5p/miR-183-5p-X-Linked IAP (XIAP) Signal in Bone Marrow Mesenchymal Stem Cell (BMSC). J. Biomater. Tissue Eng. 2022, 12, 2427–2431. [Google Scholar] [CrossRef]
- Jie, C.; Lin, G.; Jie, N.; Ping, H.; Kai, H.; Ying-Li, S. MiR-183 Regulates ITGB1P Expression and Promotes Invasion of Endometrial Stromal Cells. Biomed. Res. Int. 2015, 2015, 340218. [Google Scholar]
- Zhu, Y.; Li, Q. Mitf involved in shell pigmentation by activating tyrosinase-mediated melanin synthesis in Pacific oyster (Crassostrea gigas). Gene 2024, 897, 148086. [Google Scholar] [CrossRef]
- Yu, F.; Qu, B.; Lin, D.; Deng, Y.; Huang, R.; Zhong, Z. Pax3 Gene Regulated Melanin Synthesis by Tyrosinase Pathway in Pteria penguin. Int. J. Mol. Sci. 2018, 19, 3700. [Google Scholar] [CrossRef]
- Kang, D.; Kim, H. Functional relation of agouti signaling proteins (ASIPs) to pigmentation and color change in the starry flounder, Platichthys stellatus. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2024, 291, 111524. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Song, H.; Pan, X.; Liu, Y.; Jiang, Y.; Yang, Y.; Mou, X.; Liu, C.; Hu, Y.; Zhou, S. Expression analysis of mc1r gene relating to body color variation in Amphilophus citrinellus. Chin. J. Zool. 2019, 54, 45–56. [Google Scholar]
- Motohiro, M.; Hiroyuki, T.; Akiko, S.; Tetsuaki, K.; Ikuko, W.; Hikaru, K.; Yusuke, N.; Kiyoshi, N.; Shin-Ichi, H.; Takashi, S. A gene regulatory network combining Pax3/7, Sox10 and Mitf generates diverse pigment cell types in medaka and zebrafish. Development 2023, 19, dev202114. [Google Scholar]
- Yin, H.; Luo, M.; Luo, W.; Wang, L.; Zhu, W.; Fu, J.; Dong, Z. miR-196a regulates the skin pigmentation of koi carp (Cyprinus carpio L.) by targeting transcription factormitfa. Aquac. Res. 2021, 52, 229–236. [Google Scholar] [CrossRef]
- Yan, B.; Liu, B.; Zhu, C.D.; Li, K.L.; Yue, L.J.; Zhao, J.L.; Gong, X.L.; Wang, C.H. microRNA regulation of skin pigmentation in fish. J. Cell Sci. 2013, 126, 3401–3408. [Google Scholar]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef]
- Suzuki, N.; Mutai, H.; Miya, F.; Tsunoda, T.; Terashima, H.; Morimoto, N.; Matsunaga, T. A case report of reversible generalized seizures in a patient with Waardenburg syndrome associated with a novel nonsense mutation in the penultimate exon of SOX10. BMC Pediatr. 2018, 18, 171. [Google Scholar] [CrossRef] [PubMed]

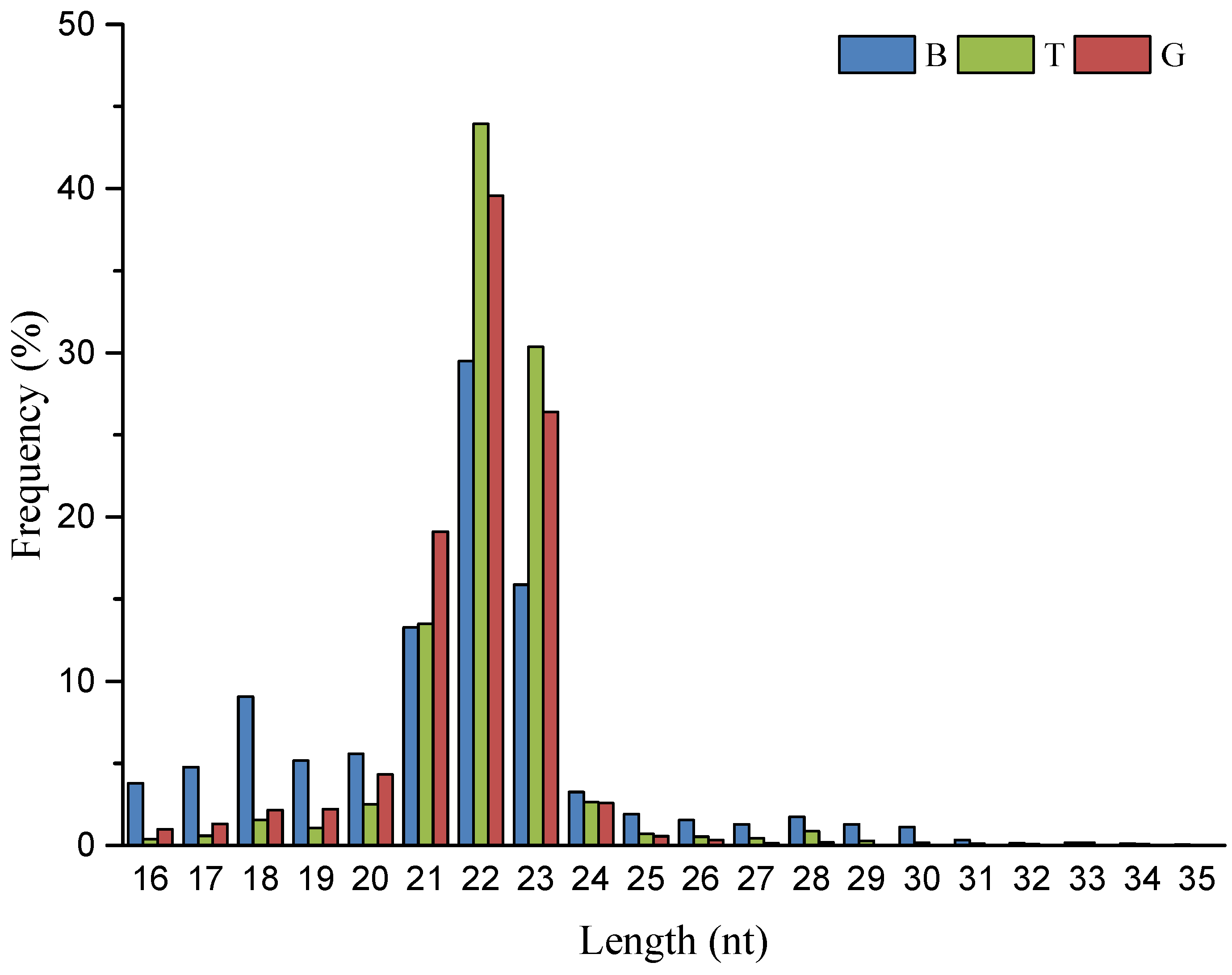
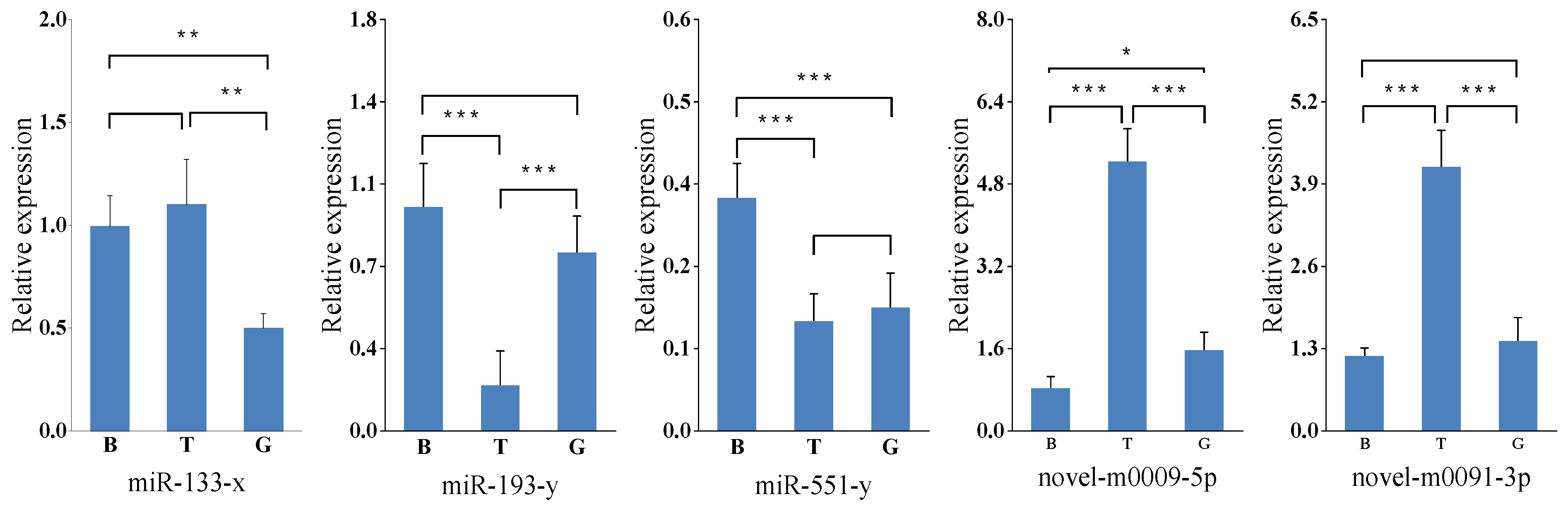



| miRNAs ID | Primer | Sequence (5′-3′) |
|---|---|---|
| miRNA RT-primer | RT | TTACCTAGCGTATCGTTGACAGCTTTTTTTTTTTTTTTTTTTTVN |
| miR-4492-y | Forward | CGGGGCTGGGCTCGCGCC |
| miR-183-x | Forward | TATGGCACTGGTAGAATTCACT |
| miR-451-x | Forward | AAACCGTTACCATTACTGAGTTT |
| miR-1335-y | Forward | CGGCTGAGGTGGGATCCC |
| miR-6937-x | Forward | TGGCTCTAAGGGCTGGGTC |
| miR-133-x | Forward | AGCTGGTAAAATGGAACCAAAT |
| novel-m0009-5p | Forward | GTGAATCCTTGGACCCATTGTCA |
| miR-551-y | Forward | GCGACCCATCCTTTGTTTCTG |
| miR-193-y | Forward | AACTGGCCTACAAAGTCCCAGT |
| novel-m0091-3p | Forward | GTTCAAATCCGGATGCCCCCT |
| U6 | Forward | CTCGCTTCGGCAGCACA |
| Universal Primer | Reverse | TTACCTAGCGTATCGTTGAC |
| miRNA | B | miRNA | T | miRNA | G |
|---|---|---|---|---|---|
| miR-26-x | 159,339 | miR-199-x | 130,621 | miR-199-x | 120,493 |
| miR-199-x | 127,301 | miR-26-x | 121,902 | miR-100-x | 85,094 |
| miR-181-x | 94,227 | miR-181-x | 83,262 | miR-26-x | 79,633 |
| miR-205-x | 82,376 | miR-21-x | 74,605 | miR-181-x | 61,932 |
| miR-22-y | 47,845 | miR-10-x | 68,839 | let-7-x | 57,923 |
| let-7-x | 46,787 | miR-100-x | 64,030 | miR-205-x | 55,605 |
| miR-21-x | 45,663 | miR205-x | 62,212 | miR-21-x | 55,018 |
| miR-200-y | 38,329 | let-7-x | 46,821 | miR-146-x | 49,708 |
| miR-203-y | 32,515 | miR-146-x | 43,975 | miR-10-x | 44,171 |
| miR-199-y | 30,214 | miR-22-y | 34,915 | miR-200-y | 39,404 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, G.; Mu, X.; Liu, Y.; Liu, C.; Wang, X.; Yang, Y.; Song, H. Identification and Characterization of microRNAs in Morphological Color Change of Polychromatic Midas Cichlids (Amphilophus citrinellus). Fishes 2024, 9, 194. https://doi.org/10.3390/fishes9060194
Wu G, Mu X, Liu Y, Liu C, Wang X, Yang Y, Song H. Identification and Characterization of microRNAs in Morphological Color Change of Polychromatic Midas Cichlids (Amphilophus citrinellus). Fishes. 2024; 9(6):194. https://doi.org/10.3390/fishes9060194
Chicago/Turabian StyleWu, Guoqiang, Xidong Mu, Yi Liu, Chao Liu, Xuejie Wang, Yexin Yang, and Hongmei Song. 2024. "Identification and Characterization of microRNAs in Morphological Color Change of Polychromatic Midas Cichlids (Amphilophus citrinellus)" Fishes 9, no. 6: 194. https://doi.org/10.3390/fishes9060194





