Genetic Characterization of Endangered Sterlet (Acipenser ruthenus, Linnaeus 1758) Gene Bank Broodstock, Natural and Cultured Populations in Hungary
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling and DNA Isolation
2.2. PCR and Microsatellite Analysis
2.3. Statistical Analysis
3. Results
3.1. Microsatellite Analysis
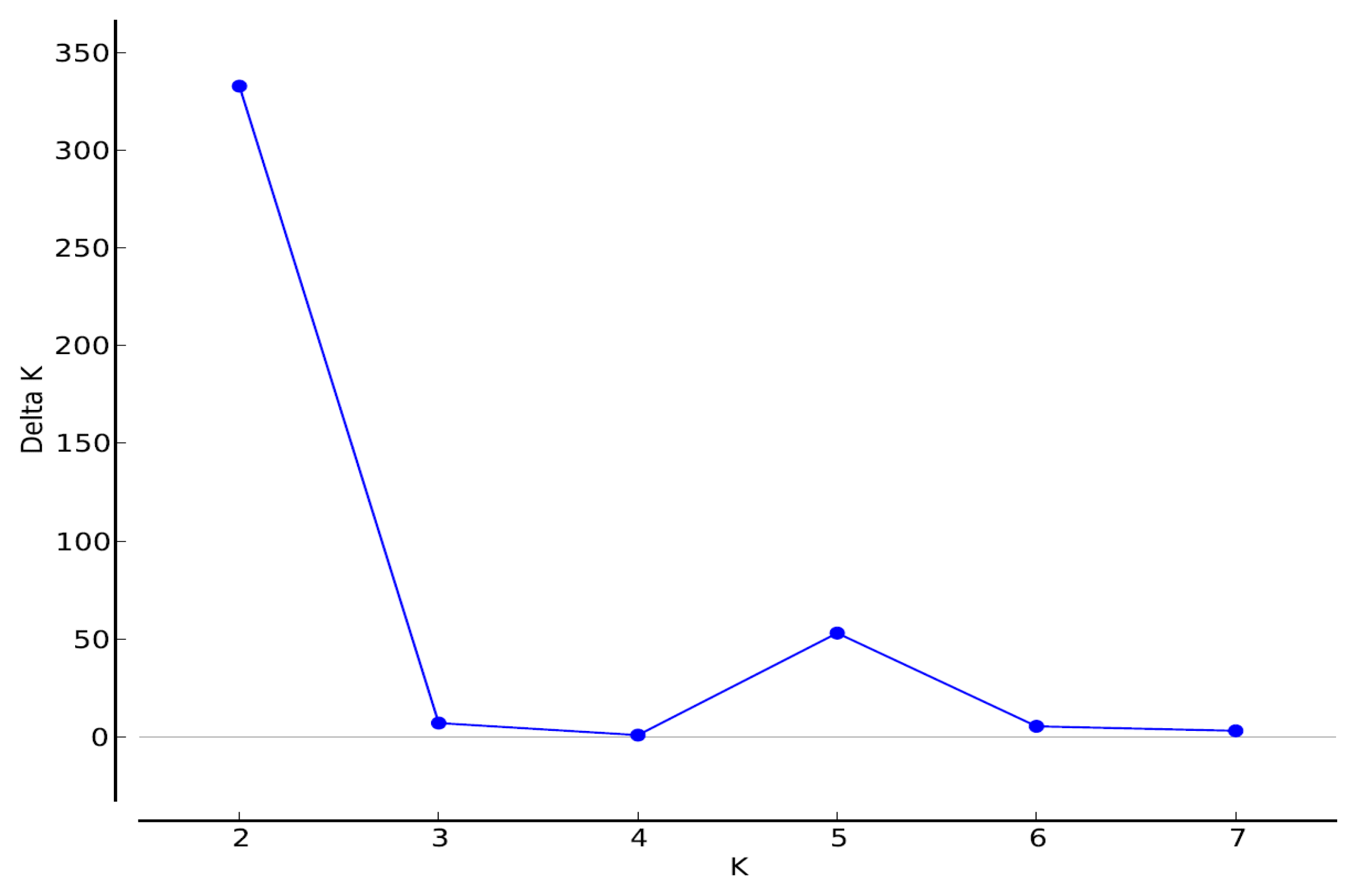
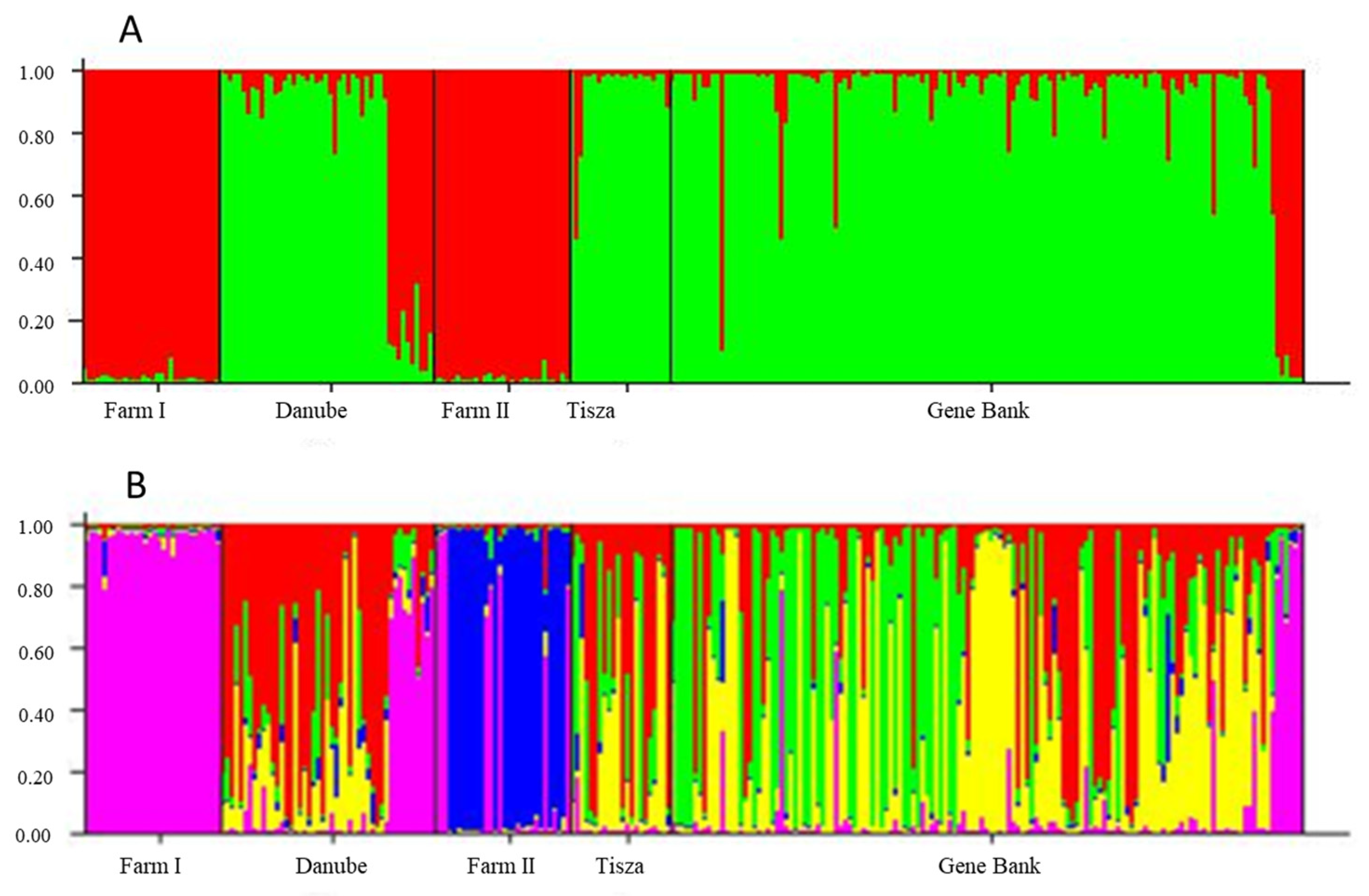
3.2. Structure Results
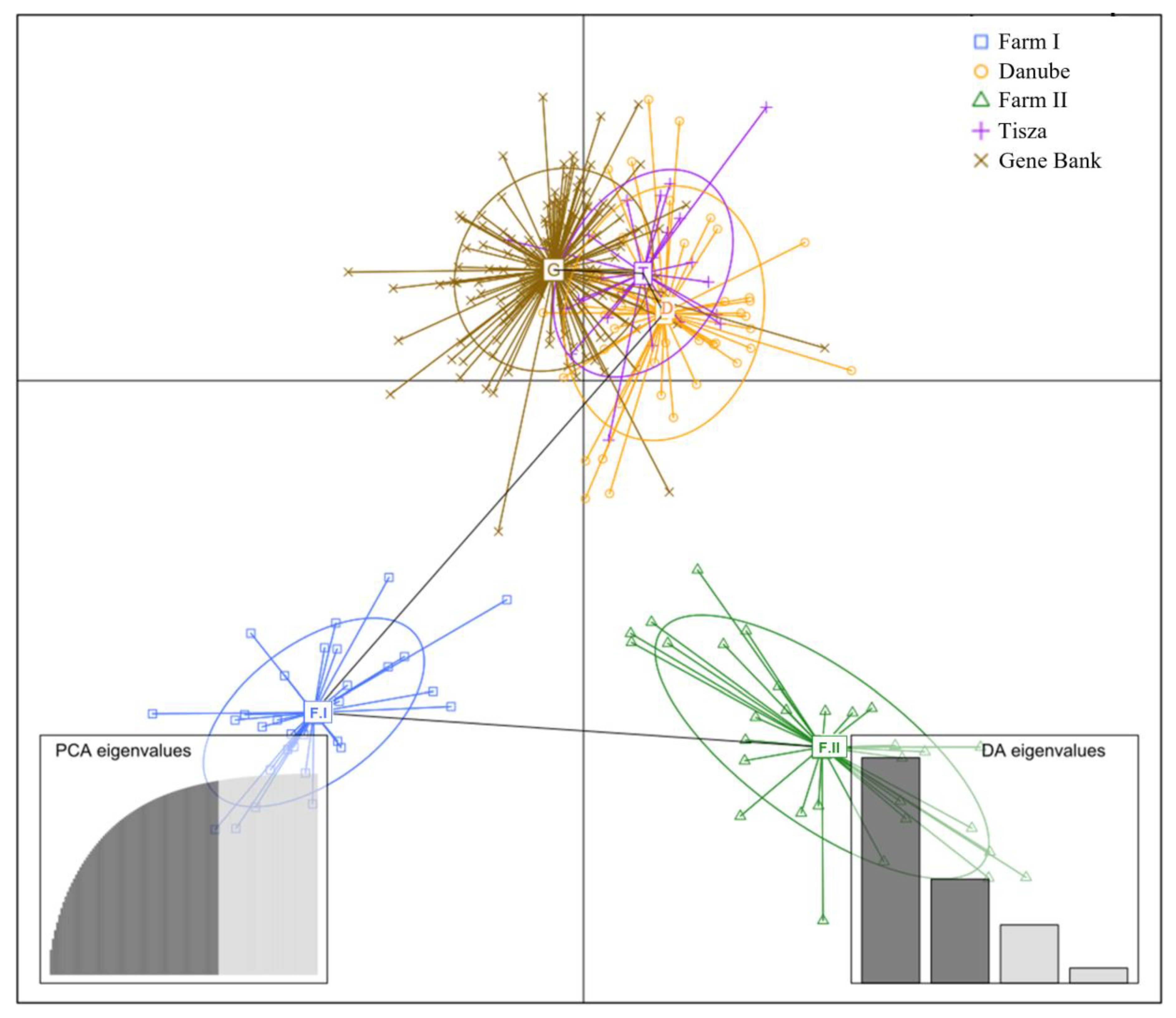
3.3. Principal Component Analysis (PCA) Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reinartz, R.; Lippold, S.; Lieckfeldt, D.; Ludwig, A. Population Genetic Analyses of Acipenser ruthenus as a Prerequisite for the Conservation of the Uppermost Danube Population. J. Appl. Ichtyol. 2011, 27, 477–483. [Google Scholar] [CrossRef]
- Guti, G. Ecological Impacts of the Gabčikovo River Barrage System on Fish in the Szigetköz Floodplain in Hungary. Int. Ver. Für Theor. Und Angew. Limnol. Verhandlungen 1998, 26, 2251–2254. [Google Scholar] [CrossRef]
- Guti, G. Past and Present Status of Sturgeons in Hungary and Problems Involving their Conservation. Riv. Syst. 2008, 18, 61–79. [Google Scholar] [CrossRef]
- Rónyai, A. Overview of Sturgeons Related Activities in HAKI. Presented at Visit of WWF Delegation to HAKI, Szarvas, Hungary. 2012; Unpublished Results. [Google Scholar] [CrossRef]
- Gessner, J.; Freyhof, J.; Kottelat, M.; Friedrich, T. Acipenser ruthenus. The IUCN Red List of Threatened Species. 2022. Available online: https://www.iucnredlist.org/species/227/135062526 (accessed on 9 April 2024).
- Guti, G.; Gaebele, T. Long-Term Changes of Sterlet (Acipenser ruthenus) Population in the Hungarian Section of the Danube. Opusc. Zool. Bp. 2009, 40, 17–25. [Google Scholar]
- Bemis, W.E.; Kynard, B. Sturgeon Rivers: An Introduction to Acipenseriform Biogeography and Life History. Environ. Biol. Fishes 1997, 48, 167–183. [Google Scholar] [CrossRef]
- Friedrich, T.; Reinartz, R.; Gessner, J. Sturgeon Re-Introduction in the Upper and Middle Danube River Basin. J. Appl. Ichtyol. 2019, 35, 1059–1068. [Google Scholar] [CrossRef]
- Fazekas, G.; Kovács, G.; Bogár, K.; Kovács, B. Genetic Analysis of Sterlet (Acipenser ruthenus) Related to Restocking Program in Hungary. Res. J. Agricult. Sci. 2018, 50, 166–170. [Google Scholar]
- Neuburg, J.; Friedrich, T. First Description of a Remnant Population of Sterlet (Acipenser ruthenus, LINNAEUS 1758) in the Eastern Austrian Danube. Nat. Conserv. 2023, 75, 126473. [Google Scholar] [CrossRef]
- Mikheev, P.B.; Kazarinov, S.N.; Melnikova, A.G.; Ponosov, S.V.; Petrenko, N.G.; Nikiforov, A.I.; Puzik, A.Y.; Elchenkova, O.N. Artificial Enhancement of Sturgeon Stock in Freshwater Reservoirs: A Case Study on Sterlet Acipenser ruthenus of the Kama Reservoir. Aquac. Fish. 2024, 9, 287–294. [Google Scholar] [CrossRef]
- Strat, D.; Gheorghe, I.F. Conservation Status and Effectiveness of the National and International Policies for the Protection and Conservation of Sturgeons in the Danube River and Black Sea Basin. Diversity 2023, 15, 568. [Google Scholar] [CrossRef]
- Holčík, J.; Klindová, A.; Masár, J.; Mészáros, J. Sturgeons in the Slovakian Rivers of the Danube River Basin: An Overview of Their Current Status and Proposal for Their Conservation and Restoration. J. Appl. Ichtyol. 2006, 22, 17–22. [Google Scholar] [CrossRef]
- Pekárik, L.; Ĝ Iamporová-Zať Ovičová, Z.; Arendt, D.; Ĝ Iampor, F. Current Stocking Program of the Sterlet (Acipenser ruthenus, L.) Can Negatively Shape Its Genetic Variability in the Middle Danube. Knowl. Manag. Aquat. Ecosyst. 2019, 420, 19. [Google Scholar] [CrossRef]
- Ljubobratović, U.; Bogár, K.; Káldy, J.; Fazekas, G.; Vass, N.; Feledi, T.; Kovács, G. Optimizing the Gonadoliberin Dosage and Evaluating the Egg Quality in the Preseason and Seasonal Artificial Reproduction of Pond-reared Sterlet Acipenser ruthenus. Anim. Reprod. Sci. 2022, 247, 107097. [Google Scholar] [CrossRef] [PubMed]
- Kovács, B.; Kovács, G.; Fazekas, G. A Tokfajok Genetikai Háttere és Erőforrásai In A Tokalakúak Biológiája és Tenyésztése; Urbányi, B., Horváth, Á., Eds.; Vármédia Print kft: Gödöllő, Hungary, 2019; pp. 77–107. [Google Scholar]
- Kohlmann, K.; Kersten, P.; Gessner, J.; Onara, D.; Taflan, E.; Suciu, R. New Microsatellite Multiplex PCR Sets for Genetic Studies of the Sterlet Sturgeon, Acipenser ruthenus. Env. Biotech. 2017, 13, 11–17. [Google Scholar] [CrossRef]
- McQuown, E.C.; Sloss, B.L.; Sheehan, R.J.; Rodzen, J.; Tranah, G.J.; May, B. Microsatellite Analysis of Genetic Variation in Sturgeon: New Primer Sequences for Scaphirhynchus and Acipenser. Trans. Am. Fish. Soc. 2000, 129, 1380–1388. [Google Scholar] [CrossRef]
- Welsh, A.B.; Blumberg, M.; May, B. Identification of Microsatellite Loci in Lake Sturgeon, Acipenser fulvescens, and Their Variability in Green Sturgeon. A. medirostris. Molec. Ecol. Notes 2003, 3, 47–55. [Google Scholar] [CrossRef]
- May, B.; Krueger, C.C.; Kincaid, H.L. Genetic Variation at Microsatellite Loci in Sturgeon: Primer Sequence Homology in Acipenser and Scaphirhynchus. Can. J. Fish. Aquat. Sci. 1997, 54, 1542–1547. [Google Scholar] [CrossRef]
- Zane, L.; Patarnello, T.; Ludwig, A.; Fontana, F.; Congiu, L. Isolation and Characterization of Microsatellites in the Adriatic Sturgeon (Acipenser naccarii). Molec. Ecol. Not. 2002, 2, 586–588. [Google Scholar] [CrossRef]
- Henderson-Arzapalo, A.; King, T.L. Novel Microsatellite Markers for Atlantic sturgeon (Acipenser oxyrinchus) Population Delineation and Broodstock Management. Molec. Ecol. Notes 2002, 2, 437–439. [Google Scholar] [CrossRef]
- Shimizu, M.; Kosaka, N.; Shimada, T.; Nagahata, T.; Iwasaki, H.; Nagai, H.; Shiba, T.; Emi, M. Universal Fluorescent Labeling (UFL) Method for Automated Microsatellite Analysis. DNA Res. 2002, 9, 173–178. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenALEx 6.5: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research-an Update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Park, S.D.E. The Excel Microsatellite Toolkit. Trypanotolerance in West African Cattle and the Population Genetic Effects of Selection. Ph.D. Thesis, University of Dublin, Dublin, Ireland, 2001. [Google Scholar]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. MICRO-CHECKER: Software for Identifying and Correcting Genotyping Errors in Microsatellite Data. Molec. Ecol. Notes. 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Belkhir, K.P.; Borsa, P.; Chikhi, L.; Raufaste, N.; Bonhomme, F. 1996–2004 GENETIX, Version 4.05. Available online: http://kimura.univ-montp2.fr/genetix/ (accessed on 9 April 2024).
- Piry, S.; Luikart, G.; Cornuet, J.-M. BOTTLENECK: A Computer Program for Detecting Recent Reductions in the Effective Size Using Allele Frequency Data. J. Heredity 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.J.; Ovenden, J.R. NeEstimator v2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Res. 2014, 14, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software STRUCTURE: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Jombart, T. Adegenet: A R Package for the Multivariate Analysis of Genetic Markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarczyk, D.; Fopp-Bayat, D. Assemblage of Spawning Pairs Based on Their Individual Genetic Profiles—As Tool for Maintaining Genetic Variation within Sturgeon Populations. Aquac. Res. 2013, 44, 677. [Google Scholar] [CrossRef]
- Fopp-Bayat, D.; Kuzniar, P.; Kolman, R.; Liszewski, T.; Kucinski, M. Genetic Analysis of Six Sterlet Populations—Recommendations for The Plan of Restitution in the Dniester River. Iran.S J. Fish Sci. 2015, 14, 634–645. [Google Scholar]
- Østbye, K.; Næsje, T.F.; Bernatchez, L.; Sandlund, O.T.; Hindar, K. Morphological Divergence and Origin of Sympatric Populations of European Whitefish (Coregonus lavaretus L.) in Lake Femund, Norway. J Evol Biol. 2005, 18, 683–702. [Google Scholar] [CrossRef] [PubMed]
| PCR Duplex Set | Annealing Temperature | Locus | Dye | Primer Sequence 5′-3′ | Allele Size Range (bp) | Number of Alleles | PIC Value |
|---|---|---|---|---|---|---|---|
| 1. | 57 °C | AfuG 41 | NED | TGACGCACAGTAGTATTATTTATG | 208–274 | 24 | 0.92 |
| TGATGTTTGCTGAGGCTTTTC | |||||||
| An20 | AATAACAATCATTACATGAGGCT | 162–194 | 15 | 0.88 | |||
| TGGTCAGTTGTTTTTTTATTGAT | |||||||
| 2. | 57 °C | AfuG 51 | VIC | ATAATAATGAGCGTGCTTTCTGTT | 247–259 | 4 | 0.12 |
| ATTCCGCTTGCGACTTATTTA | |||||||
| Aru12 | AAATAGCATGTTCCCCAGCA | 187–194 | 3 | 0.40 | |||
| TCCATTGCACTTTTCCTTCTTT | |||||||
| 3. | 57 °C | Spl-163 | PET | TGCTTGTAAACTGCCCCACT | 206–242 | 17 | 0.83 |
| CCACATGCAGTTTGAGCTGC | |||||||
| Aru26 | AAAGCAACAACTCCACCAGG | 156–188 | 8 | 0.48 | |||
| TGCCTTGTCTACTGTCCGAA | |||||||
| 4. | 60 °C | LS-68 | NED | TTATTGCATGGTGTAGCTAAAC | 177–245 | 17 | 0.91 |
| AGCCCAACACAGACAATATC | |||||||
| Aru13 | TCCACTTTATTCCGTTGTGG | 90–150 | 15 | 0.79 | |||
| AGACCGGAATCAAACCCAG | |||||||
| 5. | 60 °C | AoxD161 | VIC | GTTTGAAATGATTGAGAAAATGC | 122–162 | 11 | 0.70 |
| TGAGACAGACACTCTAGTTAAACAGC | |||||||
| Aru19 | GCGTGGTGTAAGTGAACCCT | 206–208 | 2 | 0.21 | |||
| CTTCAATTGTGCTTGGCTCA | |||||||
| 6. | 60 °C | Aru50 | PET | TGGAAACCAAATTAATTCACAAAA | 116–142 | 6 | 0.47 |
| TGGGATCCTCTGTAGAACAGTCT | |||||||
| Aru18 | CCTGGAACACGTCCAGTTTT | 158–164 | 4 | 0.15 | |||
| TGGGTGAATGTTTTGGTGTG |
| Population | Na | Ne | Ho | He | Fis |
|---|---|---|---|---|---|
| Gene Bank | 9.08 | 4.473 | 0.547 | 0.557 | 0.015 |
| Danube | 8.00 | 4.686 | 0.539 | 0.588 | 0.090 |
| Tisza | 5.83 | 3.393 | 0.595 | 0.517 | −0.163 |
| Farm I | 5.33 | 3.628 | 0.465 | 0.568 | 0.127 |
| Farm II | 5.83 | 3.028 | 0.494 | 0.531 | 0.005 |
| Populations | Number of Individuals | Estimated Ne | Parametric 95% Confidence Interval for Ne |
|---|---|---|---|
| Gene Bank | 138 | 107.5 | 91.7–109.7 |
| Danube | 47 | 68.7 | 50.9–100.9 |
| Tisza | 22 | Infinite | 108–Infinite |
| Farm I | 30 | 38 | 23.7–76.3 |
| Farm II | 30 | 21.5 | 15.5–30 |
| Gene Bank | Danube | Tisza | Farm I | Farm II | |
|---|---|---|---|---|---|
| Gene Bank | 0.000 | ||||
| Danube | 0.016 | 0.000 | |||
| Tisza | 0.016 | 0.017 | 0.000 | ||
| Farm I | 0.067 | 0.066 | 0.091 | 0.000 | |
| Farm II | 0.099 | 0.091 | 0.119 | 0.068 | 0.000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fazekas, G.; Kovács, G.; Sándor, Z.J.; Bogár, K.; Géczi, A.; Kovács, B. Genetic Characterization of Endangered Sterlet (Acipenser ruthenus, Linnaeus 1758) Gene Bank Broodstock, Natural and Cultured Populations in Hungary. Fishes 2024, 9, 201. https://doi.org/10.3390/fishes9060201
Fazekas G, Kovács G, Sándor ZJ, Bogár K, Géczi A, Kovács B. Genetic Characterization of Endangered Sterlet (Acipenser ruthenus, Linnaeus 1758) Gene Bank Broodstock, Natural and Cultured Populations in Hungary. Fishes. 2024; 9(6):201. https://doi.org/10.3390/fishes9060201
Chicago/Turabian StyleFazekas, Gyöngyvér, Gyula Kovács, Zsuzsanna J. Sándor, Katalin Bogár, Aliz Géczi, and Balázs Kovács. 2024. "Genetic Characterization of Endangered Sterlet (Acipenser ruthenus, Linnaeus 1758) Gene Bank Broodstock, Natural and Cultured Populations in Hungary" Fishes 9, no. 6: 201. https://doi.org/10.3390/fishes9060201







