Abstract
The subfamily Rasborinae is a species-rich group of freshwater fish related to zebrafish; however, its taxonomy remains unclear. We present the complete mitogenome and corresponding polyadenylated mitotranscriptome of Rasbora rasbora (Hamilton 1822) based on long-read and high-coverage Oxford Nanopore Technology sequencing. The mitogenome size, gene content, and gene organization correspond to the typical vertebrate composition, and the mitogenome generates 10 polyadenylated mRNAs. Two alternative polyadenylation sites of ND5 mRNA were detected, one with a 596 nt 3′untranslated region corresponding to the antisense ND6 gene. Polyadenylation also generates seven of the mRNA UAA stop codons. Complete mitogenome sequences, excluding the control region, were carefully aligned for RNA-coding and protein-coding features using 54 available species of the subfamily Rasborinae. The phylogenetic analyses based on maximum likelihood, Bayesian inference, and neighbor-joining tree building methods confirm the transfer of R. rasbora into the Sumatrana species group. The overall phylogeny of the subfamily Rasborinae supports with high confidence some previously observed changes within this subfamily, as well as contradicts some conclusions set by previous studies.
Keywords:
long-read sequencing; nanopore; mitochondrial DNA; mitochondrial RNA; molecular phylogeny; polyadenylation; Rasbora Key Contribution:
Complete mitogenome sequence analyses revealed new poly(A) mRNA features, and molecular phylogeny challenged current taxonomy of subfamily Rasborinae.
1. Introduction
The subfamily Rasborinae is a species-rich group of teleosts. According to Eschmeyer’s Catalog of Fishes (California Academy of Sciences), it contains 11 valid genera with 124 valid species [1]. However, the classification within this subfamily remains largely unresolved. Multiple species have been renamed, redescribed, or merged over the years, while some species remain uncertain as valid, cryptic, or unclassified [2]. Therefore, the number of the species in each of the genera, and the genera themselves, remains unclear [3]. Recognizing the relationships within the Rasborinae subfamily has been a matter of interest for decades, from morphological to time-calibrated phylogenetic studies. The classification is, however, complicated due to high species diversity with a large amount of cryptic diversity [3,4,5,6]. Some of the Rasbora species have been assigned into groups, which have later been updated and revised [3,6,7]. Tang and co-workers [3] reconstructed the systematics of this group using mitochondrial Cytb and COI, along with nuclear opsin and recombination activating gene 1 (RAG1). Sholiah and co-workers [4,5] also revisited the relationships within this subfamily using molecular operational taxonomic units (MOTUs), and divided the subfamily Rasborinae into four different clades.
The complete mitochondrial genome (mitogenome; mtDNA) represents an appropriate molecular marker in inferring phylogenetic relationships in metazoans, including teleosts [8,9]. Vertebrate mitogenomes are in general multicopy, inherited maternally, do not recombine, have a stable gene content and organization, and are available in databases from numbers of species, which makes them applicable to phylogenetic assessments [10]. Mitogenome-based phylogenies are well reported in teleosts and include pioneer studies by Nishida and co-workers [9,11,12,13,14,15] as well as within-family studies in, e.g., Gadidae [16,17] and Danionidae [18].
Common methods for obtaining a complete mitogenome sequence are based on targeting of the mitogenome separately from the nuclear genome followed by next-generation sequencing [19,20,21]. Even though short-read sequencing enables deep coverage along with high read accuracy, the final assemblies are prone to error due to the inability to resolve low-complexity regions or repeats [22]. Oxford Nanopore Technology (ONT) enables mitogenome sequencing without any enrichment while maintaining deep coverage. Aside from reduced error rate with the new chemistry [23] and resolving repetitive sequences, an additional advantage of long reads involves avoiding potential contamination from nuclear mitochondrial pseudogenes (NUMTs) and the resulting inaccurate identification of heteroplasmy [24].
The Gangetic scissortail rasbora, Rasbora rasbora, is a small fish species with a silver-colored body with yellow fins accompanied with characteristic black spots on the tip of the caudal fin (Figure S1). NCBI Taxonomy further files R. rasbora under the Rasbora genus; however, the available sequence records for this species include only partial mitochondrial data. R. rasbora and Danio rerio (zebrafish) are distantly related species of the family Danionidae, but classified into different subfamilies. Thus, it is of urgent interest to characterize structural genomic features of R. rasbora that may contribute to elucidating the evolutionary history and sequence divergence among D. rerio strains [18,25,26].
As part of an ongoing and ONT-based whole-genome sequencing project of R. rasbora, we assembled and annotated the mitogenome from two specimens. The sequences were subjected to phylogenetic analyses to determine the taxonomic position of R. rasbora and to improve the classification and genetic resolution of the subfamily Rasborinae. We also determined the R. rasbora polyadenylated (poly(A)) mitotranscriptome and assessed sequence divergence between R. rasbora and D. rerio in RNA-coding genes.
2. Materials and Methods
2.1. Tissue Collection and Nucleic Acid Isolation
A reference specimen tissue sample of D. rerio (AB-NU; AB zebrafish line) was obtained from the zebrafish facility, Nord University. R. rasbora (specimens 1 and 2) were obtained from a commercial pet shop (Bodø, Norway), kept in a private collection, and muscle tissue was donated to the project. The tissue samples (S1-NU; S2-NU) were subsequently stored at the Nord University tissue collection at −80 °C. Approximately 40 mg of muscle tissue from the trunk was used to isolate genomic DNA using the Monarch® HMW DNA Extraction Kit for Tissue (New England BioLabs Inc., Ipswich, MA, USA) following the manufacturer’s instructions. Total RNA was extracted from the same tissue sample by phenol–chloroform extraction following the QIAzol® Handbook protocol (March 2021, Qiagen, Hilden, Germany), with the addition of incubation with isopropanol at −20 °C overnight.
2.2. DNA and RNA Sequencing
The D. rerio reference mitogenome was sequenced by Illumina NextSeq 500 technology (San Diego, CA, USA). The library was prepared using NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® (New England Biolabs, Ipswich, MA, USA) according to protocol. Paired-end (2 × 150 bp) sequencing resulted in 37,042 mapped reads (~317 times coverage). Sequencing R. rasbora using ONT was conducted as part of a whole-genome sequencing (WGS) project. The library was prepared based on Ligation Sequencing Kit V14 (SQK-LSK114; Oxford Nanopore Technologies, Oxford, UK) for the MinION branch of that protocol. Roughly 1 µg input genomic DNA was used for the library preparation. The sequencing was performed on a MinION fitted with R10.4.1 flow cells (FLO-MIN114) according to the manufacturer’s instructions (Oxford Nanopore Technologies, Oxford, UK). The mean sequence coverages of R. rasbora specimens 1 and 2 were 3772 and 1012, respectively, with average read length of 4442 nt and 7582 nt (Figure S2). An RNA-sequencing library was prepared following the Ligation Sequencing Kit V14-Direct cDNA sequencing (SQK-LSK114) involving reverse transcription. The sequencing was performed on a PromethION fitted with a R10.4.1 flow cell (FLO-PRO114M).
2.3. Mitogenome Assembly and Annotation
Dorado (version 0.5) was used for base calling using a duplex base-calling model (DNA) and simplex model (cDNA). Reads were trimmed for adapters (Porechop, version 0.2.4) followed by filtering of the duplex reads using Filtlong (version 0.2.1). Samtools (version 1.17) was used to create a consensus sequence based on comparison to R. paviana (MW232470). Gene annotation was done manually. A more detailed description of mitogenome assembly based on ONT is given in the Supplementary Materials, Note S1.
2.4. Secondary Structure Predictions of rRNAs and tRNAs
The secondary folding patterns and helical numbering of mitochondrial small-subunit ribosomal RNA (mtSSU rRNA; 12S rRNA), mitochondrial large-subunit rRNA (mtLSU rRNA; 16S rRNA), and transfer RNAs (tRNAs) were based on previously reported structures in fish mitochondria [15,27,28,29]. Illustrations in figures were generated in Adobe Illustrator 2020 (https://www.adobe.com (accessed 1 May 2024)).
2.5. Phylogenetic Analysis
A sequence alignment of complete mitogenomes (except control region; CR) that included 15,776 nucleotide positions was generated using the Clustal W alignment method (gap opening penalty 15.00, gap extension penalty 6.66 for both pairwise alignment and multiple alignment). Multiple sequence alignment blocks with gaps across the species or erroneously aligned sites were excluded. MEGA software (version 11) [30] was used for model testing, and the best-fit model was GTR+G+I model. Maximum likelihood (ML) and neighbor-joining (NJ) tree topologies were evaluated by bootstrapping with 2000 replicates. Bayesian inference (BI) of phylogeny was conducted using Mr. Bayes (version 3.2.6) [31] plugin in Geneious Prime® 2024.0.7 software (www.geneious.com (accessed 25 July 2024)). The GTR+I+G model and default settings (1.1 million generations and sampling every 200th generation) were selected.
3. Results
3.1. Mitogenome Sequencing
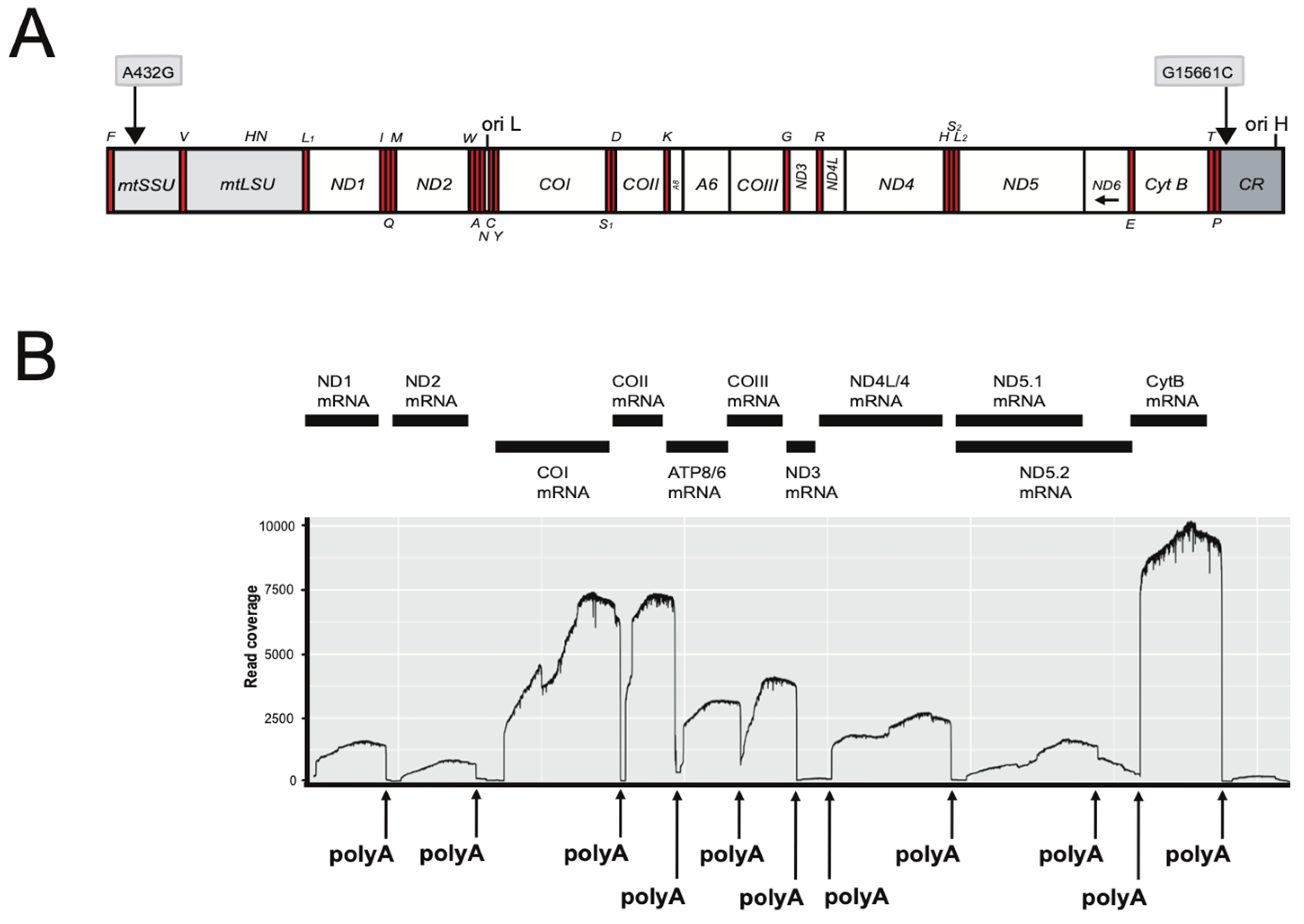
The mitogenomes of D. rerio (AB-NU; 16,596 bp) and R. rasbora (S1-NU; S2-NU; both 16,541 bp) contained the typical vertebrate composition of 22 tRNA genes, 13 protein-coding genes, two rRNA genes and a main control region, CR (see Figure 1A). The heavy strand encodes 28 genes, while the light strand accompanies the other nine genes. The D. rerio mitogenome sequence was highly similar, but not identical, to a previously reported D. rerio reference AB strain (AC024175). We noted 16 polymorphic sites that include eight non-synonymous amino acid changes in derived proteins (Table S1). Two nucleotide differences were found between the two studied R. rasbora individuals (Figure 1A). Nucleotide similarity between R. rasbora and D. rerio was determined to be 80.1%.
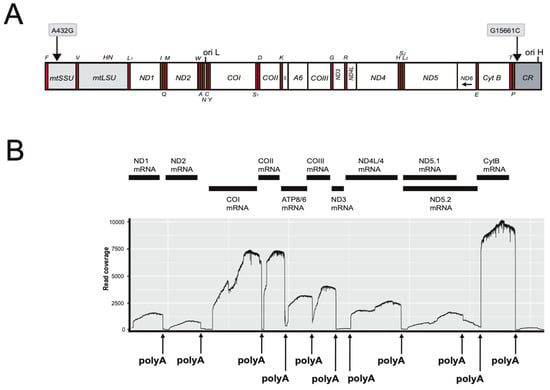
Figure 1.
Mitogenome organization and polyadenylated mRNA mapping of Rasbora rasbora. (A) Linear map presentation of the circular mitogenome. The two SNPs between specimens 1 and 2 are indicated above the gene map. Gene abbreviations: mtSSU and mtLSU, mitochondrial small- (12S) and large- (16S) subunit ribosomal RNA; ND1–6, NADH dehydrogenase subunit 1 to 6 (complex I); Cyt B, cytochrome b (complex III); COI-III, cytochrome c oxidase subunit I to III (complex IV); A6 and A8, ATPase subunit 6 and 8 (complex V). All genes, except ND6 and eight tRNA (indicated below map) are H-strand-specific. Ori H and Ori L, origins of heavy (H) and light (L) strands, respectively. (B) Schematic view of mitochondrial mRNA transcripts and corresponding ONT read mapping onto the mitogenome reference. Polyadenylation (polyA) sites are indicated (see Table 1 for details).
3.2. The Polyadenylated Mitotranscriptome of R. rasbora
RNA mapping identified ten polyadenylated mRNAs (Figure 1B): two bicistronic (ATPase8/ATPase6 and ND4L/ND4) and eight monocistronic transcripts. The ND6 mRNA appeared not to be polyadenylated. Interestingly, poly(A) generates complete UAA stop codons in seven of the mRNAs (Table 1). Two poly(A) sites were mapped downstream of the ND5 mRNA stop codon, which corresponded to a short (5 nt) and a long (596 nt) 3′ untranslated region (UTRs). The COI mRNA has an extended 3′ UTR of 72 nt (Figure 1B; Table 1). Assessment of transcripts indicate that the CytB (Complex III) and CO (Complex IV) subunits were more abundant compared to the ND (Complex I) subunits (Figure 1B). In addition to mRNAs, we also detected a polyA tail on the mtLSU rRNA (Figure S3).

Table 1.
Polyadenylation site features of R. rasbora mitochondrial mRNAs derived from ONT cDNA reads.
3.3. Comparing RNA Coding Genes of R. rasbora and D. rerio
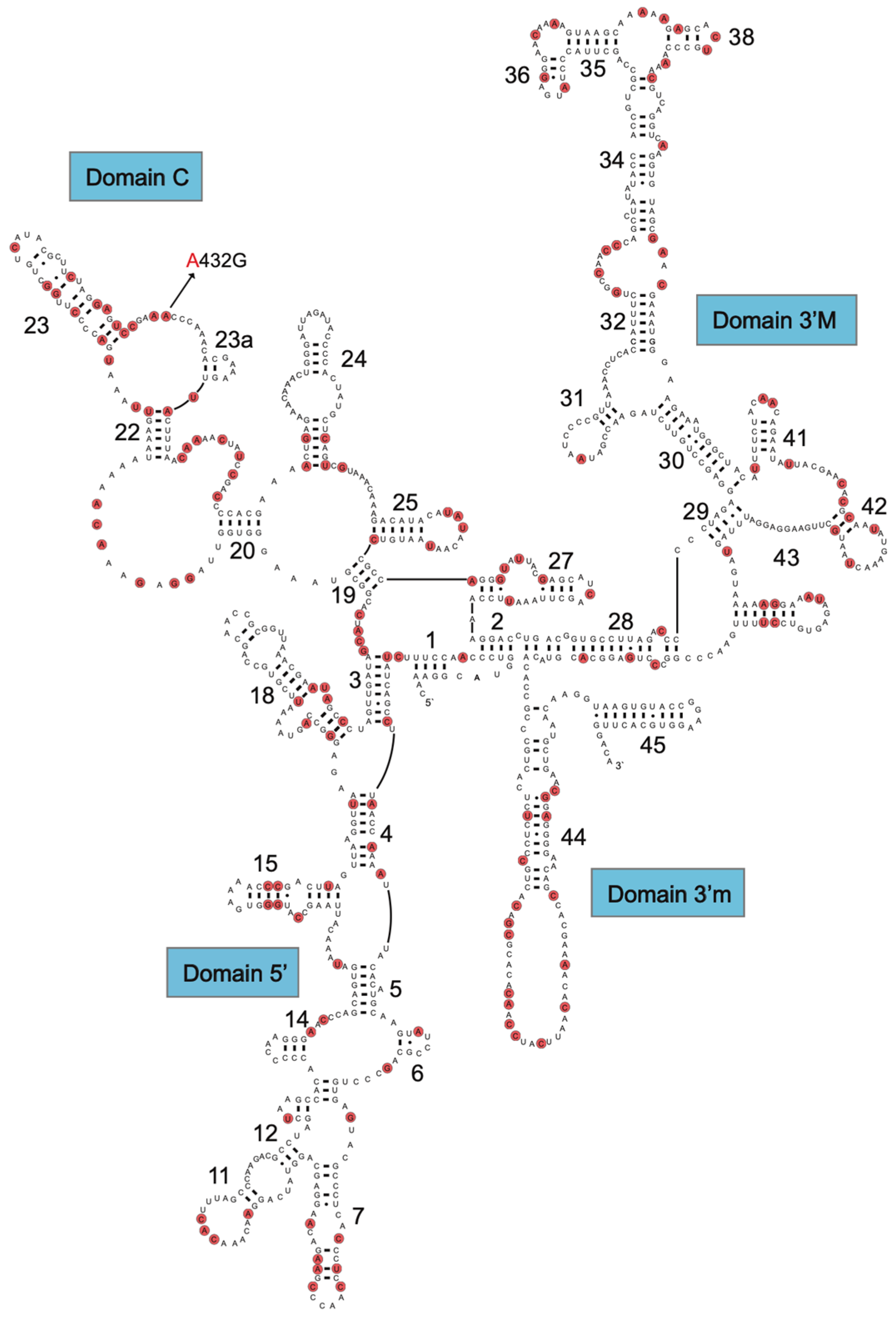
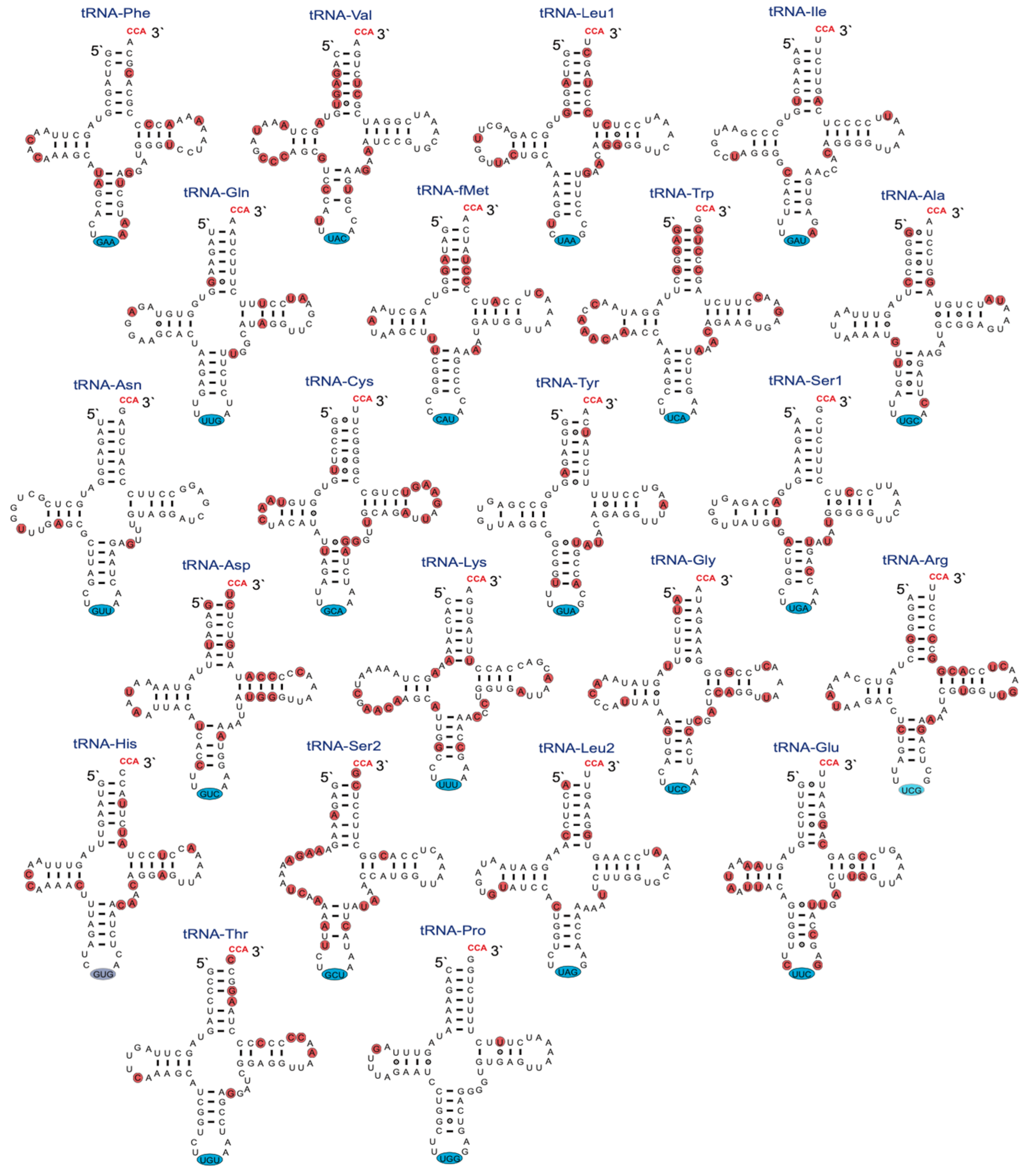
The mitochondrial structural RNA genes are important sources of information for sequence divergence and phylogenetic interpretations. Knowledge about rRNA structure folding appears essential for obtaining optimal sequence alignments, and tRNA folding appears important for correct mitogenome gene annotation. The secondary structure of the 952 nt R. rasbora mtSSU rRNA is presented in Figure 2, and variable positions compared to the corresponding D. rerio rRNA are indicated. Interestingly, despite highly conserved secondary structural features, 147 variable sites (15.4%) were noted between R. rasbora and D. rerio. Similar results were seen when comparing the mtLSU rRNA features. Here, 274 variable sites were noted between R. rasbora and D. rerio, which corresponded to a sequence divergence of 16.3% (Figure S3). The tRNAs were highly conserved in structure, but varied significantly in sequence divergence (Figure 3). Some tRNAs like tRNA-Pro (97.1%) and tRNA-Asn (95.9%) were highly conserved, while tRNA-Val (74.7%) and tRNA-Trp (74.0%) appeared much more variable in sequence divergence.
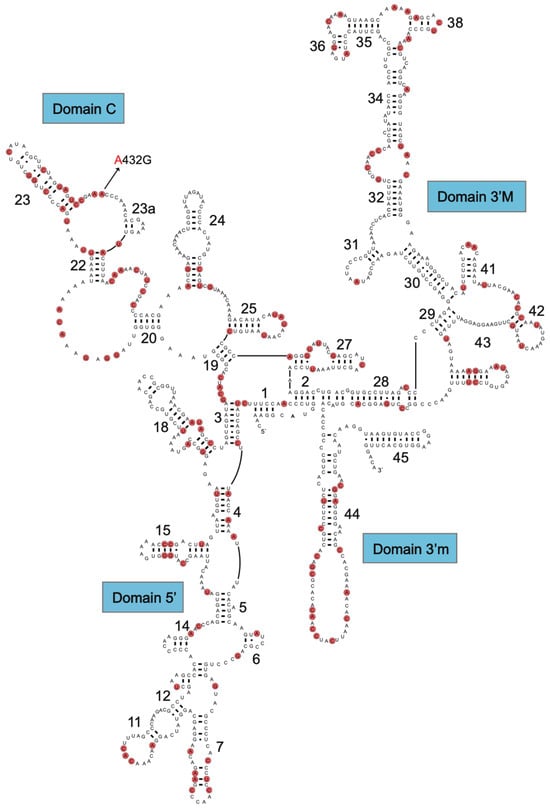
Figure 2.
Secondary structure diagram of R. rasbora mtSSU rRNA. Variable positions between R. rasbora and D. rerio are indicated by red marks.
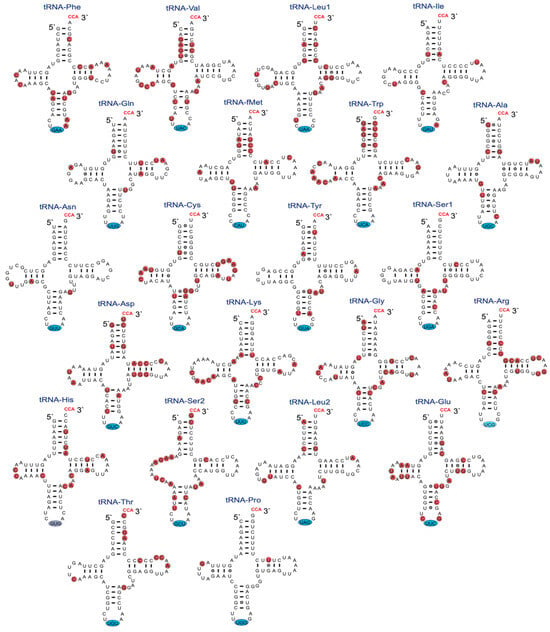
Figure 3.
Secondary structure diagrams of R. rasbora mitochondrial tRNAs. Non-template CCA (red letters at 3′ end), anticodon triplets (blue marks), and variable positions (red marks) between R. rasbora and D. rerio mt-tRNAs are indicated.
3.4. Molecular Phylogeny of Subfamily Rasborinae
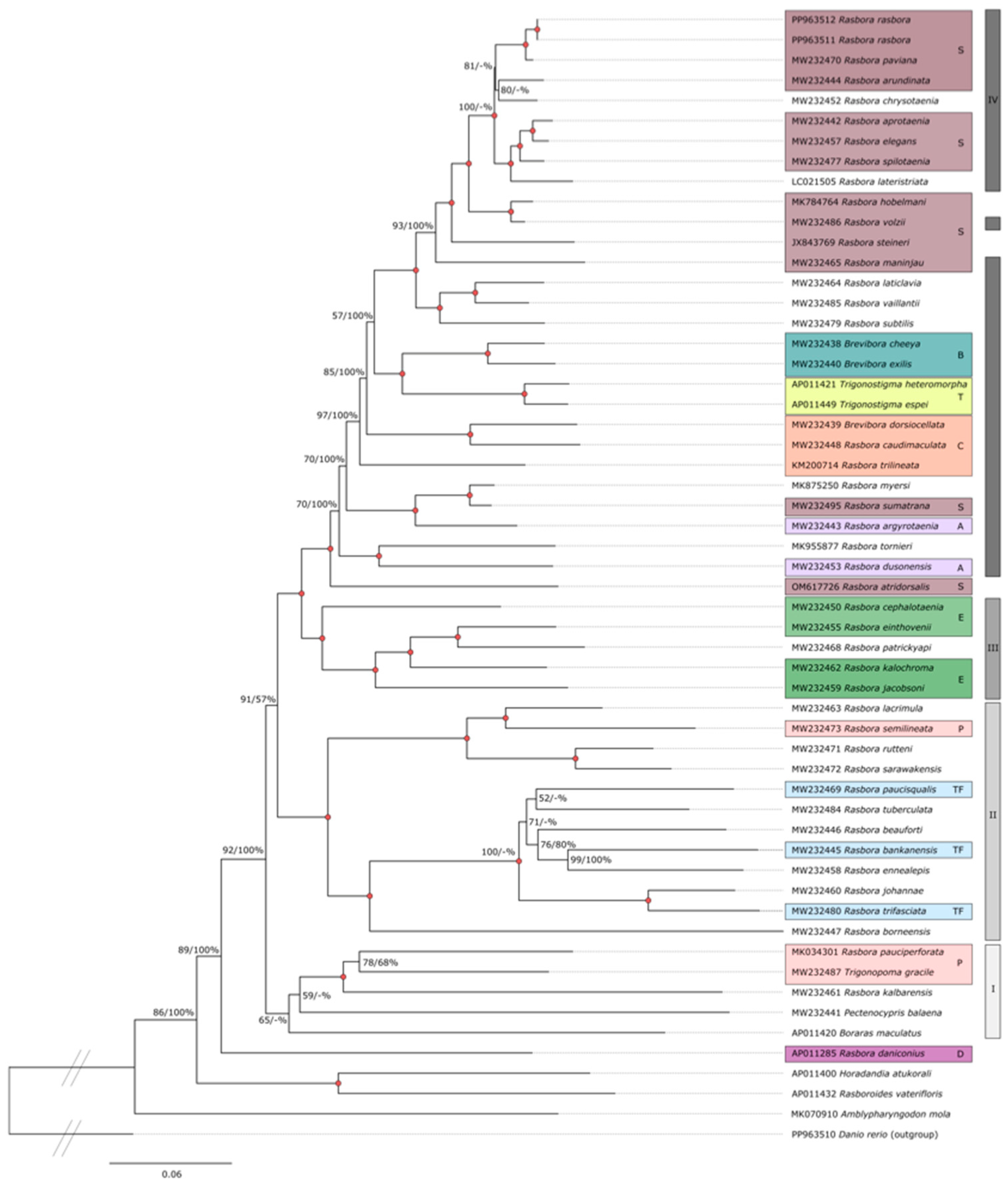
To investigate phylogenetic relationships within the subfamily Rasborinae, we performed phylogenetic analyses that included 53 complete mitogenomes (except the CR) available in the database as well as the two R. rasbora specimens and D. rerio (Table S2) The dataset represents an alignment of 15,776 nt positions that include all protein-coding genes and carefully aligned structural RNA-coding genes, but excluding the control region (CR). The 3′ (left) domain in CR appears highly variable in sequence, and two species in the dataset (Trigonostigma espei and Rasbora argyrotaenia) contain direct repeat features (Figure S4). In addition, two intergenic spacer region sequences (between genes of mtLSU rRNA and tRNA-Leu1 and genes of tRNA-Trp and tRNA-Pro) were manually removed from the alignment in three species (Horadandia atukotali, Amblypharyngodon mole, and Rasbora daniconius; Figure S4).
An ML tree that includes 43 distinct Rasbora species and representative species of the genera Brevibora, Trigonostigma, Amblypharyngodon, Boraras, Pectenocypris, Rasboroides, Horadandia, and Trigonopoma is shown in Figure 4. D. rerio was included as an outgroup in the analysis. Many individual clades within the ML tree were highly supported by bootstrap analysis (2000 replicates). In parallel, BI and NJ trees were generated from the same dataset (Figures S5 and S6). The ML, BI and NJ tree topologies were largely concurrent. Several interesting features were noted. (1) R. rasbora was confirmed to belong to the Sumatrana group in the subfamily Rasborinae, and Rasbora paviana was identified as the closest relative (98.1% overall similarity). (2) The Rasbora genus appears monophyletic within the subfamily Rasborinae, but with some notable exceptions. (3) The genera Brevibora and Trigonostigma were positioned with high bootstrap confidence within the Rasbora groups. Here, the closest relative to Brevibora dorsiocellata was Rasbora caudimaculata (bootstrap 100%), suggesting that the former is a member of the Caudimaculata group. (4) The genera Amblypharyngodon, Boraras, Pectenocypris, Horadandia, and Trigonopoma all have a more basal position in the ML, BI and NJ trees and separated from most Rasbora species. (5) Three Rasbora species (R. pauciperforata, R. kalbarensis, and R. daniconious), however, were basal. The closest relative to R. pauciperforata and R. kalbarensis was Trigonopoma gracile (bootstrap 100%, as R. taeniata [=R. gracilis] in [3]).
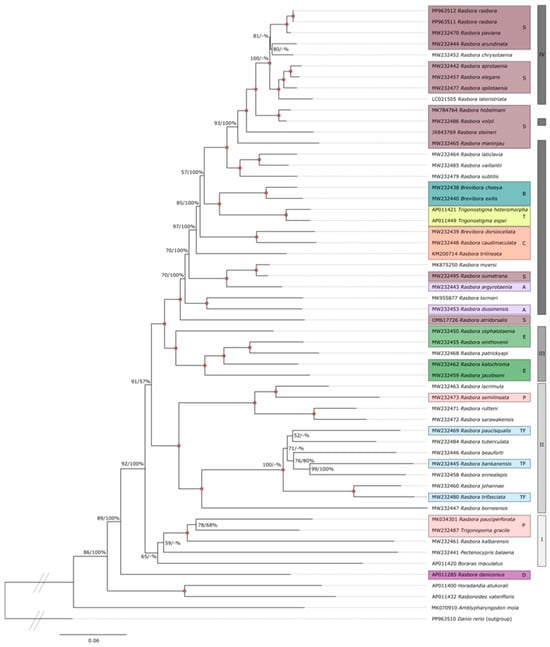
Figure 4.
ML phylogenetic tree of the subfamily Rasborinae. ML analysis was based on complete mitogenome sequences excluding the control region (15,776 nt positions) of 55 Rasborinae specimens. D. rerio was used as an outgroup. ML bootstrap values (%) from 2000 replicates with BI posterior probabilities (%) are shown at branches. Red filled circles indicate bootstrap/posterior probability value of 100% at branch point. Species name and GenBank accession numbers are shown. The different Rasborinae groups (as contained in [3,6,7,32]) are indicated by color code: Sumatrana group (S; burgundy); Brevibora (B; turquoise); Trigonostigma (T; yellow); Caudimaculata group (C; orange); Argyrotaenia group (A; pink); Einthovenii group (E; green); Pauciperforata group (P; peach); Trifaciata group (TF; light blue); Daniconius group (D; dark purple). Right vertical lines from top to bottom: Clade IV (dark gray); Clade III (gray); Clade II (lighter gray); Clade I (lightest gray).
4. Discussion
A mitogenome has been assembled for two R. rasbora specimens and applied in molecular phylogeny assessments of the subfamily Rasborinae. The sequences were generated by long-read ONT with high coverage, which give several advantages in mitogenome assembly, including the elimination of NUMT contaminants. These mitochondrial pseudogenes have been recognized in several teleost nuclear genomes [33]. RNA sequencing based on ONT generated full-length mitochondrial mRNAs that significantly improved mitotranscriptome mapping. The relative abundance of mRNA transcripts and polyadenylation sites correspond well to what has been previously reported in other fish species, such as the Atlantic cod and European monkfish [17,29]. An interesting observation was that 3′UTRs were present in two of the mRNAs (COI and ND5 mRNAs), suggesting a regulatory role in mitochondrial gene expression. The 3′UTR in COI mRNA, which is antisense to tRNA-Ser1, appears conserved in vertebrates and has been reported in both teleosts and mammals [17,29,34,35]. Studies on rats have shown that the nuclear-derived microRNA miR-181c binds to the COI mRNA 3′UTR and regulates mitochondrial function in heart and liver tissues [35,36,37]. Mapping of the Rasbora mRNA, based on ONT cDNA sequencing, yielded an approximately 600 nt ND5 mRNA 3′UTR that corresponds to the complementary ND6 gene sequence that is likely involved in antisense regulation [17]. Interestingly, it has been reported that the ND5 mRNA appears tightly regulated through RNA stability mechanisms in mouse mitochondria [34]. More studies are needed to further explore the role of ND5 mRNA 3′UTR in mRNA stability and mitochondrial gene expression.
Our study supports a Sumatrana group [3] that includes R. rasbora [3,7]. In our phylogenies, a well-supported Sumatrana also contains R. arundinata, R. maninjau as noted in [32], along with R. chrysotaenia and R. lateristriata, with the latter having its own Lateristriata group initially [6]; however, there was minimal support for this group by Tang and co-workers [3]. R. rasbora and R. steineri, which were also part of the Lateristriata species group, have been transferred to the Sumatrana group [3,6]. We suggest the same for R. lateristriata. Nevertheless, the problem within this group comes with the name itself, as R. sumatrana is placed as a sister species to R. myersi, with that group being sister to R. argyrotaenia (Argyrotaenia group). Skoliah and co-workers [4] suggested that R. sumatrana was misidentified as R. dusonensis [3,38] and R. vulgaris misidentified as R. sumatrana [3]. However, R. dusonensis appears to be a sister species to R. tornieri embedded in the same part of the tree as R. sumatrana. Renaming the group as Elegans could be an option, as the group was initially called sumatrana–elegans complex (based on Brittan’s classification [3]).
The updated Einthovenii group containing R. einthovenii, R. kalochroma and R. jacobsoni (including R. patrickyapi in our study as well) is highly supported in our study. This group surprisingly contains as a sister species R. cephalotaenia, which had little support for the Einthovenii group by Tang and co-workers [3] and was found in a different clade. The Caudimaculata group species were localized in two separate clades by [3]: R. caudimaculata and B. dorsiocellata as sister to the Sumatrana group, while R. trilineata as sister to the Trigonostigma. However, our phylogenetic hypotheses do not support that, and moreover R. trilineata appears to be an outgroup to R. caudimaculata and B. dorsiocellata based on the ML and BI analysis.
Some of the species of the Pauciperforata group [6] that were not part of the study by [3] were grouped in the same part of the tree—R. semilineata and R. beuforti (initially in that group [6]), along with species of the Trifasciata group, R. trifasciata, R. bankanensis, R. paucisqualis, and with some additional species that did not have group information available. R. pauciperforata itself, however, clusters with Pectenocypris, Boraras, Trigonopoma and R. kalbarensis. In addition, Tang and co-workers [3] recommended synonymizing Boraras and Trigonostigma with Rasbora, the same as with four new genera (Brevibora, Kottelatia, Rasbosoma, and Trigonopoma) proposed by [6] based on morphological phylogeny. We support this recommendation, as the available species from these genera are immersed within Rasbora in our study as well.
Our phylogeny also supports that R. daniconius does not group with the rest of Rasbora. Tang and co-workers (2010) suggested removing this species from the Rasbora genus (or synonymizing Pectenocypris). However, as noted by [3], sequence data of the other Daniconius group members are currently not available. Amblypharyngodon is placed as the basal sister group to all other species of the available Rasborinae, followed by Horadandia and Rasboroides. More recent phylogeny reconstructed by upcoming studies [4,5] divided the subfamily Rasborinae into four different clades, named I to IV (Figure S8), while confirming the monophyly of the genera Boraras, Pectenocypris and Trigonostigma, and supported the paraphyly of the genus Trigonopoma and the polyphyly of the genera Brevibora and Rasbora. Their time-calibrated phylogeny of Rasborinae was reconstructed based on mitogenomes and COI sequences and subsequently used to define molecular operational taxonomic units (MOTUs). The species’ groups were mainly supported within the clades, with the exception of Clade IV, where changes appeared (see Figure S8D).
5. Conclusions
The mitogenome of R. rasbora represents typical vertebrate organization, but the transcriptome analysis identified seven mRNA stop codons generated by post-transcriptional polyadenylation and two mRNAs with extended 3′ untranslated regions with regulatory potential. Mitogenome-based phylogeny was performed using complete mitogenome sequences, but excluding the control region. The latter appeared variable in sequence, leading to ambiguous alignment at the subfamily level, including direct repeat features in some species. The alignment contained RNA-coding genes and protein-coding genes, and the former were carefully optimized according to rRNA and tRNA structural features. The main findings from generally well-supported ML, BI, and NJ phylogenetic analyses were as follows. (1) R. rasbora individuals cluster within the Sumatrana group of the genus Rasbora. This is an important notion with relevance to the ongoing R. rasbora whole-genome and transcriptome sequencing. (2) The genera Brevibora and Trigonostigma cluster with high confidence within the Rasbora clade, suggesting these species are members of the Rasbora genus. (3) Three Rasbora species (R. pauciperforata, R. kalbarensis and R. daniconius) appear basal within the subfamily Rasborinae and more closely related to other genera than Rasbora. These results indicate the need for a reevaluation of the current taxonomy of the subfamily Rasborinae.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/fishes9080317/s1. Figure S1: Rasbora rasbora—Gangetic scissortail rasbora (photo by SDJ); Figure S2: Rasbora rasbora mitogenome sequencing coverage by ONT; Figure S3: Secondary structure diagram of R. rasbora mtLSU rRNA; Figure S4: Schematic presentation of mitochondrial intergenic spacers and direct repeat features in Rasborinae specimens; Figure S5: Bayesian inference (BI) phylogenetic tree of the subfamily Rasborinae; Figure S6: Neighbor-joining (NJ) phylogenetic tree of the subfamily Rasborinae; Figure S7: Taxonomy of the subfamily Rasborinae based on the classification of Tang and co-workers; Figure S8: Taxonomy of the subfamily Rasborinae based on the classification of Sholihah and co-workers; Table S1: Polymorphic sites in the mitochondrial genome of Danio rerio (AB-NU; PP963510) compared to the reference sequence AC024175; Table S2: Rasborinae species included in ML, BI, and NJ phylogenetic analyses. Note S1: Mitogenome assembly based on ONT reads.
Author Contributions
Conceptualization, S.W.P., T.E.J., I.B. and S.D.J.; methodology, S.W.P., T.E.J. and E.A.; investigation, S.W.P., T.E.J., E.A., I.B. and S.D.J.; data curation, T.E.J. and S.W.P.; writing—original draft preparation, S.W.P. and S.D.J.; writing—review and editing, S.W.P., T.E.J., E.A., I.B. and S.D.J.; visualization, S.W.P., T.E.J. and S.D.J.; supervision, S.D.J., T.E.J. and I.B.; project administration, S.D.J. and I.B.; funding acquisition, I.B., S.D.J. and S.W.P. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by general grants from Nord University and the Research Council of Norway (InnControl project (grant 275786; I. Babiak).
Institutional Review Board Statement
The study was conducted on tissue samples only (not live fish) and performed at Nord University, Norway, according to ethical guidelines stated by the Norwegian Ministry of Agriculture and Food through the Animal Welfare Act. According to guidelines, we were not required to, and therefore do not, have a specific approval or approval number.
Data Availability Statement
Mitogenome sequencing data are available in GenBank under the accession numbers PP963510 (Danio rerio AB-NU), PP963511 (Rasbora rasbora S1-NU), and PP963512 (Rasbora rasbora S2-NU).
Acknowledgments
We thank the genomics facility at Nord University for general support.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Species by Family/Subfamily. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/SpeciesByFamily.asp (accessed on 15 June 2024).
- Eschmeyer’s Catalog of Fishes: Genera, Species, References. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 15 June 2024).
- Tang, K.L.; Agnew, M.K.; Hirt, M.V.; Sado, T.; Schneider, L.M.; Freyhof, J.; Sulaiman, Z.; Swartz, E.; Vidthayanon, C.; Miya, M.; et al. Systematics of the subfamily Danioninae (Teleostei: Cypriniformes: Cyprinidae). Mol. Phylogenetics Evol. 2010, 57, 189–214. [Google Scholar] [CrossRef] [PubMed]
- Sholihah, A.; Delrieu-Trottin, E.; Sukmono, T.; Dahruddin, H.; Risdawati, R.; Elvyra, R.; Wibowo, A.; Kustiati, K.; Busson, F.; Sauri, S.; et al. Disentangling the taxonomy of the subfamily Rasborinae (Cypriniformes, Danionidae) in Sundaland using DNA barcodes. Sci. Rep. 2020, 10, 2818. [Google Scholar] [CrossRef] [PubMed]
- Sholihah, A.; Delrieu-Trottin, E.; Sukmono, T.; Dahruddin, H.; Pauzadoux, J.; Tilak, M.K.; Fitriana, Y.; Agnèse, J.F.; Condamine, F.L.; Wowor, D.; et al. Limited dispersal and in situ diversification drive the evolutionary history of Rasborinae fishes in Sundaland. J. Biogeogr. 2021, 48, 2153–2173. [Google Scholar] [CrossRef]
- Liao, T.Y.; Kullander, S.O.; Fang, F. Phylogenetic analysis of the genus Rasbora (Teleostei: Cyprinidae). Zool. Scr. 2010, 39, 155–176. [Google Scholar] [CrossRef]
- Kottelat, M.; Vidthayanon, M.K.C. Boraras micros, a new genus and species of minute freshwater fish from Thailand (Teleostei: Cyprinidae). Ichthyol. Explor. Freshw. 1993, 4, 161–176. [Google Scholar]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Miya, M.; Nishida, M. Use of mitogenomic information in teleostean molecular phylogenetics: A tree-based exploration under the maximum-parsimony optimality criterion. Mol. Phylogenetics Evol. 2000, 17, 437–455. [Google Scholar] [CrossRef]
- Iwasaki, W.; Fukunaga, T.; Isagozawa, R.; Yamada, K.; Maeda, Y.; Satoh, T.P.; Sado, T.; Mabuchi, K.; Takeshima, H.; Miya, M.; et al. MitoFish and MitoAnnotator: A mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol. Biol. Evol. 2013, 30, 2531–2540. [Google Scholar] [CrossRef] [PubMed]
- Inoue, J.G.; Miya, M.; Tsukamoto, K.; Nishida, M. A mitogenomic perspective on the basal teleostean phylogeny: Resolving higher-level relationships with longer DNA sequences. Mol. Phylogenetics Evol. 2001, 20, 275–285. [Google Scholar] [CrossRef]
- Miya, M.; Takeshima, H.; Endo, H.; Ishiguro, N.D.; Inoue, J.G.; Mukai, T.; Satoh, T.P.; Yamaguchi, M.; Kawaguchi, A.; Mabuchi, K.; et al. Major patterns of higher phylogenies: A new perspective based on 100 complete mitochondrial DNA sequences. Mol. Phylogenetics Evol. 2003, 26, 121–138. [Google Scholar] [CrossRef]
- Satoh, T.P.; Sado, T.; Mayden, R.L.; Hanzawa, N.; Nakamura, K.; Nishida, M.; Miya, M. Mitogenomic evolution and interrelationships of the Cypriniformes (Actinopterygii: Ostariophysi): The first evidence toward resolution of higher-level relationships of the world’s largest freshwater fish clade based on 59 whole mitogenome sequences. J. Mol. Evol. 2006, 63, 826–841. [Google Scholar] [CrossRef] [PubMed]
- Miya, M.; Pietsch, T.W.; Orr, J.W.; Arnold, R.J.; Satoh, T.P.; Shedlock, A.M.; Ho, H.C.; Shimazaki, M.; Yabe, M.; Nishida, M. Evolutionary history of anglerfishes (Teleostei: Lophiiformes): A mitogenomic perspective. BMC Evol. Biol. 2010, 10, 58. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.P.; Miya, M.; Mabuchi, K.; Nishida, M. Structure and variation of the mitochondrial genome of fishes. BMC Genom. 2016, 17, 719. [Google Scholar] [CrossRef]
- Breines, R.; Ursvik, A.; Nymark, M.; Johansen, S.D.; Coucheron, D.H. Complete mitochondrial genome sequences of the Arctic Ocean codfishes Arctogadus glacialis and Boreogadus saida reveal oriL and tRNA gene duplications. Polar Biol. 2008, 31, 1245–1252. [Google Scholar] [CrossRef]
- Coucheron, D.H.; Nymark, M.; Breines, R.; Karlsen, B.O.; Andreassen, M.; Jørgensen, T.E.; Moum, T.; Johansen, S.D. Characterization of mitochondrial mRNA in codfish reveals unique features compared to mammals. Curr. Genet. 2011, 57, 213–222. [Google Scholar] [CrossRef]
- Flynn, T.; Signal, B.; Johnson, S.L.; Gemmell, N.J. Mitochondrial genome diversity among six laboratory zebrafish (Danio rerio) strains. Mitochondrial DNA A 2016, 27, 4364–4371. [Google Scholar] [CrossRef] [PubMed]
- Quispe-Tintaya, W.; White, R.R.; Popov, V.N.; Vijg, J.; Maslov, A.Y. Fast mitochondrial DNA isolation from mammalian cells for next-generation sequencing. Biotechniques 2013, 55, 133–136. [Google Scholar] [CrossRef]
- Bekaert, B.; Ellerington, R.; Van den Abbeele, L.; Decorte, R. In-solution hybridization for the targeted enrichment of the whole mitochondrial genome. Methods Mol. Biol. 2016, 1420, 173–183. [Google Scholar] [CrossRef]
- Senovska, A.; Drozdova, E.; Vaculik, O.; Pardy, F.; Brzobohata, K.; Fialova, D.; Smerda, J.; Kos, P. Cost-effective straightforward method for captured whole mitogenome sequencing of ancient DNA. Forensic Sci. Int. 2021, 319, 110638. [Google Scholar] [CrossRef]
- Zascavage, R.R.; Hall, C.L.; Thorson, K.; Mahmoud, M.; Sedlazeck, F.J.; Planz, J.V. Approaches to whole mitochondrial genome sequencing in the Oxford Nanopore MinION. Curr. Protoc. Hum. Genet. 2019, 104, e94. [Google Scholar] [CrossRef]
- Nanopore Sequencing Accuracy. Available online: https://nanoporetech.com/platform/accuracy (accessed on 15 June 2024).
- Maude, H.; Davidson, M.; Charitakis, N.; Diaz, L.; Bowers, W.H.T.; Gradovich, E.; Andrew, T.; Huntley, D. NUMT confounding biases mitochondrial heteroplasmy calls in favor of the reference. Front. Cell Dev. Biol. 2019, 7, 201. [Google Scholar] [CrossRef]
- Deng, Y.; Qian, Y.; Meng, M.; Jiang, H.; Dong, Y.; Fang, C.; He, S.; Yang, L. Extensive sequence divergence between the reference genomes of two zebrafish strains, Tuebingen and AB. Mol. Ecol. Resour. 2021, 22, 2148–2157. [Google Scholar] [CrossRef]
- Chernyavskaya, Y.; Zhang, X.; Liu, J.; Blackburn, J. Long-read sequencing of the zebrafish genome reorganizes genomic architecture. BMC Genom. 2022, 23, 116. [Google Scholar] [CrossRef]
- Bakke, I.; Johansen, S. Characterization of mitochondrial ribosomal RNA genes in gadiformes: Sequence variations, secondary structural features, and phylogenetic implications. Mol. Phylogenetics Evol. 2002, 25, 87–100. [Google Scholar] [CrossRef]
- Jørgensen, T.E.; Karlsen, B.O.; Emblem, Å.; Breines, R.; Andreassen, M.; Rounge, T.B.; Nederbragt, A.J.; Jakobsen, K.S.; Nymark, M.; Ursvik, A.; et al. Mitochondrial genome variation of Atlantic cod. BMC Res. Notes 2018, 11, 397. [Google Scholar] [CrossRef]
- Dubin, A.; Jørgensen, T.E.; Jakt, L.M.; Johansen, S.D. The mitochondrial transcriptome of the anglerfish Lophius piscatorius. BMC Res. Notes 2019, 12, 800. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Lumbantobing, D.N. Four new species of Rasbora of the Sumatrana group (Teleostei: Cyprinidae) from norther Sumatra, Indonesia. Zootaxa 2014, 3764, 1–25. [Google Scholar] [CrossRef]
- Antunes, A.; Ramos, M.J. Discovery of a large number of previously unrecognized mitochondrial pseudogenes in fish genomes. Genomics 2005, 86, 708–717. [Google Scholar] [CrossRef][Green Version]
- Bai, Y.; Shakeley, R.M.; Attardi, G. Tight control of respiration by NADH dehydrogenase ND5 subunit gene expression in mouse mitochondria. Mol. Cell Biol. 2000, 20, 805–815. [Google Scholar] [CrossRef]
- Das, S.; Ferlito, M.; Kent, O.A.; Fox-Talbot, K.; Wang, R.; Liu, D.; Raghavachari, N.; Yang, Y.; Wheelan, S.J.; Murphy, E.; et al. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ. Res. 2012, 110, 1596–1603. [Google Scholar] [CrossRef]
- Das, S.; Bedja, D.; Campbell, N.; Dunkerly, B.; Chenna, V.; Maitra, A.; Steenbergen, C. miR-181c regulates the mitochondrial genome, bioenergetics, and propensity for heart failure in vivo. PLoS ONE 2014, 9, e96820. [Google Scholar] [CrossRef]
- Akiyoshi, K.; Boersma, G.J.; Johnson, M.D.; Velasquez, F.C.; Dunkerly-Eyring, B.; O’Brien, S.; Yamaguchi, A.; Steenbergen, C.; Tamashiro, K.L.K.; Das, S. Role of miR-181c in diet-induced obesity through regulation of lipid synthesis in liver. PLoS ONE 2021, 16, e0256973. [Google Scholar] [CrossRef]
- Collins, R.A.; Armstrong, K.F.; Meier, R.; Yi, Y.; Brown, S.D.; Cruickshank, R.H.; Keeling, S.; Johnston, C. Barcoding and border biosecurity: Identifying cyprinid fishes in the aquarium trade. PLoS ONE 2012, 7, e28381. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).