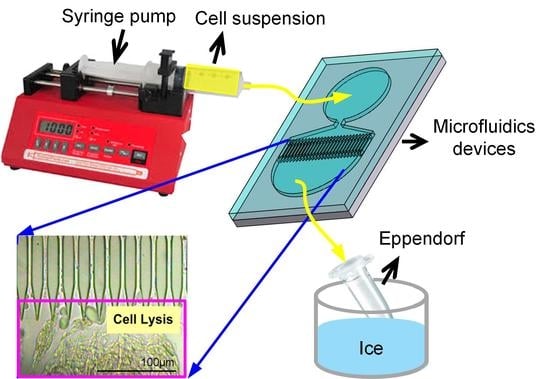
Chemical-Free Extraction of Functional Mitochondria Using a Microfluidic Device
Abstract
:1. Introduction
2. Materials and Methods
2.1. Design of the Microfluidic Mitochondria Extraction Device
2.2. Fabrication of the Microfluidic Device
2.3. Cell Culture and Mitochondria Extraction
2.4. Protein Assay
2.5. SDS-PAGE and Western Blot
2.6. Flow Cytometry
2.7. Examination of Mitochondrial Morphology
3. Results
3.1. Device Structure Selection
3.2. Chip-Based Microfluidic Device
3.3. Pressure Drop and Shear Stress
3.4. Working Concentration of the Cell Suspension
3.5. Function of Device-Extracted Mitochondria
3.6. Mitochondrial Ultrastructure of Device-Extracted Mitochondria
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Desler, C.; Rasmussen, L.J. Mitochondria in biology and medicine. Mitochondrion 2012, 12, 472–476. [Google Scholar] [CrossRef] [PubMed]
- McBride, H.M.; Neuspiel, M.; Wasiak, S. Mitochondria: More than just a powerhouse. Curr. Biol. 2006, 16, R551–560. [Google Scholar] [CrossRef] [PubMed]
- Voet, D.; Voet, J.G.; Pratt, C.W. Fundamentals of Biochemistry, 2nd ed.; John Wiley and Sons, Inc.: New York, NY, USA, 2006. [Google Scholar]
- Richter, C.; Park, J.; Ames, B.N. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc. Natl. Acad. Sci. USA 1988, 85, 6465–6467. [Google Scholar] [CrossRef] [PubMed]
- Pourahmad, J.; Hosseini, M.J. Toxicity of Arsenic (III) on Isolated Liver Mitochondria: A New Mechanistic Approach. Iran. J. Pharm. Res. 2012, 11, 703–704. [Google Scholar] [PubMed]
- Giang, A.H.; Raymond, T.; Brookes, P.; de Mesy Bentley, K.; Schwarz, E.; O’Keefe, R.; Eliseev, R. Mitochondrial Dysfunction and permeability transition in osteosarcoma cells showing the warburg effect. J. Biol. Chem. 2013, 288, 33303–33311. [Google Scholar] [CrossRef] [PubMed]
- Knott, A.B.; Perkins, G.; Schwarzenbacher, R.; Bossy-Wetzel, E. Mitochondrial fragmentation in neurodegeneration. Nat. Rev. Neurosci. 2008, 9, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Spees, J.L.; Olson, S.D.; Whitney, M.J.; Prockop, D.J. Mitochondrial transfer between cells can rescue aerobic respiration. Cell Biol. 2006, 103, 1283–1288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elliott, R.L.; Jiang, X.P.; Head, J.F. Mitochondria organelle transplantation: Introduction of normal epithelial mitochondria into human cancer cells inhibits proliferation and increases drug sensitivity. Breast Cancer Res. Treat. 2012, 136, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Masuzawa, A.; Black, K.M.; Pacak, C.A.; Ericsson, M.; Barnett, R.J.; Drumm, C.; Seth, P.; Bloch, D.B.; Levitsky, S.; Cowan, D.B.; et al. Transplantation of autologously derived mitochondria protects the heart from ischemia-reperfusion injury. Am. J. Physiol. Heart Circ. Physiol. 2013, 304, H966–H982. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.; Kwon, K.W.; Park, M.C.; Lee, S.H.; Kim, S.M.; Suh, K.Y. Soft Lithography for Microfluidics: A Review. Biochip J. 2008, 2, 1–11. [Google Scholar]
- Qin, D.; Xia, Y.; Whitesides, G.M. Soft lithography for micro- and nanoscale patterning. Nat. Protoc. 2010, 5, 491–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kido, H.; Micic, M.; Smith, D.; Zoval, J.; Norton, J.; Madou, M. A novel compact disk-like centrifugal microfluidics system for cell lysis and sample homogenization. Colloids Surf. B Biointerfaces 2007, 58, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.Y.; Wo, A.M.; Lo, Y.J.; Chen, K.C.; Lin, C.M.; Yang, C.R. Three dimensional electrode array for cell lysis via electroporation. Biosens. Bioelectron. 2006, 22, 568–574. [Google Scholar] [CrossRef] [PubMed]
- Jha, S.K.; Ra, G.S.; Joo, G.S.; Kim, Y.S. Electrochemical cell lysis on a miniaturized flow-through device. Curr. Appl. Phys. 2009, 9, e301–e303. [Google Scholar] [CrossRef]
- Ikeda, N.; Tanaka, N.; Yanagida, Y.; Hatsuzawa, T. On-chip single-cell lysis for extracting intracellular material. Jpn. J. Appl. Phys. 2007, 46, 6410. [Google Scholar] [CrossRef]
- Liu, C. Recent Developments in Polymer MEMS. Adv. Mater. 2007, 19, 3783–3790. [Google Scholar] [CrossRef]
- Nisar, A.; Afzulpurkar, N.; Mahaisavariya, B.; Tuantranont, A. MEMS-based, micropumps in drug delivery and biomedical applications. Sens. Actuators B Chem. 2008, 130, 917–942. [Google Scholar] [CrossRef]
- Chen, Y.C.; Chen, G.Y.; Lin, Y.C.; Wang, G.J. A lab-on-a-chip capillary network for red blood cell hydrodynamics. Microfluid. Nnaofluid. 2010, 9, 585–591. [Google Scholar] [CrossRef]
- Li, Y.; Park, J.S.; Deng, J.H.; Bai, Y. Cytochrome c oxidase subunit IV is essential for assembly and respiratory function of the enzyme complex. J. Bioenerg. Biomembr. 2006, 38, 283–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terada, K.; Kanazawa, M.; Yano, M.; Hanson, B.; Hoogenraad, N.; Mori, M. Participation of the import receptor Tom20 in protein import into mammalian mitochondria: Analyses in vitro and in cultured cells. FEBS Lett. 1997, 403, 309–312. [Google Scholar] [CrossRef]
- Ramirez-Aguilar, S.J.; Keuthe, M.; Rocha, M.; Fedyaev, V.V.; Kramp, K.; Gupta, K.J.; Rasmusson, A.G.; Schulze, W.X.; van Dongen, J.T. The composition of plant mitochondrial supercomplexes changes with oxygen availability. J. Biol. Chem. 2011, 286, 43045–43053. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.C.; Liu, K.H.; Chuang, C.S.; Su, H.L.; Wei, Y.H.; Kuo, S.J.; Liu, C.S. Treatment of human cells derived from MERRF syndrome by peptide-mediated mitochondrial delivery. Cytotherapy 2013, 15, 1580–1596. [Google Scholar] [CrossRef] [PubMed]
- Corcelli, A.; Saponetti, M.S.; Zaccagnino, P.; Lopalco, P.; Mastrodonato, M.; Liquori, G.E.; Lorusso, M. Mitochondria isolated in nearly isotonic KCl buffer: Focus on cardiolipin and organelle morphology. Biochim. Biophys. Acta 2010, 1798, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Gross, V.S.; Greenberg, H.K.; Baranov, S.V.; Carlson, G.M.; Stavrovskaya, I.G.; Lazarev, A.V.; Kristal, B.S. Isolation of functional mitochondria from rat kidney and skeletal muscle without manual homogenization. Anal. Biochem. 2011, 418, 213–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hornig-Do, H.T.; Günther, G.; Bust, M.; Lehnartz, P.; Bosio, A.; Wiesnera, R.J. Isolation of functional pure mitochondria by superparamagnetic microbeads. Anal. Biochem. 2009, 389, 1–5. [Google Scholar] [CrossRef] [PubMed]






| n | m | ΔP (mPa) | σ (mPa) |
|---|---|---|---|
| 30 | 0 | 6.73 × 10−5 | 1.722 × 10−6 |
| 30 | 5 | 8.40 × 10−5 | 2.15 × 10−6 |
| 30 | 10 | 10.49 × 10−5 | 2.69 × 10−6 |
| 30 | 15 | 13.99 × 10−5 | 3.58 × 10−6 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsiao, Y.-H.; Li, C.-W.; Chang, J.-C.; Chen, S.-T.; Liu, C.-S.; Wang, G.-J. Chemical-Free Extraction of Functional Mitochondria Using a Microfluidic Device. Inventions 2018, 3, 68. https://doi.org/10.3390/inventions3040068
Hsiao Y-H, Li C-W, Chang J-C, Chen S-T, Liu C-S, Wang G-J. Chemical-Free Extraction of Functional Mitochondria Using a Microfluidic Device. Inventions. 2018; 3(4):68. https://doi.org/10.3390/inventions3040068
Chicago/Turabian StyleHsiao, Yu-Han, Ching-Wen Li, Jui-Chih Chang, Sung-Tzu Chen, Chin-San Liu, and Gou-Jen Wang. 2018. "Chemical-Free Extraction of Functional Mitochondria Using a Microfluidic Device" Inventions 3, no. 4: 68. https://doi.org/10.3390/inventions3040068
APA StyleHsiao, Y.-H., Li, C.-W., Chang, J.-C., Chen, S.-T., Liu, C.-S., & Wang, G.-J. (2018). Chemical-Free Extraction of Functional Mitochondria Using a Microfluidic Device. Inventions, 3(4), 68. https://doi.org/10.3390/inventions3040068