The Development of a Personalised Training Framework: Implementation of Emerging Technologies for Performance
Abstract
:1. Introduction
- (1)
- To what training will my athlete best respond?
- (2)
- How well is my athlete adapting to training?
- (3)
- When should I change the training stimulus (i.e., has the athlete reached their adaptive ceiling for this training modality)?
- (4)
- How long will it take for a certain adaptation to occur?
- (5)
- How well is my athlete tolerating the current training load?
- (6)
- What load can my athlete handle today?
2. A Personalised Medicine Approach to Performance?
3. The Use of Genetic Information within the Personalised Training Process
4. Novel Markers of Exercise Adaptation and Recovery
4.1. Epigenetic Modifications—Novel Markers of Exercise Adaptation and Fatigue
4.1.1. Methylation
4.1.2. Histone Modifications
4.1.3. miRNA
4.1.4. Utilisation of Epigenetic Markers within Training Programmes
4.1.5. Practical Perspectives
4.1.6. Section Summary
4.2. Cell-Free DNA (cfDNA): A Novel Marker of Exercise Adaptation?
5. The Microbiome, Exercise and Elite Performance
6. Pharmacogenomics—Personalised Sports Nutrition?
7. The Integration of Other “omes”
8. The Use of Technology in the Personalised Training Process
9. Prediction, Data Mining & Machine Learning
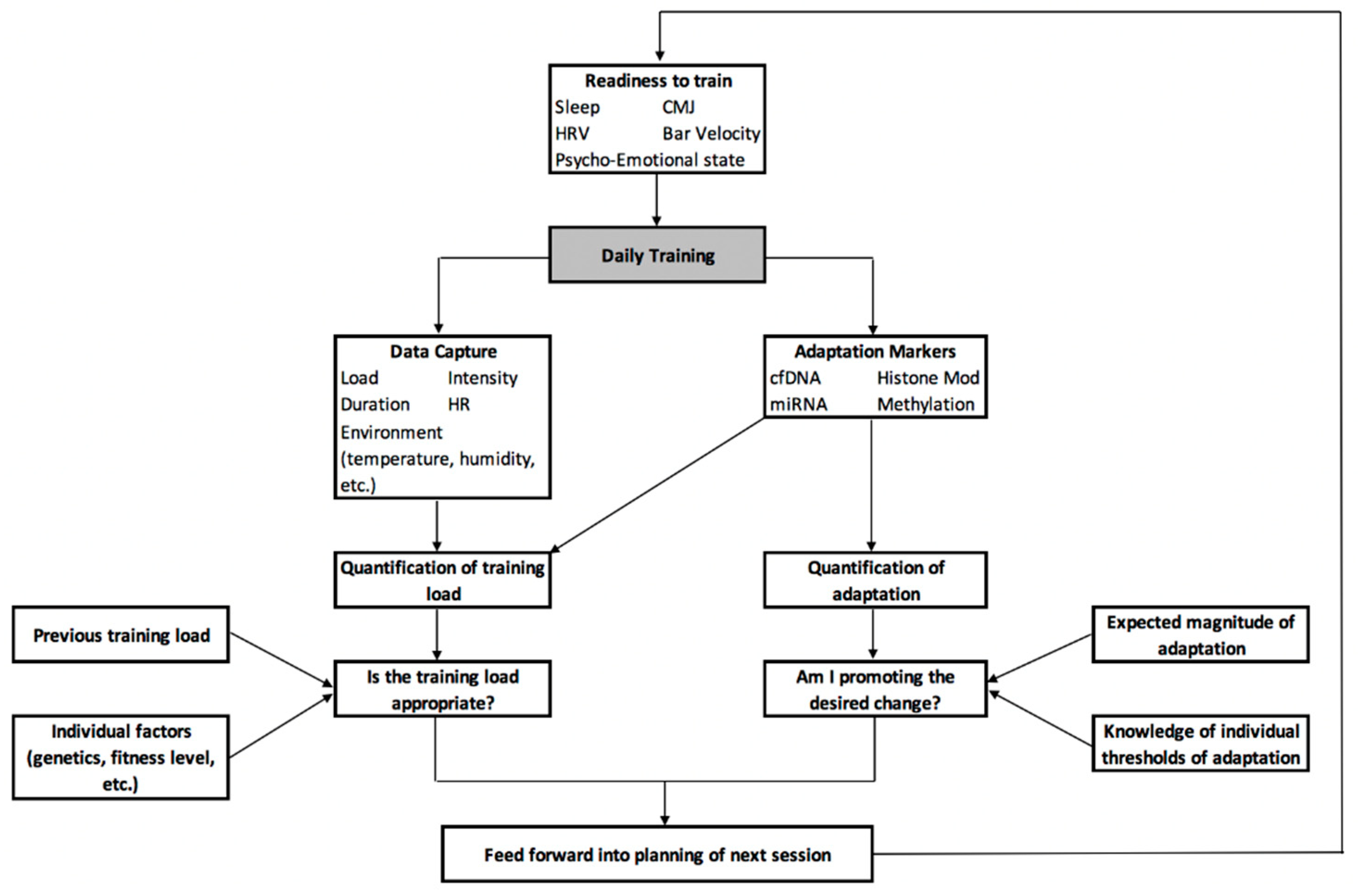
10. A Centralised Framework for the Development of a Personalised Training Process
11. Summary
Author Contributions
Funding
Conflicts of Interest
References
- Pickering, C.; Kiely, J. Understanding personalized training responses: Can genetic assessment help? Open Sports Sci. J. 2017, 10. [Google Scholar] [CrossRef]
- Pickering, C.; Kiely, J. Are the current guidelines on caffeine use in sport optimal for everyone? Inter-individual variation in caffeine ergogenicity, and a move towards personalised sports nutrition. Sports Med. 2018, 48, 7–16. [Google Scholar] [CrossRef]
- Pickering, C.; Kiely, J. Can the ability to adapt to exercise be considered a talent—And if so, can we test for it? Sports Med. Open 2017, 3, 43. [Google Scholar] [CrossRef]
- Montalvo, A.M.; Tse-Dinh, Y.C.; Liu, Y.; Swartzon, M.; Hechtman, K.S.; Myer, G.D. Precision sports medicine: The future of advancing health and performance in youth and beyond. Strength Cond. J. 2017, 39, 48–58. [Google Scholar] [CrossRef]
- Gabbett, T.J. The training-injury prevention paradox: Should athletes be training smarter and harder? Br. J. Sports Med. 2016, 50, 273–280. [Google Scholar] [CrossRef]
- Saw, A.E.; Main, L.C.; Gastin, P.B. Monitoring the athlete training response: Subjective self-reported measures trump commonly used objective measures: A systematic review. Br. J. Sports Med. 2016, 50, 281–291. [Google Scholar] [CrossRef]
- Peake, J.; Kerr, G.K.; Sullivan, J.P. A critical review of consumer wearables, mobile applications and equipment for providing biofeedback, monitoring stress and sleep in physically active populations. Front. Physiol. 2018, 9, 743. [Google Scholar] [CrossRef] [PubMed]
- Jones, N.; Kiely, J.; Suraci, B.; Collins, D.J.; De Lorenzo, D.; Pickering, C.; Grimaldi, K.A. A genetic-based algorithm for personalized resistance training. Biol. Sport 2016, 33, 117. [Google Scholar] [CrossRef]
- Pickering, C.; Kiely, J.; Suraci, B.; Collins, D. The magnitude of Yo-Yo test improvements following an aerobic training intervention are associated with total genotype score. PLoS ONE 2018, 13, e0207597. [Google Scholar] [CrossRef] [PubMed]
- Moraes, V.N.; Trapé, A.A.; Ferezin, L.P.; Gonçalves, T.C.; Monteiro, C.P.; Junior, C.B. Association of ACE ID and ACTN3 C> T genetic polymorphisms with response to a multicomponent training program in physical performance in women from 50 to 70 years. Sci. Sports 2018, 33, 282–290. [Google Scholar] [CrossRef]
- Buchheit, M. Houston, we still have a problem. Int. J. Sports Physiol. Perform. 2017, 12, 1111–1114. [Google Scholar] [CrossRef] [PubMed]
- McNamee, M.J.; Coveney, C.M.; Faulkner, A.; Gabe, J. Ethics, evidence based sports medicine, and the use of platelet rich plasma in the English premier league. Health Care Anal. 2018, 26, 344–361. [Google Scholar] [CrossRef] [PubMed]
- Green, E.D.; Guyer, M.S.; Institute, N.H. Charting a course for genomic medicine from base pairs to bedside. Nature 2011, 470, 204. [Google Scholar] [CrossRef]
- Manolio, T.A.; Green, E.D. Leading the way to genomic medicine. Am. J. Med. Genet. C Semin. Med. Genet. 2014, 166, 1–7. [Google Scholar] [CrossRef]
- Ashley, E.A. The precision medicine initiative: A new national effort. JAMA 2015, 313, 2119–2120. [Google Scholar] [CrossRef] [PubMed]
- Ashley, E.A. Towards precision medicine. Nat. Rev. Genet. 2016, 17, 507. [Google Scholar] [CrossRef]
- Wainwright, C.E.; Elborn, J.S.; Ramsey, B.W.; Marigowda, G.; Huang, X.; Cipolli, M.; Colombo, C.; Davies, J.C.; De Boeck, K.; Flume, P.A.; et al. Lumacaftor–ivacaftor in patients with cystic fibrosis homozygous for Phe508del CFTR. New. Engl. J. Med. 2015, 373, 220–231. [Google Scholar] [CrossRef]
- Hofker, M.; Wijmenga, C. A supersized list of obesity genes. Nat. Genet. 2009, 41, 139. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. New. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Pine, A.C.; Fioretti, F.F.; Brooke, G.N.; Bevan, C.L. Advances in genetics: Widening our understanding of prostate cancer. F1000Research 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Relling, M.V.; Evans, W.E. Pharmacogenomics in the clinic. Nature 2015, 526, 343. [Google Scholar] [CrossRef] [PubMed]
- International Warfarin Pharmacogenetics Consortium. Estimation of the warfarin dose with clinical and pharmacogenetic data. New Engl. J. Med. 2009, 360, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Yip, V.L.; Alfirevic, A.; Pirmohamed, M. Genetics of immune-mediated adverse drug reactions: A comprehensive and clinical review. Clin. Rev. Allergy Immunol. 2015, 48, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.M.; Sarkar, I.N. Exploring the potential of direct-to-consumer genomic test data for predicting adverse drug events. AMIA Summits Transl. Sci. Proc. 2018, 2018, 247–256. [Google Scholar]
- Thomas, F.; Desmedt, C.; Aftimos, P.; Awada, A. Impact of tumor sequencing on the use of anticancer drugs. Curr. Opin. Oncol. 2014, 26, 347–356. [Google Scholar] [CrossRef]
- Damodaran, S.; Berger, M.F.; Roychowdhury, S. Clinical tumor sequencing: Opportunities and challenges for precision cancer medicine. Am. Soc. Clin. Oncol. Educ. Book 2015, e175–e182. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Laetsch, T.W.; Kummar, S.; DuBois, S.G.; Lassen, U.N.; Demetri, G.D.; Nathenson, M.; Doebele, R.C.; Farago, A.F.; Pappo, A.S.; et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. New Engl. J. Med. 2018, 378, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Caudle, W.M.; Bammler, T.K.; Lin, Y.; Pan, S.; Zhang, J. Using ‘omics’ to define pathogenesis and biomarkers of Parkinson’s disease. Expert Rev. Neurother. 2010, 10, 925–942. [Google Scholar] [CrossRef] [PubMed]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef]
- Taylor-Robinson, D.; Kee, F. Precision public health—The Emperor’s new clothes. Int. J. Epidemiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Marcon, A.R.; Bieber, M.; Caulfield, T. Representing a “revolution”: How the popular press has portrayed personalized medicine. Genet. Med. 2018, 20, 950. [Google Scholar] [CrossRef] [PubMed]
- Rey-López, J.P.; Sá, T.H.; Rezende, L.F. Why precision medicine is not the best route to a healthier world. Revista Saude Publica 2018, 52, 12. [Google Scholar] [CrossRef]
- Joyner, M.J.; Paneth, N. Promises, promises, and precision medicine. J. Clin. Investig. 2019, 129, 946–948. [Google Scholar] [CrossRef] [PubMed]
- Maron, B.J.; Maron, M.S.; Semsarian, C. Genetics of hypertrophic cardiomyopathy after 20 years: Clinical perspectives. J. Am. Coll. Cardiol. 2012, 60, 705–715. [Google Scholar] [CrossRef] [PubMed]
- Posthumus, M.; September, A.V.; O’Cuinneagain, D.; van der Merwe, W.; Schwellnus, M.P.; Collins, M. The COL5A1 gene is associated with increased risk of anterior cruciate ligament ruptures in female participants. Am. J. Sports Med. 2009, 37, 2234–2240. [Google Scholar] [CrossRef] [PubMed]
- Ordovas, J.M.; Ferguson, L.R.; Tai, E.S.; Mathers, J.C. Personalised nutrition and health. BMJ 2018, 361. [Google Scholar] [CrossRef] [PubMed]
- Guest, N.; Horne, J.; Vanderhout, S.; El-Sohemy, A. Sport nutrigenomics: Personalized nutrition for athletic performance. Front. Nutr. 2019, 6, 8. [Google Scholar] [CrossRef]
- Bouchard, C.; An, P.; Rice, T.; Skinner, J.S.; Wilmore, J.H.; Gagnon, J.; Pérusse, L.; Leon, A.S.; Rao, D.C. Familial aggregation of VO2max response to exercise training: Results from the HERITAGE Family Study. J. Appl. Physiol. 1999, 87, 1003–1008. [Google Scholar] [CrossRef]
- Skinner, J.S.; Jaskólski, A.; Jaskólska, A.; Krasnoff, J.; Gagnon, J.; Leon, A.S.; Rao, D.C.T.; Wilmore, J.H.; Bouchard, C. Age, sex, race, initial fitness, and response to training: The HERITAGE Family Study. J. Appl. Physiol. 2001, 90, 1770–1776. [Google Scholar] [CrossRef]
- Bouchard, C.; Rankinen, T.; Chagnon, Y.C.; Rice, T.; Pérusse, L.; Gagnon, J.; Borecki, I.; An, P.; Leon, A.S.; Skinner, J.S. Genomic scan for maximal oxygen uptake and its response to training in the HERITAGE Family Study. J. Appl. Physiol. 2000, 88, 551–559. [Google Scholar] [CrossRef]
- Bouchard, C. Exercise genomics—A paradigm shift is needed: A commentary. Br. J. Sports Med. 2015, 49, 1492–1496. [Google Scholar] [CrossRef]
- Rankinen, T.; Fuku, N.; Wolfarth, B.; Wang, G.; Sarzynski, M.A.; Alexeev, D.G.; Ahmetov, I.I.; Boulay, M.R.; Cieszczyk, P.; Eynon, N.; et al. No evidence of a common DNA variant profile specific to world class endurance athletes. PLoS ONE 2016, 11, e0147330. [Google Scholar] [CrossRef] [PubMed]
- Webborn, N.; Williams, A.; McNamee, M.; Bouchard, C.; Pitsiladis, Y.; Ahmetov, I.; Ashley, E.; Byrne, N.; Camporesi, S.; Collins, M.; et al. Direct-to-consumer genetic testing for predicting sports performance and talent identification: Consensus statement. Br. J. Sports Med. 2015, 49, 1486–1491. [Google Scholar] [CrossRef] [PubMed]
- Camporesi, S.; McNamee, M.J. Ethics, genetic testing, and athletic talent: Children’s best interests, and the right to an open (athletic) future. Physiol. Genom. 2016, 48, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Vlahovich, N.; Fricker, P.A.; Brown, M.A.; Hughes, D. Ethics of genetic testing and research in sport: A position statement from the Australian Institute of Sport. Br. J. Sports Med. 2017, 51, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Varley, I.; Patel, S.; Williams, A.G.; Hennis, P.J. The current use, and opinions of elite athletes and support staff in relation to genetic testing in elite sport within the UK. Biol. Sport 2018, 35, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Del Coso, J.; Valero, M.; Salinero, J.J.; Lara, B.; Díaz, G.; Gallo-Salazar, C.; Ruiz-Vicente, D.; Areces, F.; Puente, C.; Carril, J.C.; et al. ACTN3 genotype influences exercise-induced muscle damage during a marathon competition. Eur. J. Appl. Physiol. 2017, 117, 409–416. [Google Scholar] [CrossRef]
- Del Coso, J.; Valero, M.; Salinero, J.J.; Lara, B.; Gallo-Salazar, C.; Areces, F. Optimum polygenic profile to resist exertional rhabdomyolysis during a marathon. PLoS ONE 2017, 12, e0172965. [Google Scholar] [CrossRef]
- Del Coso, J.; Salinero, J.J.; Lara, B.; Gallo-Salazar, C.; Areces, F.; Puente, C.; Herrero, D. ACTN3 X-allele carriers had greater levels of muscle damage during a half-ironman. Eur J. Appl. Physiol. 2017, 117, 151–158. [Google Scholar] [CrossRef]
- Del Coso, J.; Salinero, J.J.; Lara, B.; Gallo-Salazar, C.; Areces, F.; Herrero, D.; Puente, C. Polygenic profile and exercise-induced muscle damage by a competitive half-ironman. J. Strength Cond. Res. 2018. [Google Scholar] [CrossRef]
- Collins, M.; Raleigh, S.M. Genetic risk factors for musculoskeletal soft tissue injuries. Med. Sport Sci. 2009, 54, 136–149. [Google Scholar] [PubMed]
- September, A.V.; Schwellnus, M.P.; Collins, M. Tendon and ligament injuries: The genetic component. Br. J. Sports Med. 2007, 41, 241–246. [Google Scholar] [CrossRef] [PubMed]
- MacArthur, D.G.; North, K.N. A gene for speed? the evolution and function of α-actinin-3. Bioessays 2004, 26, 786–795. [Google Scholar] [CrossRef] [PubMed]
- Vincent, B.; De Bock, K.; Ramaekers, M.; Van den Eede, E.; Van Leemputte, M.; Hespel, P.; Thomis, M.A. ACTN3 (R577X) genotype is associated with fiber type distribution. Physiol. Genom. 2007, 32, 58–63. [Google Scholar] [CrossRef] [PubMed]
- MacArthur, D.G.; Seto, J.T.; Chan, S.; Quinlan, K.G.; Raftery, J.M.; Turner, N.; Nicholson, M.D.; Kee, A.J.; Hardeman, E.C.; Gunning, P.W.; et al. An Actn3 knockout mouse provides mechanistic insights into the association between α-actinin-3 deficiency and human athletic performance. Hum. Mol. Genet. 2008, 17, 1076–1086. [Google Scholar] [CrossRef] [PubMed]
- Eynon, N.; Hanson, E.D.; Lucia, A.; Houweling, P.J.; Garton, F.; North, K.N.; Bishop, D.J. Genes for elite power and sprint performance: ACTN3 leads the way. Sports Med. 2013, 43, 803–817. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Yang, Y.; Li, X.; Zhou, F.; Gao, C.; Li, M.; Gao, L. The association of sport performance with ACE and ACTN3 genetic polymorphisms: A systematic review and meta-analysis. PLoS ONE 2013, 8, e54685. [Google Scholar] [CrossRef] [PubMed]
- Ahmetov, I.I.; Egorova, E.S.; Gabdrakhmanova, L.J.; Fedotovskaya, O.N. Genes and athletic performance: An update. Med. Sport Sci. 2016, 61, 41–54. [Google Scholar]
- Houweling, P.J.; Papadimitriou, I.D.; Seto, J.T.; Pérez, L.M.; Del Coso, J.; North, K.N.; Lucia, A.; Eynon, N. Is evolutionary loss our gain? The role of ACTN3 p. Arg577Ter (R577X) genotype in athletic performance, ageing, and disease. Hum. Mutat. 2018, 39, 1774–1787. [Google Scholar] [CrossRef]
- Yang, N.; MacArthur, D.G.; Gulbin, J.P.; Hahn, A.G.; Beggs, A.H.; Easteal, S.; North, K. ACTN3 genotype is associated with human elite athletic performance. Am. J. Hum. Genet. 2003, 73, 627–631. [Google Scholar] [CrossRef]
- Scott, R.A.; Irving, R.; Irwin, L.; Morrison, E.; Charlton, V.; Austin, K.; Tladi, D.; Deeason, M.; Headley, S.A.; Kolkhorst, F.W.; et al. ACTN3 and ACE genotypes in elite Jamaican and US sprinters. Med. Sci. Sports Exerc. 2010, 42, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Pickering, C.; Kiely, J. ACTN3: More than just a gene for speed. Front. Physiol. 2017, 8, 1080. [Google Scholar] [CrossRef] [PubMed]
- Del Coso, J.; Hiam, D.; Houweling, P.; Pérez, L.M.; Eynon, N.; Lucía, A. More than a ‘speed gene’: ACTN3 R577X genotype, trainability, muscle damage, and the risk for injuries. Eur. J. Appl. Physiol. 2019, 119, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Schindeler, A.; McDonald, M.M.; Seto, J.T.; Houweling, P.J.; Lek, M.; Hogarth, M.; Morse, A.R.; Raftery, J.M.; Balasuriya, D.; et al. α-Actinin-3 deficiency is associated with reduced bone mass in human and mouse. Bone 2011, 49, 790–798. [Google Scholar] [CrossRef] [PubMed]
- Delmonico, M.J.; Kostek, M.C.; Doldo, N.A.; Hand, B.D.; Walsh, S.; Conway, J.M.; Carignan, C.R.; Roth, S.M.; Hurley, B.F. Alpha-actinin-3 (ACTN3) R577X polymorphism influences knee extensor peak power response to strength training in older men and women. J. Gerontol. A Biol. Sci. Med. Sci. 2007, 62, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Wagle, J.P.; Carroll, K.M.; Cunanan, A.J.; Wetmore, A.; Taber, C.B.; DeWeese, B.H.; Sato, K.; Stuart, C.A.; Stone, M.H. Preliminary investigation into the effect of ACTN3 and ACE polymorphisms on muscle and performance characteristics. J. Strength Cond. Res. 2018. [Google Scholar] [CrossRef]
- Kikuchi, N.; Nakazato, K. Effective utilization of genetic information for athletes and coaches: Focus on ACTN3 R577X polymorphism. J. Exerc. Nutr. Biochem. 2015, 19, 157. [Google Scholar] [CrossRef]
- Karanikolou, A.; Wang, G.; Pitsiladis, Y. Letter to the editor: A genetic-based algorithm for personalized resistance training. Biol. Sport 2017, 34, 31. [Google Scholar] [CrossRef]
- Jones, N.; Kiely, J.; Suraci, B.; Collins, D.J.; De Lorenzo, D.; Pickering, C.; Grimaldi, K.A. A response to letter to the editor: A genetic-based algorithm for personalized resistance training. Biol. Sport 2017, 34, 35. [Google Scholar] [CrossRef]
- Monnerat-Cahli, G.; Paulúcio, D.; Neto, R.M.; Silva, R.; Pompeu, F.A.; Budowle, B.; Santos, C.G. Letter to the editor: Are the doors opened to a genetic-based algorithm for personalized resistance training? Biol. Sport 2017, 34, 27. [Google Scholar] [CrossRef]
- Ehlert, T.; Simon, P.; Moser, D.A. Epigenetics in sports. Sports Med. 2013, 43, 93–110. [Google Scholar] [CrossRef] [PubMed]
- Ling, C.; Rönn, T. Epigenetic adaptation to regular exercise in humans. Drug Discov. Today 2014, 19, 1015–1018. [Google Scholar] [CrossRef] [PubMed]
- Moran, C.N.; Pitsiladis, Y.P. Tour de France Champions born or made: Where do we take the genetics of performance? J. Sports Sci. 2017, 35, 1411–1419. [Google Scholar] [CrossRef] [PubMed]
- Widmann, M.; Nieß, A.M.; Munz, B. Physical exercise and epigenetic modifications in skeletal muscle. Sports Med. 2019, 49, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Voisin, S.; Eynon, N.; Yan, X.; Bishop, D.J. Exercise training and DNA methylation in humans. Acta Physiol. 2015, 213, 39–59. [Google Scholar] [CrossRef] [PubMed]
- Polakovičová, M.; Musil, P.; Laczo, E.; Hamar, D.; Kyselovič, J. Circulating microRNAs as potential biomarkers of exercise response. Int. J. Mol. Sci. 2016, 17, 1553. [Google Scholar] [CrossRef] [PubMed]
- Håkansson, K.E.; Sollie, O.; Simons, K.H.; Quax, P.H.; Jensen, J.; Nossent, A.Y. Circulating small noncoding RNAs as biomarkers for recovery after exhaustive or repetitive exercise. Front. Physiol. 2018, 9, 1136. [Google Scholar] [CrossRef]
- Ling, C.; Del Guerra, S.; Lupi, R.; Rönn, T.; Granhall, C.; Luthman, H.; Masiello, P.; Marchetti, P.; Groop, L.; Del Prato, S. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia 2008, 51, 615–622. [Google Scholar] [CrossRef]
- Pareja-Galeano, H.; Sanchis-Gomar, F.; García-Giménez, J.L. Physical exercise and epigenetic modulation: Elucidating intricate mechanisms. Sports Med. 2014, 44, 429–436. [Google Scholar] [CrossRef]
- Seaborne, R.A.; Strauss, J.; Cocks, M.; Shepherd, S.; O’Brien, T.D.; Someren, K.A.; Bell, P.G.; Murgatroyd, C.; Morton, J.P.; Stewart, C.E.; et al. Human Skeletal Muscle Possesses an Epigenetic Memory of Hypertrophy. Sci. Rep. 2018, 8, 1898. [Google Scholar] [CrossRef]
- McKinsey, T.A.; Zhang, C.L.; Olson, E.N. Control of muscle development by dueling HATs and HDACs. Curr. Opin. Genet. Devel. 2001, 11, 497–504. [Google Scholar] [CrossRef]
- Egan, B.; Zierath, J.R. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013, 17, 162–184. [Google Scholar] [CrossRef] [PubMed]
- Berdeaux, R.; Goebel, N.; Banaszynski, L.; Takemori, H.; Wandless, T.; Shelton, G.D.; Montminy, M. SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nat. Med. 2007, 13, 597. [Google Scholar] [CrossRef] [PubMed]
- Pandorf, C.E.; Haddad, F.; Wright, C.; Bodell, P.W.; Baldwin, K.M. Differential epigenetic modifications of histones at the myosin heavy chain genes in fast and slow skeletal muscle fibers and in response to muscle unloading. Am. J. Physiol. Cell Physiol. 2009, 297, C6–C16. [Google Scholar] [CrossRef] [PubMed]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- McCarthy, J.J.; Esser, K.A. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J. Appl. Physiol. 2007, 102, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Davidsen, P.K.; Gallagher, I.J.; Hartman, J.W.; Tarnopolsky, M.A.; Dela, F.; Helge, J.W.; Timmons, J.A.; Phillips, S.M. High responders to resistance exercise training demonstrate differential regulation of skeletal muscle microRNA expression. J. Appl. Physiol. 2010, 110, 309–317. [Google Scholar] [CrossRef]
- D’Souza, R.F.; Bjørnsen, T.; Zeng, N.; Aasen, K.M.; Raastad, T.; Cameron-Smith, D.; Mitchell, C.J. MicroRNAs in muscle: Characterizing the powerlifter phenotype. Front. Physiol. 2017, 8, 383. [Google Scholar] [CrossRef]
- Horak, M.; Zlamal, F.; Iliev, R.; Kucera, J.; Cacek, J.; Svobodova, L.; Hlavonova, Z.; Kalina, T.; Slaby, O.; Bienertova-Vasku, J. Exercise-induced circulating microRNA changes in athletes in various training scenarios. PLoS ONE 2018, 13, e0191060. [Google Scholar] [CrossRef]
- Nielsen, S.; Scheele, C.; Yfanti, C.; Åkerström, T.; Nielsen, A.R.; Pedersen, B.K.; Laye, M. Muscle specific microRNAs are regulated by endurance exercise in human skeletal muscle. J. Physiol. 2010, 588, 4029–4037. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.P.; Lamon, S.; Boon, H.; Wada, S.; Güller, I.; Brown, E.L.; Chibalin, A.V.; Zierath, J.R.; Snow, R.J.; Stepto, N.; et al. Regulation of miRNAs in human skeletal muscle following acute endurance exercise and short-term endurance training. J. Physiol. 2013, 591, 4637–4653. [Google Scholar] [CrossRef] [PubMed]
- Aoi, W.; Ichikawa, H.; Mune, K.; Tanimura, Y.; Mizushima, K.; Naito, Y.; Yoshikawa, T. Muscle-enriched microRNA miR-486 decreases in circulation in response to exercise in young men. Front. Physiol. 2013, 4, 80. [Google Scholar] [CrossRef] [PubMed]
- Domańska-Senderowska, D.; Jastrzębski, Z.; Kiszałkiewicz, J.; Brzeziański, M.; Pastuszak-Lewandoska, D.; Radzimińki, Ł.; Brzezianska-Lasota, E.; Jegier, A. Expression analysis of selected classes of circulating exosomal miRNAs in soccer players as an indicator of adaptation to physical activity. Biol. Sport 2017, 34, 331. [Google Scholar] [CrossRef] [PubMed]
- Bye, A.; Røsjø, H.; Aspenes, S.T.; Condorelli, G.; Omland, T.; Wisløff, U. Circulating microRNAs and aerobic fitness–the HUNT-Study. PLoS ONE 2013, 8, e57496. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, B.; Rolfes, F.; Schelleckes, K.; Mewes, M.; Thorwesten, L.; Krüger, M.; Klose, A.; Brand, S.M. Longer work/rest intervals during high-intensity interval training (HIIT) lead to elevated levels of miR-222 and miR-29c. Front. Physiol. 2018, 9, 395. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xiao, J.; Zhu, H.; Wei, X.; Platt, C.; Damilano, F.; Xiao, C.; Bezzerides, V.; Bostrom, P.; Che, L.; et al. miR-222 is necessary for exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell Metab. 2015, 21, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.; Sutherland, L.B.; Thatcher, J.E.; DiMaio, J.M.; Naseem, R.H.; Marshall, W.S.; Hill, J.A.; Olson, E.N. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13027–13032. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, B.; Schelleckes, K.; Nedele, J.; Thorwesten, L.; Klose, A.; Lenders, M.; Kruger, M.; Brand, E.; Brand, S.M. Dose-response of High-Intensity Training (HIT) on atheroprotective miRNA-126 levels. Front. Physiol. 2017, 8, 349. [Google Scholar] [CrossRef]
- Gomes, C.P.; Oliveira-Jr, G.P.; Madrid, B.; Almeida, J.A.; Franco, O.L.; Pereira, R.W. Circulating miR-1, miR-133a, and miR-206 levels are increased after a half-marathon run. Biomarkers 2014, 19, 585–589. [Google Scholar] [CrossRef]
- de Gonzalo-Calvo, D.; Dávalos, A.; Montero, A.; García-González, Á.; Tyshkovska, I.; González-Medina, A.; Soares, S.M.; Martínez-Camblor, P.; Casas-Agustench, P.; Rabadán, M.; et al. Circulating inflammatory miRNA signature in response to different doses of aerobic exercise. J. Appl. Physiol. 2015, 119, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Dávalos, A.; Goedeke, L.; Smibert, P.; Ramírez, C.M.; Warrier, N.P.; Andreo, U.; Cirera-Salinas, D.; Rayner, K.; Suresh, U.; Pastor-Pareja, J.C.; et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 9232–9237. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Garzon, R.; Sun, H.; Ladner, K.J.; Singh, R.; Dahlman, J.; Cheng, A.; Hall, B.M.; Qualman, S.J.; Chandler, D.S.; et al. NF-κB–YY1–miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 2008, 14, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Baggish, A.L.; Hale, A.; Weiner, R.B.; Lewis, G.D.; Systrom, D.; Wang, F.; Wang, T.J.; Chan, S.Y. Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. J. Physiol. 2011, 589, 3983–3994. [Google Scholar] [CrossRef] [PubMed]
- Zacharewicz, E.; Lamon, S.; Russell, A.P. MicroRNAs in skeletal muscle and their regulation with exercise, ageing, and disease. Front. Physiol. 2013, 4, 266. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Sanjurjo, M.; de Gonzalo-Calvo, D.; Fernández-García, B.; Díez-Robles, S.; Martínez-Canal, Á.; Olmedillas, H.; Davalos, A.; Iglesias-Gutierrez, E. Circulating microRNA as Emerging Biomarkers of Exercise. Exerc. Sport Sci. Rev. 2018, 46, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Mooren, F.C.; Viereck, J.; Krüger, K.; Thum, T. Circulating microRNAs as potential biomarkers of aerobic exercise capacity. Am. J. Physiol. Heart Circ. Physiol. 2013, 306, H557–H563. [Google Scholar] [CrossRef]
- Baggish, A.L.; Park, J.; Min, P.K.; Isaacs, S.; Parker, B.A.; Thompson, P.D.; Troyanos, C.; D’Hemecourt, P.; Dyer, S.; Thiel, M.; et al. Rapid upregulation and clearance of distinct circulating microRNAs after prolonged aerobic exercise. J. Appl. Physiol. 2014, 116, 522–531. [Google Scholar] [CrossRef]
- Clauss, S.; Wakili, R.; Hildebrand, B.; Kääb, S.; Hoster, E.; Klier, I.; Martens, E.; Hanley, A.; Hanssen, H.; Halle, M.; et al. MicroRNAs as biomarkers for acute atrial remodeling in marathon runners (The miRathon study—A sub-study of the Munich marathon study). PLoS ONE 2016, 11, e0148599. [Google Scholar] [CrossRef]
- Terruzzi, I.; Senesi, P.; Montesano, A.; La Torre, A.; Alberti, G.; Benedini, S.; Caumo, A.; Fermo, I.; Luzi, L. Genetic polymorphisms of the enzymes involved in DNA methylation and synthesis in elite athletes. Physiol. Genom. 2011, 43, 965–973. [Google Scholar] [CrossRef]
- Lokk, K.; Modhukur, V.; Rajashekar, B.; Märtens, K.; Mägi, R.; Kolde, R.; Koltsina, M.; Nilsson, T.K.; Vilo, J.; Salumets, A.; et al. DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome Biol. 2014, 15, 3248. [Google Scholar] [CrossRef] [PubMed]
- Langie, S.A.; Moisse, M.; Declerck, K.; Koppen, G.; Godderis, L.; Vanden Berghe, W.; Drury, S.; De Boever, P. Salivary DNA methylation profiling: Aspects to consider for biomarker identification. Basic Clin. Pharmacol. Toxicol. 2017, 121, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Breitbach, S.; Tug, S.; Simon, P. Circulating cell-free DNA. Sports Med. 2012, 42, 565–586. [Google Scholar] [CrossRef] [PubMed]
- Swarup, V.; Rajeswari, M.R. Circulating (cell-free) nucleic acids—A promising, non-invasive tool for early detection of several human diseases. FEBS Lett. 2007, 581, 795–799. [Google Scholar] [CrossRef] [PubMed]
- Atamaniuk, J.; Vidotto, C.; Tschan, H.; Bachl, N.; Stuhlmeier, K.M.; Müller, M.M. Increased concentrations of cell-free plasma DNA after exhaustive exercise. Clin. Chem. 2004, 50, 1668–1670. [Google Scholar] [CrossRef] [PubMed]
- Atamaniuk, J.; Stuhlmeier, K.M.; Vidotto, C.; Tschan, H.; Dossenbach-Glaninger, A.; Mueller, M.M. Effects of ultra-marathon on circulating DNA and mRNA expression of pro-and anti-apoptotic genes in mononuclear cells. Eur. J. Appl. Physiol. 2008, 104, 711–717. [Google Scholar] [CrossRef] [PubMed]
- Atamaniuk, J.; Vidotto, C.; Kinzlbauer, M.; Bachl, N.; Tiran, B.; Tschan, H. Cell-free plasma DNA and purine nucleotide degradation markers following weightlifting exercise. Eur. J. Appl. Physiol. 2010, 110, 695–701. [Google Scholar] [CrossRef] [PubMed]
- Fatouros, I.G.; Destouni, A.; Margonis, K.; Jamurtas, A.Z.; Vrettou, C.; Kouretas, D.; Mastorakos, G.; Mitrakou, A.; Taxildaris, K.; Kanavakis, E.; et al. Cell-free plasma DNA as a novel marker of aseptic inflammation severity related to exercise overtraining. Clin. Chem. 2006, 52, 1820–1824. [Google Scholar] [CrossRef] [PubMed]
- Andreatta, M.V.; Curty, V.M.; Coutinho, J.V.; Santos, M.Â.; Vassallo, P.F.; de Sousa, N.F.; Barauna, V.G. cfDNA as an Earlier Predictor of Exercise-Induced Performance Decrement Related to Muscle Damage. Int. J. Sports Physiol. Perform. 2018, 13, 953–956. [Google Scholar] [CrossRef]
- Haller, N.; Tug, S.; Breitbach, S.; Jörgensen, A.; Simon, P. Increases in circulating cell-free DNA during aerobic running depend on intensity and duration. Int. J. Sports Physiol. Perform. 2017, 12, 455–462. [Google Scholar] [CrossRef]
- Lewis, N.A.; Collins, D.; Pedlar, C.R.; Rogers, J.P. Can clinicians and scientists explain and prevent unexplained underperformance syndrome in elite athletes: An interdisciplinary perspective and 2016 update. BMJ Open Sport Exerc. Med. 2015, 1, e000063. [Google Scholar] [CrossRef] [PubMed]
- Haller, N.; Helmig, S.; Taenny, P.; Petry, J.; Schmidt, S.; Simon, P. Circulating, cell-free DNA as a marker for exercise load in intermittent sports. PLoS ONE 2018, 13, e0191915. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T.; et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014, 32, 834. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.J.; Chiu, C.C.; Li, Y.P.; Huang, W.C.; Te Huang, Y.; Huang, C.C.; Chuang, H.L. Effect of intestinal microbiota on exercise performance in mice. J. Strength Cond. Res. 2015, 29, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Marley, M.G.; Meganathan, R.; Bentley, R. Menaquinone (vitamin K2) biosynthesis in Escherichia coli: Synthesis of o-succinylbenzoate does not require the decarboxylase activity of the ketoglutarate dehydrogenase complex. Biochemistry 1986, 25, 1304–1307. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Stilling, R.M.; Dinan, T.G.; Cryan, J.F. Microbial genes, brain & behaviour-epigenetic regulation of the gut-brain axis. Genes Brain Behav. 2014, 13, 69–86. [Google Scholar] [PubMed]
- Clark, A.; Mach, N. Exercise-induced stress behavior, gut-microbiota-brain axis and diet: A systematic review for athletes. J. Int. Soc. Sports Nutr. 2016, 13, 43. [Google Scholar] [CrossRef]
- Mach, N.; Fuster-Botella, D. Endurance exercise and gut microbiota: A review. J. Sport Health Sci. 2017, 6, 179–197. [Google Scholar] [CrossRef]
- Clarke, S.F.; Murphy, E.F.; O’Sullivan, O.; Lucey, A.J.; Humphreys, M.; Hogan, A.; Hayes, P.; O’Reilly, M.; Jeffrey, I.B.; Wood-Martin, R.; et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut 2014, 63, 1913–1920. [Google Scholar] [CrossRef]
- Moreno-Pérez, D.; Bressa, C.; Bailén, M.; Hamed-Bousdar, S.; Naclerio, F.; Carmona, M.; Perez, M.; Gonzalez-Soltero, R.; Montalvo-Lominchar, M.G.; Carabana, C.; et al. Effect of a protein supplement on the gut microbiota of endurance athletes: A randomized, controlled, double-blind pilot study. Nutrients 2018, 10, 337. [Google Scholar] [CrossRef] [PubMed]
- Chassard, C.; Lacroix, C. Carbohydrates and the human gut microbiota. Curr. Opin. Clin. Nutr. Metabol. Care 2013, 16, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Navarro, M.; Muñoz, G.; Salinero, J.J.; Muñoz-Guerra, J.; Fernández-Álvarez, M.; Plata, M.D.; Del Coso, J. Urine caffeine concentration in doping control samples from 2004 to 2015. Nutrients 2019, 11, 286. [Google Scholar] [CrossRef] [PubMed]
- Grgic, J.; Grgic, I.; Pickering, C.; Schoenfeld, B.; Bishop, D.; Pedisic, Z. Wake up and smell the coffee: Caffeine supplementation and exercise performance—an umbrella review of 21 published meta-analyses. Br. J. Sports Med. 2019. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Gonzalez, F.J.; Kalow, W.; Tang, B.K. Biotransformation of caffeine, paraxanthine, theobromine and theophylline by cDNA-expressed human CYP1A2 and CYP2E1. Pharmacogenetics 1992, 2, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Sachse, C.; Brockmöller, J.; Bauer, S.; Roots, I. Functional significance of a C→ A polymorphism in intron 1 of the cytochrome P450 CYP1A2 gene tested with caffeine. Br. J. Clin. Pharmacol. 1999, 47, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Fulton, J.; Dinas, P.; Carrillo, A.; Edsall, J.; Ryan, E.; Ryan, E. Impact of genetic variability on physiological responses to caffeine in humans: A systematic review. Nutrients 2018, 10, 1373. [Google Scholar] [CrossRef]
- Womack, C.J.; Saunders, M.J.; Bechtel, M.K.; Bolton, D.J.; Martin, M.; Luden, N.D.; Dunham, W.; Hancock, M. The influence of a CYP1A2 polymorphism on the ergogenic effects of caffeine. J. Int. Soc. Sports Nutr. 2012, 9, 7. [Google Scholar] [CrossRef]
- Guest, N.; Corey, P.; Vescovi, J.; El-Sohemy, A. Caffeine, CYP1A2 genotype, and endurance performance in athletes. Med. Sci. Sports Exerc. 2018, 50, 1570–1578. [Google Scholar] [CrossRef]
- Loy, B.D.; O’Connor, P.J.; Lindheimer, J.B.; Covert, S.F. Caffeine is ergogenic for adenosine A2A receptor gene (ADORA2A) T allele homozygotes: A pilot study. J. Caffeine Res. 2015, 5, 73–81. [Google Scholar] [CrossRef]
- Cornelis, M.C.; El-Sohemy, A.; Campos, H. Genetic polymorphism of the adenosine A2A receptor is associated with habitual caffeine consumption. Am. J. Clin. Nutr. 2007, 86, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Alsene, K.; Deckert, J.; Sand, P.; de Wit, H. Association between A 2a receptor gene polymorphisms and caffeine-induced anxiety. Neuropsychopharmacology 2003, 28, 1694. [Google Scholar] [CrossRef] [PubMed]
- Byrne, E.M.; Johnson, J.; McRae, A.F.; Nyholt, D.R.; Medland, S.E.; Gehrman, P.R.; Heath, A.C.; Madden, P.A.; Montgomery, G.W.; Chenevix-Trench, G.; et al. A genome-wide association study of caffeine-related sleep disturbance: Confirmation of a role for a common variant in the adenosine receptor. Sleep 2012, 35, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Heibel, A.B.; Perim, P.H.; Oliveira, L.F.; McNaughton, L.R.; Saunders, B. Time to optimize supplementation: Modifying factors influencing the individual responses to extracellular buffering agents. Front. Nutr. 2018, 5, 35. [Google Scholar] [CrossRef]
- Pickering, C. Caffeine, CYP1A2 genotype, and sports performance: Is timing important? Ir. J. Med. Sci. 2019, 188, 349–350. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Pasquali, C.; Appel, R.D.; Ou, K.; Golaz, O.; Sanchez, J.C.; Yan, J.X.; Gooley, A.A.; Hughes, G.; Humphery-Smith, I.; et al. From proteins to proteomes: Large scale protein identification by two-dimensional electrophoresis and amino acid analysis. Biotechnology 1996, 14, 61. [Google Scholar] [CrossRef] [PubMed]
- Timmons, J.A. Variability in training-induced skeletal muscle adaptation. J. Appl. Physiol. 2011, 110, 846–853. [Google Scholar] [CrossRef]
- Petriz, B.A.; Gomes, C.P.; Rocha, L.A.; Rezende, T.M.; Franco, O.L. Proteomics applied to exercise physiology: A cutting-edge technology. J. Cell Physiol. 2012, 227, 885–898. [Google Scholar] [CrossRef]
- Wittwer, M.; Billeter, R.; Hoppeler, H.; Flück, M. Regulatory gene expression in skeletal muscle of highly endurance-trained humans. Acta Physiol. Scand. 2004, 180, 217–227. [Google Scholar] [CrossRef]
- Stepto, N.K.; Coffey, V.G.; Carey, A.L.; Ponnampalam, A.P.; Canny, B.J.; Powell, D.; Hawley, J.A. Global gene expression in skeletal muscle from well-trained strength and endurance athletes. Med. Sci. Sports Exerc. 2009, 41, 546–565. [Google Scholar] [CrossRef]
- Burniston, J.G.; Hoffman, E.P. Proteomic responses of skeletal and cardiac muscle to exercise. Expert Rev. Proteom. 2011, 8, 361–377. [Google Scholar] [CrossRef] [PubMed]
- Islam, H.; Edgett, B.A.; Bonafiglia, J.T.; Shulman, T.; Ma, A.; Quadrilatero, J.; Simpson, C.A.; Gurd, B.J. Repeatability of exercise-induced changes in mRNA expression and technical considerations for qPCR analysis in human skeletal muscle. Exp. Physiol. 2019, 104, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Starnes, J.W.; Parry, T.L.; O’Neal, S.K.; Bain, J.R.; Muehlbauer, M.J.; Honcoop, A.; Ilaiwy, A.; Christopher, P.M.; Patterson, C.; Willis, M.S. Exercise-induced alterations in skeletal muscle, heart, liver, and serum metabolome identified by non-targeted metabolomics analysis. Metabolites 2017, 7, 40. [Google Scholar] [CrossRef] [PubMed]
- Friso, S.; Choi, S.W.; Girelli, D.; Mason, J.B.; Dolnikowski, G.G.; Bagley, P.J.; Oliveri, O.; Jacques, P.F.; Rosenberg, I.H.; Corrocher, R.; et al. A common mutation in the 5, 10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc. Natl. Acad. Sci. USA 2002, 99, 5606–5611. [Google Scholar] [CrossRef] [PubMed]
- Alves, R.D.; Dane, A.D.; Harms, A.; Strassburg, K.; Seifar, R.M.; Verdijk, L.B.; Kersten, S.; Berger, R.; Hankemeir, T.; Vreeken, R.J. Global profiling of the muscle metabolome: Method optimization, validation and application to determine exercise-induced metabolic effects. Metabolomics 2015, 11, 271–285. [Google Scholar] [CrossRef]
- Wing, C. Designing pre-season training programs using global positioning systems: A systematic approach. Strength Cond. J. 2018. [Google Scholar] [CrossRef]
- Sperlich, B.; Düking, P.; Holmberg, H.C. A SWOT analysis of the use and potential misuse of implantable monitoring devices by athletes. Front. Physiol. 2017, 8, 629. [Google Scholar] [CrossRef]
- McGuigan, M.R.; Cormack, S.J.; Gill, N.D. Strength and power profiling of athletes: Selecting tests and how to use the information for program design. Strength Cond. J. 2013, 35, 7–14. [Google Scholar] [CrossRef]
- Düking, P.; Hotho, A.; Holmberg, H.C.; Fuss, F.K.; Sperlich, B. Comparison of non-invasive individual monitoring of the training and health of athletes with commercially available wearable technologies. Front. Physiol. 2016, 7, 71. [Google Scholar] [CrossRef]
- Düking, P.; Achtzehn, S.; Holmberg, H.C.; Sperlich, B. Integrated framework of load monitoring by a combination of smartphone applications, wearables and point-of-care testing provides feedback that allows individual responsive adjustments to activities of daily living. Sensors 2018, 18, 1632. [Google Scholar] [CrossRef]
- Düking, P.; Holmberg, H.C.; Sperlich, B. Instant biofeedback provided by wearable sensor technology can help to optimize exercise and prevent injury and overuse. Front. Physiol. 2017, 8, 167. [Google Scholar] [CrossRef] [PubMed]
- Haugen, T.A.; Breitschädel, F.; Samozino, P. Power-force-velocity profiling of sprinting athletes: Methodological and practical considerations when using timing gates. J. Strength Cond. Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Mero, A.; Komi, P.V. Force-, EMG-, and elasticity-velocity relationships at submaximal, maximal and supramaximal running speeds in sprinters. Eur. J. Appl. Physiol. Occup. Physiol. 1986, 55, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Nagahara, R.; Morin, J.B. Sensor insole for measuring temporal variables and vertical force during sprinting. Proc. Inst. Mech. Eng. Part P J. Sports Eng. Technol. 2018, 232, 369–374. [Google Scholar] [CrossRef]
- Romero-Franco, N.; Jiménez-Reyes, P.; Castaño-Zambudio, A.; Capelo-Ramírez, F.; Rodríguez-Juan, J.J.; González-Hernández, J.; Toscano-Bendala, F.J.; Cuadrado-Penafiel, V.; Balsalobre-Fernandez, C. Sprint performance and mechanical outputs computed with an iPhone app: Comparison with existing reference methods. Eur. J. Sport Sci. 2017, 17, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Cross, M.R.; Lahti, J.; Brown, S.R.; Chedati, M.; Jimenez-Reyes, P.; Samozino, P.; Eriksrud, O.; Morin, J.B. Training at maximal power in resisted sprinting: Optimal load determination methodology and pilot results in team sport athletes. PLoS ONE 2018, 13, e0195477. [Google Scholar] [CrossRef]
- Bezodis, I.N.; Kerwin, D.G.; Salo, A.I. Lower-limb mechanics during the support phase of maximum-velocity sprint running. Med. Sci. Sports Exerc. 2008, 40, 707–715. [Google Scholar] [CrossRef]
- Dellaserra, C.L.; Gao, Y.; Ransdell, L. Use of integrated technology in team sports: A review of opportunities, challenges, and future directions for athletes. J. Strength Cond. Res. 2014, 28, 556–573. [Google Scholar] [CrossRef]
- Marin, F.; Fradet, L.; Lepetit, K.; Hansen, C.; Mansour, K.B. Inertial measurement unit in biomechanics and sport biomechanics: Past, present, future. SBS Conf. Proc. Arch. 2016, 33, 1422–1444. [Google Scholar]
- Morin, J.B.; Samozino, P. Interpreting power-force-velocity profiles for individualized and specific training. Int. J. Sports Physiol. Perform. 2016, 11, 267–272. [Google Scholar] [CrossRef]
- Cross, M.R.; Brughelli, M.; Samozino, P.; Brown, S.R.; Morin, J.B. Optimal loading for maximizing power during sled-resisted sprinting. Int. J. Sports Physiol. Perform. 2017, 12, 1069–1077. [Google Scholar] [CrossRef] [PubMed]
- Halson, S.L. Monitoring training load to understand fatigue in athletes. Sports Med. 2014, 44, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Campbell, B.I.; Bove, D.; Ward, P.; Vargas, A.; Dolan, J. Quantification of training load and training response for improving athletic performance. Strength Cond. J. 2017, 39, 3–13. [Google Scholar] [CrossRef]
- Sands, W.A.; Kavanaugh, A.A.; Murray, S.R.; McNeal, J.R.; Jemni, M. Modern techniques and technologies applied to training and performance monitoring. Int. J. Sports Physiol. Perform. 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, M. Physiological limits to exercise performance in the heat. J. Sci. Med. Sport 2008, 11, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Born, D.P.; Hoppe, M.W.; Lindner, N.; Freiwald, J.; Holmberg, H.C.; Sperlich, B. Adaptive mechanisms and behavioural recommendations: Playing football in heat, cold and high altitude conditions. Sportverletz Sportschaden 2014, 28, 17–23. [Google Scholar] [PubMed]
- Gabbett, T.J. Quantifying the physical demands of collision sports: Does microsensor technology measure what it claims to measure? J. Strength Cond. Res. 2013, 27, 2319–2322. [Google Scholar] [CrossRef]
- Blanch, P.; Gabbett, T.J. Has the athlete trained enough to return to play safely? The acute: Chronic workload ratio permits clinicians to quantify a player’s risk of subsequent injury. Br. J. Sports Med. 2016, 50, 471–475. [Google Scholar] [CrossRef]
- Gabbett, T.J.; Hulin, B.T.; Blanch, P.; Whiteley, R. High training workloads alone do not cause sports injuries: How you get there is the real issue. Br. J. Sports Med. 2016, 50, 444. [Google Scholar] [CrossRef]
- Hulin, B.T.; Gabbett, T.J.; Caputi, P.; Lawson, D.W.; Sampson, J.A. Low chronic workload and the acute: Chronic workload ratio are more predictive of injury than between-match recovery time: A two-season prospective cohort study in elite rugby league players. Br. J. Sports Med. 2016, 50, 1008–1012. [Google Scholar] [CrossRef]
- Hulin, B.T.; Gabbett, T.J.; Lawson, D.W.; Caputi, P.; Sampson, J.A. The acute: Chronic workload ratio predicts injury: High chronic workload may decrease injury risk in elite rugby league players. Br. J. Sports Med. 2016, 50, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Medina, L.; González-Badillo, J.J. Velocity loss as an indicator of neuromuscular fatigue during resistance training. Med. Sci. Sports Exerc. 2011, 43, 1725–1734. [Google Scholar] [CrossRef] [PubMed]
- Jovanović, M.; Flanagan, E.P. Researched applications of velocity based strength training. J. Aust. Strength Cond. 2014, 22, 58–69. [Google Scholar]
- Gathercole, R.; Sporer, B.; Stellingwerff, T.; Sleivert, G. Alternative countermovement-jump analysis to quantify acute neuromuscular fatigue. Int. J. Sports Physiol. Perform. 2015, 10, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Plews, D.J.; Laursen, P.B.; Stanley, J.; Kilding, A.E.; Buchheit, M. Training adaptation and heart rate variability in elite endurance athletes: Opening the door to effective monitoring. Sports Med. 2013, 43, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Plews, D.J.; Laursen, P.B.; Meur, Y.L.; Hausswirth, C.; Kilding, A.E.; Buchheit, M. Monitoring training with heart-rate variability: How much compliance is needed for valid assessment? Int. J. Sports Physiol. Perform. 2014, 9, 783–790. [Google Scholar] [CrossRef]
- Buchheit, M. Monitoring training status with HR measures: Do all roads lead to Rome? Front. Physiol. 2014, 5, 73. [Google Scholar] [CrossRef]
- Sperlich, B.; Holmberg, H.C. The responses of elite athletes to exercise: An all-day, 24-h integrative view is required! Front. Physiol. 2017, 8, 564. [Google Scholar] [CrossRef]
- Leeder, J.; Glaister, M.; Pizzoferro, K.; Dawson, J.; Pedlar, C. Sleep duration and quality in elite athletes measured using wristwatch actigraphy. J. Sports Sci. 2012, 30, 541–545. [Google Scholar] [CrossRef]
- Bird, S.P. Sleep, recovery, and athletic performance: A brief review and recommendations. Strength Cond. J. 2013, 35, 43–47. [Google Scholar] [CrossRef]
- Ferrie, J.E.; Shipley, M.J.; Akbaraly, T.N.; Marmot, M.G.; Kivimäki, M.; Singh-Manoux, A. Change in sleep duration and cognitive function: Findings from the Whitehall II Study. Sleep 2011, 34, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Irwin, M.; McClintick, J.; Costlow, C.; Fortner, M.; White, J.; Gillin, J.C. Partial night sleep deprivation reduces natural killer and cellular immune responses in humans. FASEB J. 1996, 10, 643–653. [Google Scholar] [CrossRef] [PubMed]
- Kampakis, S. Predictive modelling of football injuries. arXiv 2016, arXiv:1609.07480. [Google Scholar]
- Larruskain, J.; Celorrio, D.; Barrio, I.; Odriozola, A.; Gil, S.M.; Fernandez-Lopez, J.R.; Nozal, R.; Ortuzar, I.; Lekue, J.A.; Aznar, J.K. Genetic variants and hamstring injury in soccer: An association and validation study. Med. Sci. Sports Exerc. 2018, 50, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Kampakis, S. Comparison of Machine Learning Methods for Predicting the Recovery Time of Professional Football Players after an Undiagnosed Injury. Ph.D.Thesis, University College London, London, UK, 2013. [Google Scholar]
- Borisov, O.; Kulemin, N.; Ahmetov, I.; Generozov, E. A novel multilocus genetic model can predict muscle fibers composition. In Proceedings of the 11th International Symposium on Computer Science in Sport (IACSS 2017); Lames, M., Saupe, D., Wiemeyer, J., Eds.; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Gonzalez, K.; Sasangohar, F.; Mehta, R.K.; Lawley, M.; Erraguntla, M. Measuring fatigue through Heart Rate Variability and activity recognition: A scoping literature review of machine learning techniques. Proc. Hum. Factors Ergon. Soc. Ann Meet. 2017, 61, 1748–1752. [Google Scholar] [CrossRef]
- Vandewiele, G.; Geurkink, Y.; Lievens, M.; Ongenae, F.; De Turck, F.; Boone, J. Enabling training personalization by predicting the session rate of perceived exertion (sRPE). In Proceedings of the Machine Learning and Data Mining for Sports Analytics ECML/PKDD 2017 Workshop, Skopje, Macedonia, 18 September 2017; Available online: https://biblio.ugent.be/publication/8537058 (accessed on 30 September 2017).
- McCullagh, J. Data mining in sport: A neural network approach. Int. J. Sports Sci. Eng. 2010, 4, 131–138. [Google Scholar]
- Mezyk, E.; Unold, O. Machine learning approach to model sport training. Comput. Hum. Behav. 2011, 27, 1499–1506. [Google Scholar] [CrossRef]
- Fister, I.; Ljubič, K.; Suganthan, P.N.; Perc, M.; Fister, I. Computational intelligence in sports: Challenges and opportunities within a new research domain. Appl. Math. Comput. 2015, 262, 178–186. [Google Scholar] [CrossRef]
- Ofoghi, B.; Zeleznikow, J.; MacMahon, C.; Raab, M. Data mining in elite sports: A review and a framework. Meas. Phys. Educ. Exerc. Sci. 2013, 17, 171–186. [Google Scholar] [CrossRef]
- Ripatti, S.; Tikkanen, E.; Orho-Melander, M.; Havulinna, A.S.; Silander, K.; Sharma, A.; Guiducci, C.; Perola, M.; Jula, A.; Sinisalo, J.; et al. A multilocus genetic risk score for coronary heart disease: Case-control and prospective cohort analyses. Lancet 2010, 376, 1393–1400. [Google Scholar] [CrossRef]
- Khera, A.V.; Chaffin, M.; Aragam, K.G.; Haas, M.E.; Roselli, C.; Choi, S.H.; Natarajan, P.; Lander, E.S.; Lubitz, S.A.; Ellinor, P.T.; et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018, 50, 1219. [Google Scholar] [CrossRef] [PubMed]
- Sajda, P. Machine learning for detection and diagnosis of disease. Annu. Rev. Biomed. Eng. 2006, 8, 537–565. [Google Scholar] [CrossRef] [PubMed]
- Delen, D.; Walker, G.; Kadam, A. Predicting breast cancer survivability: A comparison of three data mining methods. Artif. Intell. Med. 2005, 34, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, K.; Rani, B.K.; Govrdhan, A. Applications of data mining techniques in healthcare and prediction of heart attacks. Int. J. Comput. Sci. Eng. 2010, 2, 250–255. [Google Scholar]
- Soni, J.; Ansari, U.; Sharma, D.; Soni, S. Predictive data mining for medical diagnosis: An overview of heart disease prediction. Int. J. Comput. Appl. 2011, 17, 43–48. [Google Scholar] [CrossRef]
- Kraft, J.A.; Laurent, M.L.; Green, J.M.; Helm, J.; Roberts, C.; Holt, S. Examination of coach and player perceptions of recovery and exertion. J. Strength Cond. Res. 2018. [Google Scholar] [CrossRef]
- Jacob, Y.; Cripps, A.; Evans, T.; Chivers, P.T.; Joyce, C.; Anderton, R.S. Identification of genetic markers for skill and athleticism in sub-elite Australian football players: A pilot study. J. Sports Med. Phys. Fit. 2018, 58, 241–248. [Google Scholar]
- Leźnicka, K.; Niewczas, M.; Kurzawski, M.; Cięszczyk, P.; Safranow, K.; Ligocka, M.; Białecka, M. The association between COMT rs4680 and OPRM1 rs1799971 polymorphisms and temperamental traits in combat athletes. Personal. Individ. Diff. 2018, 124, 105–110. [Google Scholar] [CrossRef]
- Ashfield-Watt, P.A.; Pullin, C.H.; Whiting, J.M.; Clark, Z.E.; Moat, S.J.; Newcombe, R.G.; Burr, M.L.; Lewis, M.J.; Powers, H.J.; McDowell, I.F. Methylenetetrahydrofolate reductase 677C→ T genotype modulates homocysteine responses to a folate-rich diet or a low-dose folic acid supplement: A randomized controlled trial. Am. J. Clin. Nutr. 2002, 76, 180–186. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human genetics shape the gut microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef]
- Varley, I.; Hughes, D.C.; Greeves, J.P.; Stellingwerff, T.; Ranson, C.; Fraser, W.D.; Sale, C. The association of novel polymorphisms with stress fracture injury in Elite Athletes: Further insights from the SFEA cohort. J. Sci. Med. Sport 2018, 21, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Cust, E.E.; Sweeting, A.J.; Ball, K.; Robertson, S. Machine and deep learning for sport-specific movement recognition: A systematic review of model development and performance. J. Sports Sci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Timmons, J.A.; Knudsen, S.; Rankinen, T.; Koch, L.G.; Sarzynski, M.; Jensen, T.; Keller, P.; Scheele, C.; Vollaard, N.B.; Nielsen, S.; et al. Using molecular classification to predict gains in maximal aerobic capacity following endurance exercise training in humans. J. Appl. Physiol. 2011, 108, 1487–1496. [Google Scholar] [CrossRef] [PubMed]
- Kiely, J. Periodization theory: Confronting an inconvenient truth. Sports Med. 2018, 48, 753–764. [Google Scholar] [CrossRef]
- Gaffney-Stomberg, E.; Lutz, L.J.; Shcherbina, A.; Ricke, D.O.; Petrovick, M.; Cropper, T.L.; Cable, S.J.; McClung, J.P. Association between single gene polymorphisms and bone biomarkers and response to calcium and vitamin D supplementation in young adults undergoing military training. J. Bone Min. Res. 2017, 32, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Westerman, K.; Reaver, A.; Roy, C.; Ploch, M.; Sharoni, E.; Nogal, B.; Sinclair, D.A.; Katz, D.L.; Blumberg, J.B.; Blander, G. Longitudinal analysis of biomarker data from a personalized nutrition platform in healthy subjects. Sci. Rep. 2018, 8, 14685. [Google Scholar] [CrossRef]
- Lindsay, A.; Costello, J.T. Realising the potential of urine and saliva as diagnostic tools in sport and exercise medicine. Sports Med. 2017, 47, 11–31. [Google Scholar] [CrossRef]
- Goodlin, G.T.; Roos, A.K.; Roos, T.R.; Hawkins, C.; Beache, S.; Baur, S.; Kim, S.K. Applying personal genetic data to injury risk assessment in athletes. PLoS ONE 2015, 10, e0122676. [Google Scholar] [CrossRef]
- Carey, D.L.; Crow, J.; Ong, K.L.; Blanch, P.; Morris, M.E.; Dascombe, B.J.; Crossley, K.M. Optimizing preseason training loads in Australian Football. Int. J. Sports Physiol. Perform. 2018, 13, 194–199. [Google Scholar] [CrossRef]
- Bartlett, J.D.; O’Connor, F.; Pitchford, N.; Torres-Ronda, L.; Robertson, S.J. Relationships between internal and external training load in team-sport athletes: Evidence for an individualized approach. Int. J. Sports Physiol. Perform. 2017, 12, 230–234. [Google Scholar] [CrossRef]
- Jiménez-Reyes, P.; Samozino, P.; Brughelli, M.; Morin, J.B. Effectiveness of an individualized training based on force-velocity profiling during jumping. Front. Physiol. 2017, 7, 677. [Google Scholar] [CrossRef] [PubMed]
- Vesterinen, V.; Nummela, A.; Heikura, I.; Laine, T.; Hynynen, E.; Botella, J.; Häkkinen, K. Individual endurance training prescription with heart rate variability. Med. Sci. Sports Exerc. 2016, 48, 1347–1354. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pickering, C.; Kiely, J. The Development of a Personalised Training Framework: Implementation of Emerging Technologies for Performance. J. Funct. Morphol. Kinesiol. 2019, 4, 25. https://doi.org/10.3390/jfmk4020025
Pickering C, Kiely J. The Development of a Personalised Training Framework: Implementation of Emerging Technologies for Performance. Journal of Functional Morphology and Kinesiology. 2019; 4(2):25. https://doi.org/10.3390/jfmk4020025
Chicago/Turabian StylePickering, Craig, and John Kiely. 2019. "The Development of a Personalised Training Framework: Implementation of Emerging Technologies for Performance" Journal of Functional Morphology and Kinesiology 4, no. 2: 25. https://doi.org/10.3390/jfmk4020025
APA StylePickering, C., & Kiely, J. (2019). The Development of a Personalised Training Framework: Implementation of Emerging Technologies for Performance. Journal of Functional Morphology and Kinesiology, 4(2), 25. https://doi.org/10.3390/jfmk4020025




