Aerobic Exercise Induces Alternative Splicing of Neurexins in Frontal Cortex
Abstract
1. Introduction
1.1. Beneficial Effects of Aerobic Exercise on Brain Health
1.2. Role of Neurexins in Neuroplasticity
2. Materials and Methods
2.1. Animal Care and Ethics Statement
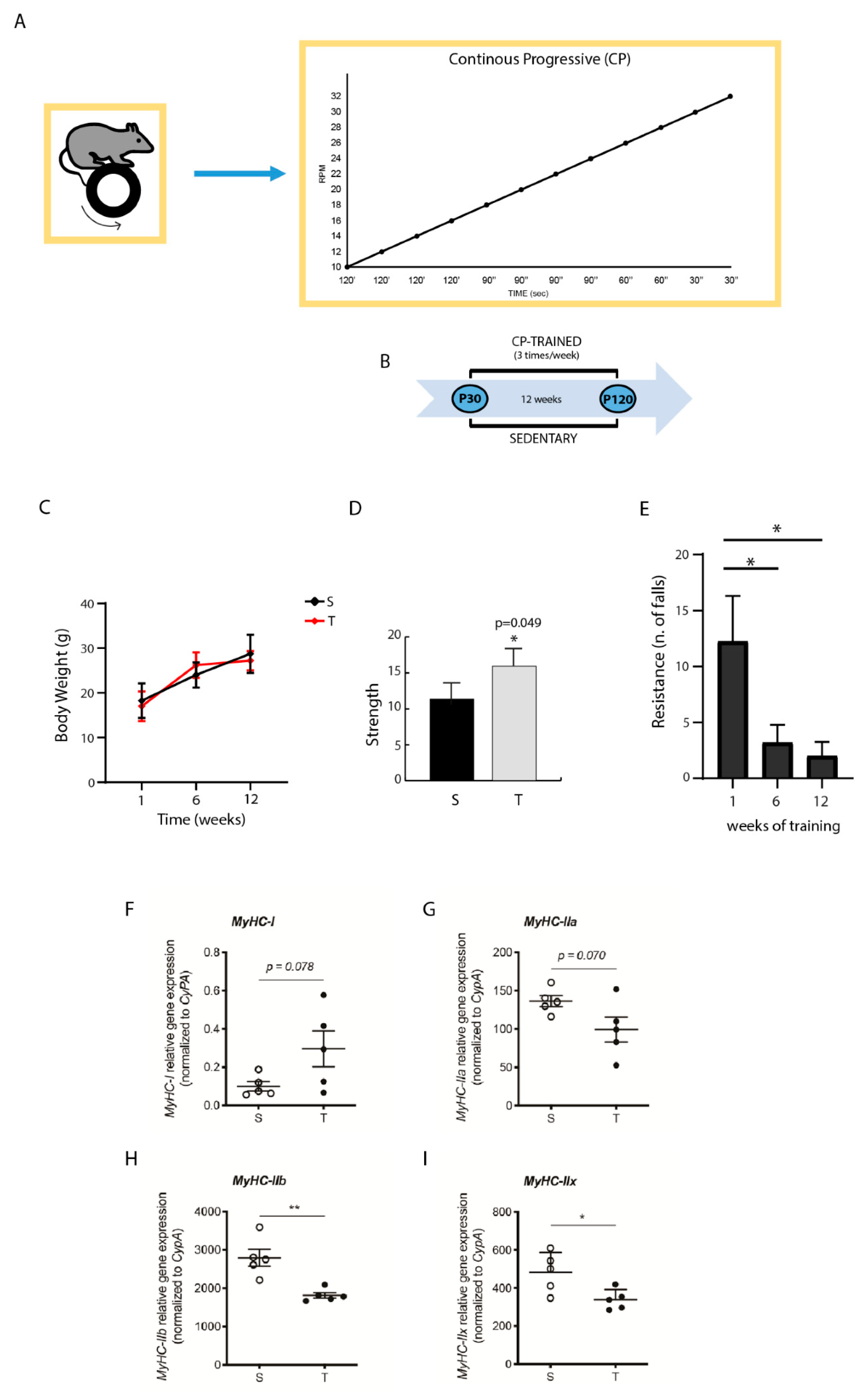
2.2. Training System and Schedule
2.3. Animals Assessment
2.4. RT-PCR Analysis and Quantitative Real-Time PCR
2.5. Statistical Analysis
3. Results
3.1. Effect of CP AE on Body Weight, Strength, and Resistance
3.2. CP AE Affects Muscle MyHC Isoform Expression
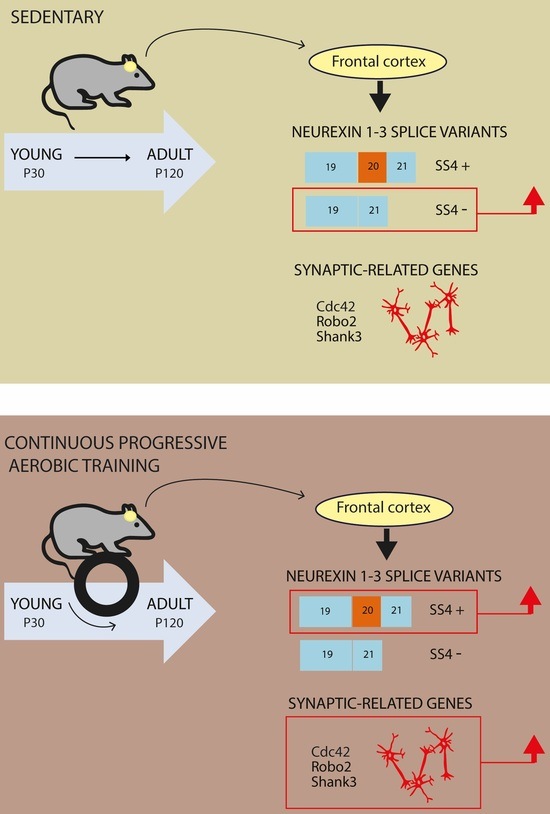
3.3. CP AE Modulates Neurexin Splicing in FC
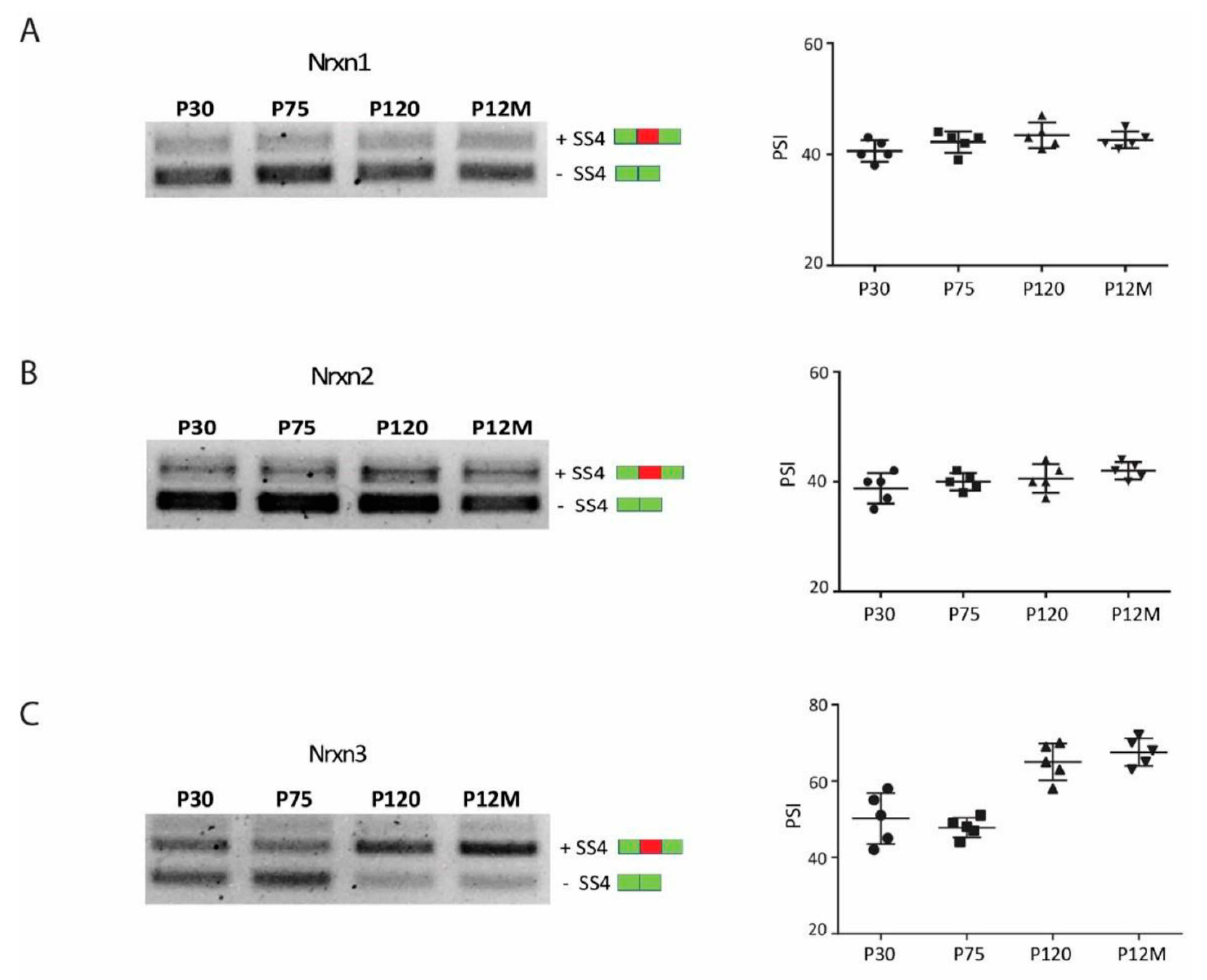
3.4. Neurexin Alternative Splicing in Postnatal Frontal Cortex
3.5. CP AE Modulates Expression of Genes Involved in Synaptic Functions in FC
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rovio, S.; Spulber, G.; Nieminen, L.J.; Niskanen, E.; Winblad, B.; Tuomilehto, J.; Nissinen, A.; Soininen, H.; Kivipelto, M. The effect of midlife physical activity on structural brain changes in the elderly. Neurobiol. Aging 2010, 31, 1927–1936. [Google Scholar] [CrossRef]
- Voss, M.W.; Heo, S.; Prakash, R.S.; Erickson, K.I.; Alves, H.; Chaddock, L.; Szabo, A.N.; Mailey, E.L.; Wójcicki, T.R.; White, S.M.; et al. The influence of aerobic fitness on cerebral white matter integrity and cognitive function in older adults: Results of a one-year exercise intervention. Hum. Brain Mapp. 2013, 34, 2972–2985. [Google Scholar] [CrossRef] [PubMed]
- Voss, M.W.; Vivar, C.; Kramer, A.F.; van Praag, H. Bridging animal and human models of exercise-induced brain plasticity. Trends Cogn. Sci. 2013, 17, 525–544. [Google Scholar] [CrossRef]
- Gelfo, F.; Mandolesi, L.; Serra, L.; Sorrentino, G.; Caltagirone, C. The Neuroprotective Effects of Experience on Cognitive Functions: Evidence from Animal Studies on the Neurobiological Bases of Brain Reserve. Neuroscience 2018, 370, 218–235. [Google Scholar] [CrossRef] [PubMed]
- Mandolesi, L.; Polverino, A.; Montuori, S.; Foti, F.; Ferraioli, G.; Sorrentino, P.; Sorrentino, G. Effects of Physical Exercise on Cognitive Functioning and Wellbeing: Biological and Psychological Benefits. Front. Psychol. 2018, 9, 509. [Google Scholar] [CrossRef]
- Kramer, A.F.; Hahn, S.; Cohen, N.J.; Banich, M.T.; McAuley, E.; Harrison, C.R.; Chason, J.; Vakil, E.; Bardell, L.; Boileau, R.A.; et al. Ageing, fitness and neurocognitive function. Nature 1999, 400, 418–419. [Google Scholar] [CrossRef]
- Colcombe, S.; Kramer, A.F. Fitness effects on the cognitive function of older adults: A meta-analytic study. Psychol. Sci. 2003, 14, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Black, J.E.; Isaacs, K.R.; Anderson, B.J.; Alcantara, A.A.; Greenough, W.T. Learning causes synaptogenesis, whereas motor activity causes angiogenesis, in cerebellar cortex of adult rats. Proc. Natl. Acad. Sci. USA 1990, 87, 5568–5572. [Google Scholar] [CrossRef] [PubMed]
- Cotman, C.W.; Berchtold, N.C. Exercise: A behavioral intervention to enhance brain health and plasticity. Trends Neurosci. 2002, 25, 295–301. [Google Scholar] [CrossRef]
- Griffin, É.W.; Mullally, S.; Foley, C.; Warmington, S.A.; O’Mara, S.M.; Kelly, A.M. Aerobic exercise improves hippocampal function and increases BDNF in the serum of young adult males. Physiol. Behav. 2011, 104, 934–941. [Google Scholar] [CrossRef] [PubMed]
- Lea-Carnall, C.A.; Trujillo-Barreto, N.J.; Montemurro, M.A.; El-Deredy, W.; Parkes, L.M. Evidence for frequency-dependent cortical plasticity in the human brain. Proc. Natl. Acad. Sci. USA 2017, 114, 8871–8876. [Google Scholar] [CrossRef] [PubMed]
- Craig, A.M.; Kang, Y. Neurexin-neuroligin signaling in synapse development. Curr. Opin. Neurobiol. 2007, 17, 43–52. [Google Scholar] [CrossRef]
- Südhof, T.C. Neuroligins and neurexins link synaptic function to cognitive disease. Nature 2008, 455, 903–911. [Google Scholar] [CrossRef] [PubMed]
- Krueger, D.D.; Tuffy, L.P.; Papadopoulos, T.; Brose, N. The role of neurexins and neuroligins in the formation, maturation, and function of vertebrate synapses. Curr. Opin. Neurobiol. 2012, 22, 412–422. [Google Scholar] [CrossRef] [PubMed]
- Ushkaryov, Y.A.; Südhof, T.C. Neurexin III alpha: Extensive alternative splicing generates membrane-bound and soluble forms. Proc. Natl. Acad. Sci. USA 1993, 90, 6410–6414. [Google Scholar] [CrossRef] [PubMed]
- Tabuchi, K.; Südhof, T.C. Structure and evolution of neurexin genes: Insight into the mechanism of alternative splicing. Genomics 2002, 79, 849–859. [Google Scholar] [CrossRef]
- Schreiner, D.; Nguyen, T.-M.; Russo, G.; Heber, S.; Patrignani, A.; Ahrné, E.; Scheiffele, P. Targeted combinatorial alternative splicing generates brain region-specific repertoires of neurexins. Neuron 2014, 84, 386–398. [Google Scholar] [CrossRef]
- Scheckel, C.; Drapeau, E.; Frias, M.A.; Park, C.Y.; Fak, J.; Zucker-Scharff, I.; Kou, Y.; Haroutunian, V.; Ma’ayan, A.; Buxbaum, J.D.; et al. Regulatory consequences of neuronal ELAV-like protein binding to coding and non-coding RNAs in human brain. eLife 2016, 5. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, M.H.; Wu, X.; Kodani, A.; Fan, J.; Doan, R.; Ozawa, M.; Ma, J.; Yoshida, N.; Reiter, J.F.; et al. Cell-Type-Specific Alternative Splicing Governs Cell Fate in the Developing Cerebral Cortex. Cell 2016, 166, 1147–1162.e15. [Google Scholar] [CrossRef]
- Cao, X.; Tabuchi, K. Functions of synapse adhesion molecules neurexin/neuroligins and neurodevelopmental disorders. Neurosci. Res. 2017, 116, 3–9. [Google Scholar] [CrossRef]
- Ichtchenko, K.; Hata, Y.; Nguyen, T.; Ullrich, B.; Missler, M.; Moomaw, C.; Südhof, T.C. Neuroligin 1: A splice site-specific ligand for beta-neurexins. Cell 1995, 81, 435–443. [Google Scholar] [CrossRef]
- Boucard, A.A.; Chubykin, A.A.; Comoletti, D.; Taylor, P.; Südhof, T.C. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron 2005, 48, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Chich, J.F.; Chapuis, C.; Henry, C.; Vidic, J.; Rezaei, H.; Noinville, S. Vesicle permeabilization by purified soluble oligomers of prion protein: A comparative study of the interaction of oligomers and monomers with lipid membranes. J. Mol. Biol. 2010, 397, 1017–1030. [Google Scholar] [CrossRef] [PubMed]
- De Wit, J.; Sylwestrak, E.; O’Sullivan, M.L.; Otto, S.; Tiglio, K.; Savas, J.N.; Yates, J.R., 3rd; Comoletti, D.; Taylor, P.; Ghosh, A. LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron 2009, 64, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Koehnke, J.; Katsamba, P.S.; Ahlsen, G.; Bahna, F.; Vendome, J.; Honig, B.; Shapiro, L.; Jin, X. Splice form dependence of beta-neurexin/neuroligin binding interactions. Neuron 2010, 67, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, T.J.; Pancaroglu, R.; Kang, Y.; Rooyakkers, A.; Craig, A.M. LRRTMs and neuroligins bind neurexins with a differential code to cooperate in glutamate synapse development. J. Neurosci. 2010, 30, 7495–7506. [Google Scholar] [CrossRef] [PubMed]
- Uemura, T.; Lee, S.-J.; Yasumura, M.; Takeuchi, T.; Yoshida, T.; Ra, M.; Taguchi, R.; Sakimura, K.; Mishina, M. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell 2010, 141, 1068–1079. [Google Scholar] [CrossRef]
- Matsuda, K.; Yuzaki, M. Cbln family proteins promote synapse formation by regulating distinct neurexin signaling pathways in various brain regions. Eur. J. Neurosci. 2011, 33, 1447–1461. [Google Scholar] [CrossRef]
- Ehrmann, I.; Gazzara, M.R.; Pagliarini, V.; Dalgliesh, C.; Kheirollahi-Chadegani, M.; Xu, Y.; Cesari, E.; Danilenko, M.; Maclennan, M.; Lowdon, K.; et al. A SLM2 Feedback Pathway Controls Cortical Network Activity and Mouse Behavior. Cell Rep. 2016, 17, 3269–3280. [Google Scholar] [CrossRef]
- Traunmüller, L.; Gomez, A.M.; Nguyen, T.-M.; Scheiffele, P. Control of neuronal synapse specification by a highly dedicated alternative splicing program. Science 2016, 352, 982–986. [Google Scholar] [CrossRef]
- Ullrich, B.; Ushkaryov, Y.A.; Südhof, T.C. Cartography of neurexins: More than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron 1995, 14, 497–507. [Google Scholar] [CrossRef]
- Ichtchenko, K.; Nguyen, T.; Südhof, T.C. Structures, alternative splicing, and neurexin binding of multiple neuroligins. J. Biol. Chem. 1996, 271, 2676–2682. [Google Scholar] [CrossRef]
- Ehrmann, I.; Dalgliesh, C.; Liu, Y.; Danilenko, M.; Crosier, M.; Overman, L.; Arthur, H.M.; Lindsay, S.; Clowry, G.J.; Venables, J.P.; et al. The tissue-specific RNA binding protein T-STAR controls regional splicing patterns of neurexin pre-mRNAs in the brain. PLoS Genet. 2013, 9, e1003474. [Google Scholar] [CrossRef] [PubMed]
- Treutlein, B.; Gokce, O.; Quake, S.R.; Südhof, T.C. Cartography of neurexin alternative splicing mapped by single-molecule long-read mRNA sequencing. Proc. Natl. Acad. Sci. USA 2014, 111, E1291–E1299. [Google Scholar] [CrossRef] [PubMed]
- Shapiro-Reznik, M.; Jilg, A.; Lerner, H.; Earnest, D.J.; Zisapel, N. Diurnal rhythms in neurexins transcripts and inhibitory/excitatory synapse scaffold proteins in the biological clock. PLoS ONE 2012, 7, e37894. [Google Scholar] [CrossRef]
- Jenkins, A.K.; Paterson, C.; Wang, Y.; Hyde, T.M.; Kleinman, J.E.; Law, A.J. Neurexin 1 (NRXN1) splice isoform expression during human neocortical development and aging. Mol. Psychiatry 2016, 21, 701–706. [Google Scholar] [CrossRef]
- Patzke, H.; Ernsberger, U. Expression of neurexin Ialpha splice variants in sympathetic neurons: Selective changes during differentiation and in response to neurotrophins. Mol. Cell. Neurosci. 2000, 15, 561–572. [Google Scholar] [CrossRef]
- Zeng, Y.; Zhao, D.; Xie, C.-W. Neurotrophins enhance CaMKII activity and rescue amyloid-β-induced deficits in hippocampal synaptic plasticity. J. Alzheimers Dis. 2010, 21, 823–831. [Google Scholar] [CrossRef]
- Rozic, G.; Lupowitz, Z.; Piontkewitz, Y.; Zisapel, N. Dynamic changes in neurexins’ alternative splicing: Role of Rho-associated protein kinases and relevance to memory formation. PLoS ONE 2011, 6, e18579. [Google Scholar] [CrossRef]
- Fuccillo, M.V.; Földy, C.; Gökce, Ö.; Rothwell, P.E.; Sun, G.L.; Malenka, R.C.; Südhof, T.C. Single-Cell mRNA Profiling Reveals Cell-Type-Specific Expression of Neurexin Isoforms. Neuron 2015, 87, 326–340. [Google Scholar] [CrossRef]
- Cariati, I.; Scimeca, M.; Tancredi, V.; D’Amico, A.G.; Pallone, G.; Palmieri, M.; Frank, C.; D’Arcangelo, G. Effects of different continuous aerobic training protocols in a heterozygous mouse model of Niemann-Pick type C disease. J. Funct. Morphol. Kinesiol. 2020, 5, 53. [Google Scholar] [CrossRef]
- Pallone, G.; Palmieri, M.; Cariati, I.; Bei, R.; Masuelli, L.; D’arcangelo, G.; Tancredi, V. Different continuous training modalities result in distinctive effects on muscle structure, plasticity and function. Biomed. Rep. 2020, 12. [Google Scholar] [CrossRef]
- Iijima, T.; Wu, K.; Witte, H.; Hanno-Iijima, Y.; Glatter, T.; Richard, S.; Scheiffele, P. SAM68 regulates neuronal activity-dependent alternative splicing of neurexin-1. Cell 2011, 147, 1601–1614. [Google Scholar] [CrossRef]
- Nguyen, T.-M.; Schreiner, D.; Xiao, L.; Traunmüller, L.; Bornmann, C.; Scheiffele, P. An alternative splicing switch shapes neurexin repertoires in principal neurons versus interneurons in the mouse hippocampus. eLife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Pette, D.; Staron, R.S. Mammalian skeletal muscle fiber type transitions. Int. Rev. Cytol. 1997, 170, 143–223. [Google Scholar] [CrossRef] [PubMed]
- Gunning, P.; Hardeman, E. Multiple mechanisms regulate muscle fiber diversity. FASEB J. 1991, 5, 3064–3070. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S.; Reggiani, C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar] [CrossRef]
- Weyn-Vanhentenryck, S.M.; Feng, H.; Ustianenko, D.; Duffié, R.; Yan, Q.; Jacko, M.; Martinez, J.C.; Goodwin, M.; Zhang, X.; Hengst, U.; et al. Precise temporal regulation of alternative splicing during neural development. Nat. Commun. 2018, 9, 2189. [Google Scholar] [CrossRef] [PubMed]
- Farini, D.; Cesari, E.; Weatheritt, R.J.; La Sala, G.; Naro, C.; Pagliarini, V.; Bonvissuto, D.; Medici, V.; Guerra, M.; Di Pietro, C.; et al. A Dynamic Splicing Program Ensures Proper Synaptic Connections in the Developing Cerebellum. Cell Rep. 2020, 31, 107703. [Google Scholar] [CrossRef]
- Schwamborn, J.C.; Püschel, A.W. The sequential activity of the GTPases Rap1B and Cdc42 determines neuronal polarity. Nat. Neurosci. 2004, 7, 923–929. [Google Scholar] [CrossRef]
- Nishimura, T.; Yamaguchi, T.; Kato, K.; Yoshizawa, M.; Nabeshima, Y.; Ohno, S.; Hoshino, M.; Kaibuchi, K. PAR-6-PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat. Cell Biol. 2005, 7, 270–277. [Google Scholar] [CrossRef]
- Garvalov, B.K.; Flynn, K.C.; Neukirchen, D.; Meyn, L.; Teusch, N.; Wu, X.; Brakebusch, C.; Bamburg, J.R.; Bradke, F. Cdc42 regulates cofilin during the establishment of neuronal polarity. J. Neurosci. 2007, 27, 13117–13129. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.H.; Wang, H.; Soderling, S.H.; Yasuda, R. Loss of Cdc42 leads to defects in synaptic plasticity and remote memory recall. eLife 2014, 3. [Google Scholar] [CrossRef]
- Kreienkamp, H.-J. Scaffolding proteins at the postsynaptic density: Shank as the architectural framework. Handb. Exp. Pharmacol. 2008, 365–380. [Google Scholar] [CrossRef]
- Wang, X.; McCoy, P.A.; Rodriguiz, R.M.; Pan, Y.; Je, H.S.; Roberts, A.C.; Kim, C.J.; Berrios, J.; Colvin, J.S.; Bousquet-Moore, D.; et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum. Mol. Genet. 2011, 20, 3093–3108. [Google Scholar] [CrossRef] [PubMed]
- Ypsilanti, A.R.; Chedotal, A. Roundabout receptors. Adv. Neurobiol. 2014, 8, 133–164. [Google Scholar] [CrossRef]
- Shen, K.; Cowan, C.W. Guidance molecules in synapse formation and plasticity. Cold Spring Harb. Perspect. Biol. 2010, 2, a001842. [Google Scholar] [CrossRef]
- Gibson, E.M.; Purger, D.; Mount, C.W.; Goldstein, A.K.; Lin, G.L.; Wood, L.S.; Inema, I.; Miller, S.E.; Bieri, G.; Zuchero, J.B.; et al. Neuronal activity promotes oligodendrogenesis and adaptive myelination in the mammalian brain. Science 2014, 344, 1252304. [Google Scholar] [CrossRef]
- Koropouli, E.; Kolodkin, A.L. Semaphorins and the dynamic regulation of synapse assembly, refinement, and function. Curr. Opin. Neurobiol. 2014, 27, 1–7. [Google Scholar] [CrossRef]
- Hendrickson, M.L.; Rao, A.J.; Demerdash, O.N.A.; Kalil, R.E. Expression of nestin by neural cells in the adult rat and human brain. PLoS ONE 2011, 6, e18535. [Google Scholar] [CrossRef]
- Ferreira, A.F.B.; Real, C.C.; Rodrigues, A.C.; Alves, A.S.; Britto, L.R.G. Moderate exercise changes synaptic and cytoskeletal proteins in motor regions of the rat brain. Brain Res. 2010, 1361, 31–42. [Google Scholar] [CrossRef]
- Fang, Z.H.; Lee, C.H.; Seo, M.K.; Cho, H.; Lee, J.G.; Lee, B.J.; Park, S.W.; Kim, Y.H. Effect of treadmill exercise on the BDNF-mediated pathway in the hippocampus of stressed rats. Neurosci. Res. 2013, 76, 187–194. [Google Scholar] [CrossRef]
- Raj, B.; Blencowe, B.J. Alternative Splicing in the Mammalian Nervous System: Recent Insights into Mechanisms and Functional Roles. Neuron 2015, 87, 14–27. [Google Scholar] [CrossRef]
- Basso, J.C.; Shang, A.; Elman, M.; Karmouta, R.; Suzuki, W.A. Acute Exercise Improves Prefrontal Cortex but not Hippocampal Function in Healthy Adults. J. Int. Neuropsychol. Soc. 2015, 21, 791–801. [Google Scholar] [CrossRef]
- Cefis, M.; Prigent-Tessier, A.; Quirié, A.; Pernet, N.; Marie, C.; Garnier, P. The effect of exercise on memory and BDNF signaling is dependent on intensity. Brain Struct. Funct. 2019, 224, 1975–1985. [Google Scholar] [CrossRef]
- Ito-Ishida, A.; Miyazaki, T.; Miura, E.; Matsuda, K.; Watanabe, M.; Yuzaki, M.; Okabe, S. Presynaptically released Cbln1 induces dynamic axonal structural changes by interacting with GluD2 during cerebellar synapse formation. Neuron 2012, 76, 549–564. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Aoto, J.; Südhof, T.C. Alternative Splicing of Presynaptic Neurexins Differentially Controls Postsynaptic NMDA and AMPA Receptor Responses. Neuron 2019, 102, 993.e5–1008.e5. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-K.; Lee, S.-J.; Kim, T.-W. Treadmill exercise enhances NMDA receptor expression in schizophrenia mice. J. Exerc. Rehabil. 2014, 10, 15–21. [Google Scholar] [CrossRef]
- Holloszy, J.O. Biochemical adaptations in muscle. Effects of exercise on mitochondrial oxygen uptake and respiratory enzyme activity in skeletal muscle. J. Biol. Chem. 1967, 242, 2278–2282. [Google Scholar] [CrossRef]
- Fitts, R.H.; Booth, F.W.; Winder, W.W.; Holloszy, J.O. Skeletal muscle respiratory capacity, endurance, and glycogen utilization. Am. J. Physiol. 1975, 228, 1029–1033. [Google Scholar] [CrossRef]
- Davies, K.J.; Packer, L.; Brooks, G.A. Biochemical adaptation of mitochondria, muscle, and whole-animal respiration to endurance training. Arch. Biochem. Biophys. 1981, 209, 539–554. [Google Scholar] [CrossRef]
- Hood, D.A. Mechanisms of exercise-induced mitochondrial biogenesis in skeletal muscle. Appl. Physiol. Nutr. Metab. 2009, 34, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Ljubicic, V.; Joseph, A.-M.; Saleem, A.; Uguccioni, G.; Collu-Marchese, M.; Lai, R.Y.J.; Nguyen, L.M.-D.; Hood, D.A. Transcriptional and post-transcriptional regulation of mitochondrial biogenesis in skeletal muscle: Effects of exercise and aging. Biochim. Biophys. Acta 2010, 1800, 223–234. [Google Scholar] [CrossRef] [PubMed]


| RT-PCR Nrxns | |
| Nrxns SS4 splice variants [43] | |
| Nrxn1 (+/−SS4) | Fw: 5′-TGT TGG GAC AGA TGA CAT CGC C-3′ |
| Rv: 5′-GAG AGC TGG CCC TGG AAG GG-3′ | |
| Nrxn2 (+/−SS4) | Fw: 5′-GTG CGC TTT ACT CGA AGT GGT G-3′ |
| Rv: 5′-CCC ATT GTA GTA GAG GCC GGA C-3′ | |
| Nrxn3 (+/−SS4) | Fw: 5′-TTG TGC GCT TCA CCA GGA ATG-3′ |
| Rv: 5′-AGA GCC CAG AGA GTT GAC CTT G-3′ | |
| Nrxns Alpha/Beta [44] | |
| Nrxn1 (+/−SS4) | Fw(α): 5′-CAG CAC AAC CTG CCA AGA-3′ |
| Fw(β): 5′-CCT GGC CCT GAT CTG GAT AGT-3′ | |
| Rv(αβ): 5′-GAG AGC TGG CCC TGG AAG GG-3′ | |
| Nrxn2 (+/−SS4) | Fw(α): 5′-CAC CAC CTG CAC CGA AGA G-3′ |
| Fw(β): 5′-GTG CCC ATC GCC ATC AA-3′ | |
| Rv(αβ): 5′-CCC ATT GTA GTA GAG GCC GGA C-3′ | |
| Nrxn3 (+/−SS4) | Fw(α): 5′-CTG TGA CTGCTC CAT GAC ATC ATATT-3′ |
| Fw(β): 5′-AAGCACCACTCTGTGCCTATTTCT-3′ | |
| Rv(αβ): 5′-AGA GCC CAG AGA GTT GAC CTT G-3′ | |
| RT-qPCR Nrxns | |
| Nrxns +/−SS4 splice variants: | |
| Nrxn1 (+SS4) | Fw: 5′-TAG TTG ATG AAT GGC TAC TCG ACA AA-3′ |
| Rv: 5′-GAC TCA GTT GTC ATA GAG GAA GGC AC-3′ | |
| Nrxn1 (−SS4) | Fw: 5′-GCT ACC CTG CAG GGC GT-3′ |
| Rv: 5′-GAG GTG GAC ATC TCA GAC TGC AT-3′ | |
| Nrxn2 (+SS4) | Fw: 5′-AAT CCC CTA CCG GCT TGG T-3′ |
| Rv: 5′-CCC CCT ATC TTG ATG GCA GC-3′ | |
| Nrxn2 (−SS4) | Fw: 5′-AGA GGT ACC CGG CAG GAC-3′ |
| Rv: 5′-GAC ACC TGG CCC TGG AAG-3′ | |
| Nrxn3 (+SS4) | Fw: 5′-AGG AGT GGC TGC AGG AAA AA-3′ |
| Rv: 5′-TTG TCC TTT CCT CCG ATG GC-3′ | |
| Nrxn3 (−SS4) | Fw: 5′-GAG CAC TAT CCT ACA GGC CG-3′ |
| Rv: 5′-AGA GTT GAC CTT GGA AGA GAC G-3′ | |
| Nrxns constant region (RC) | |
| Nrxn1 (RC) | Fw: 5′-CTGGAGCTGCACATACACCA-3′ |
| Rv: 5′-TGCCACCACTCCTTGTGAAA-3′ | |
| Nrxn2 (RC) | Fw: 5′-GTAGGCTTCAGCACACACCA-3′ |
| Rv: 5′-TCGTCCGTGCCCACATTAAA-3′ | |
| Nrxn3 (RC) | Fw: 5′-CTGGACTTGGCGACTTCCTC-3′ |
| Rv: 5′-TGAAGCGCACAACGTGGTAT-3′ | |
| β-Actin | Fw: 5′-CTG TCG AGT CGC GTC CAC-3′ |
| Rv: 5′-GCT TTG CAC ATG CCG GAG-3′ | |
| RT-qPCR | |
| Cdc42 | Fw: 5′-TGCTCTGCCCTCACACAGAAAG-3′ |
| Rv: 5′-GCGGCTCTTCTTCGGTTCTG-3′ | |
| Robo2 | Fw: 5′-CGAGCTCCTCCACAGTTTGT-3′ |
| Rv: 5′-GTAGGTTCTGGCTGCCTTCT-3′ | |
| Shank3 | Fw: 5′-GCCTTCCCTGCACTCCAATA-3′ |
| Rv: 5′-GCTTGTGTCCAACCTTCACG-3′ | |
| Nestin | Fw: 5′-GCAGGAGAAGCAGGGTCTAC-3′ |
| Rv: 5′-GAGTTCTCAGCCTCCAGCAG-3′ | |
| Mhc I | Fw: 5′-AGGGCGACCTCAACGAGAT-3′ |
| Rv: 5′-CAGCAGACTCTGGAGGCTCTT-3′ | |
| Mhc IIa | Fw: 5′-CCAAGAAAGGTGCCAAGAAG-3′ |
| Rv: 5′-CGGGAGTCTTGGTTTCATTG-3′ | |
| Mhc IIb | Fw: 5′-GCTTGAAAACGAGGTGGAAA-3′ |
| Rv: 5′-CCTCCTCAGCCTGTCTCTTG-3′ | |
| Mhc IIx | Fw: 5′-CGGTGGTGGAAAGAAAGG-3′ |
| Rv: 5′-CAGGAGTCTTGGTTTCATT-3′ | |
| CypA | Fw: 5′-GTCAACCCCACCGTGTTCTT-3′ |
| Rv: 5′-CTGCTGTCTTTGGGACCTTGT-3′ | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Innocenzi, E.; Cariati, I.; De Domenico, E.; Tiberi, E.; D’Arcangelo, G.; Verdile, V.; Paronetto, M.P.; Tancredi, V.; Barchi, M.; Rossi, P.; et al. Aerobic Exercise Induces Alternative Splicing of Neurexins in Frontal Cortex. J. Funct. Morphol. Kinesiol. 2021, 6, 48. https://doi.org/10.3390/jfmk6020048
Innocenzi E, Cariati I, De Domenico E, Tiberi E, D’Arcangelo G, Verdile V, Paronetto MP, Tancredi V, Barchi M, Rossi P, et al. Aerobic Exercise Induces Alternative Splicing of Neurexins in Frontal Cortex. Journal of Functional Morphology and Kinesiology. 2021; 6(2):48. https://doi.org/10.3390/jfmk6020048
Chicago/Turabian StyleInnocenzi, Elisa, Ida Cariati, Emanuela De Domenico, Erika Tiberi, Giovanna D’Arcangelo, Veronica Verdile, Maria Paola Paronetto, Virginia Tancredi, Marco Barchi, Pellegrino Rossi, and et al. 2021. "Aerobic Exercise Induces Alternative Splicing of Neurexins in Frontal Cortex" Journal of Functional Morphology and Kinesiology 6, no. 2: 48. https://doi.org/10.3390/jfmk6020048
APA StyleInnocenzi, E., Cariati, I., De Domenico, E., Tiberi, E., D’Arcangelo, G., Verdile, V., Paronetto, M. P., Tancredi, V., Barchi, M., Rossi, P., Sette, C., & Grimaldi, P. (2021). Aerobic Exercise Induces Alternative Splicing of Neurexins in Frontal Cortex. Journal of Functional Morphology and Kinesiology, 6(2), 48. https://doi.org/10.3390/jfmk6020048