Evolving Paradigms of Recombinant Protein Production in Pharmaceutical Industry: A Rigorous Review
Abstract
1. Introduction
2. Trends in Biopharmaceutical Proteins
3. Recombinant Pharmaceutical Protein Production in Microbes
3.1. Microbial Host Selection
3.1.1. Escherichia coli
3.1.2. Yeasts
3.1.3. Other Hosts
3.2. Vector and Promoter Systems
4. Upstream Process Development
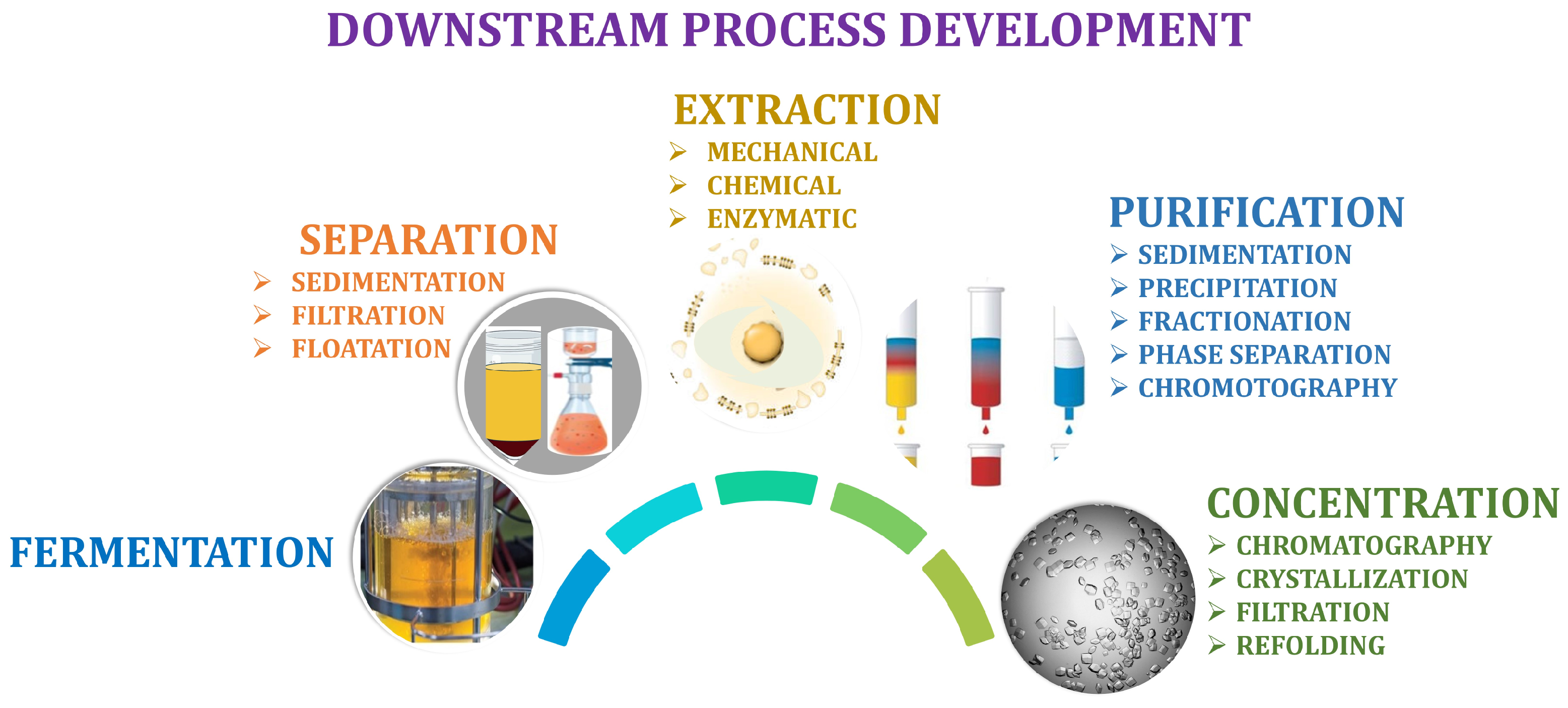
5. Downstream Process Development
5.1. Extraction
5.2. Purification
5.3. Product Concentration
6. Strategies to Overcome Low or No Expression
7. Economics of Microbial Production of Biopharmaceuticals
8. Latest Technologies in Recombinant Products
Revolutionary Technologies for Recombinant Protein Production
9. Marketed Recombinant Microbial Products
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Singh, N.; Vayer, P.; Tanwar, S.; Poyet, J.-L.; Tsaioun, K.; Villoutreix, B.O. Drug discovery and development: Introduction to the general public and patient groups. Front. Drug Discov. 2023, 3, 1201419. [Google Scholar] [CrossRef]
- Villoutreix, B.O. Post-pandemic drug discovery and development: Facing present and future challenges. Front. Drug Discov. 2021, 1, 728469. [Google Scholar] [CrossRef]
- Powell, J.S. Lasting power of new clotting proteins. Hematology 2014, 1, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Assenberg, R.; Wan, P.T.; Geisse, S.; Mayr, L.M. Advances in recombinant protein expression for use in pharmaceutical research. Curr. Opin. Struct. Biol. 2013, 23, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Savic, S.; McDermott, M.W. New monogenic diseases span the immunological disease continuum. Nat. Rev. Rheumatol. 2014, 11, 67–68. [Google Scholar] [CrossRef] [PubMed]
- Postma, P.R.; Suarez-Garcia, E.; Safi, C.; Yonathan, K.; Olivieri, G.; Barbosa, M.A.; Wijffels, R.H.; Eppink, M.H.M. Energy efficient bead milling of microalgae: Effect of bead size on disintegration and release of proteins and carbohydrates. Bioresour. Technol. 2017, 224, 670–679. [Google Scholar] [CrossRef] [PubMed]
- Trott, M. Driving Forces and Current Trends in Biopharmaceutical Analysis. Technology Networks. Available online: https://www.technologynetworks.com/biopharma/articles/driving-forces-and-current-trends-in-biopharmaceutical-analysis-341052 (accessed on 30 September 2022).
- Mordor Intelligence. Biopharmaceuticals Market Size and Share Analysis—Growth Trends and Forecast (2023–2028). 2023. Available online: https://www.mordorintelligence.com/industry-reports/global-biopharmaceuticals-market-industry/market-size (accessed on 30 September 2022).
- Goeddel, D.V.; Kleid, D.G.; Bolívar, F.; Heyneker, H.L.; Yansura, D.G.; Crea, R.; Hirose, T.; Kraszewski, A.; Itakura, K.; Riggs, A.D. Expression in Escherichia coli of chemically synthesized genes for human insulin. Proc. Natl. Acad. Sci. USA 1979, 76, 106–110. [Google Scholar] [CrossRef]
- Ebrahimi, S.B.; Samanta, D. Engineering protein-based therapeutics through structural and chemical design. Nat. Commun. 2023, 14, 2411. [Google Scholar] [CrossRef]
- Leader, B.; Baca, Q.J.; Golan, D.E. Protein therapeutics: A summary and pharmacological classification. Nat. Rev. Drug Discov. 2008, 7, 21–39. [Google Scholar] [CrossRef]
- Chang, H.Y.; Qi, L.S. Reversing the central dogma: RNA-guided control of DNA in epigenetics and genome editing. Mol. Cell 2023, 83, 442–451. [Google Scholar] [CrossRef]
- Papaneophytou, C. Design of experiments as a tool for optimization in recombinant protein biotechnology: From constructs to crystals. Mol. Biotechnol. 2019, 61, 873–891. [Google Scholar] [CrossRef]
- Markova, E.A.; Shaw, R.E.; Reynolds, C.S. Prediction of strain engineerings that amplify recombinant protein secretion through the machine learning approach MaLPHAS. Eng. Biol. 2022, 6, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, N.K.; Shrivastava, A. Recent developments in bioprocessing of recombinant proteins: Expression hosts and process development. Front. Bioeng. Biotechnol. 2019, 7, 420. [Google Scholar] [CrossRef] [PubMed]
- Pouresmaeil, M.; Azizi-Dargahlou, S. Factors involved in heterologous expression of proteins in E. coli host. Arch. Microbiol. 2023, 205, 212. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Kumar, A. Strategies for optimization of heterologous protein expression in E. coli: Roadblocks and reinforcements. Int. J. Biol. Macromol. 2018, 106, 803–822. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Shukla, P. Sophisticated cloning, fermentation, and purification technologies for an enhanced therapeutic protein production: A review. Front. Pharmacol. 2017, 8, 419. [Google Scholar] [CrossRef]
- Madhavan, A.; Arun, K.G.; Sindhu, R.; Krishnamoorthy, J.; Reshmy, R.; Sirohi, R.; Pugazhendi, A.; Awasthi, M.K.; Szakacs, G.; Binod, P. Customized yeast cell factories for biopharmaceuticals: From cell engineering to process scale up. Microb. Cell Factories 2021, 20, 124. [Google Scholar] [CrossRef]
- Terpe, K. Overview of bacterial expression systems for heterologous protein production: From molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 2006, 72, 211–222. [Google Scholar] [CrossRef]
- Gupta, S.; Shukla, P. Advanced technologies for improved expression of recombinant proteins in bacteria: Perspectives and applications. Crit. Rev. Biotechnol. 2015, 36, 1089–1098. [Google Scholar] [CrossRef]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef]
- Baghban, R.; Farajnia, S.; Rajabibazl, M.; Ghasemi, Y.; Mafi, A.; Hoseinpoor, R.; Rahbarnia, L.; Aria, M. Yeast expression systems: Overview and recent advances. Mol. Biotechnol. 2019, 61, 365–384. [Google Scholar] [CrossRef] [PubMed]
- Engel, S.R.; Dietrich, F.S.; Fisk, D.G.; Binkley, G.; Balakrishnan, R.; Costanzo, M.R.; Dwight, S.S.; Hitz, B.C.; Karra, K.; Nash, R.J.; et al. The reference genome sequence of Saccharomyces cerevisiae: Then and now. G3 Genes Genom Genet. 2014, 4, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Naghizadeh, M.M.; Plieskatt, J.; Singh, S.; Theisen, M. Cloning and Recombinant Protein Expression in Lactococcus lactis. Methods Mol. Biol. 2023, 2652, 3–20. [Google Scholar] [PubMed]
- Välimets, S.; Pedetti, P.; Virginia, L.J.; Hoang, M.N.; Sauer, M.; Peterbauer, C. Secretory Expression of Recombinant Small Laccase Genes in Gram-Positive Bacteria. Microb. Cell Factories 2023, 22, 72. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Wang, X.; Bai, Y.; Wang, Y.; Wang, Y.; Tu, T.; Qin, X.; Su, X.; Luo, H.; Yao, B.; et al. Engineering Escherichia coli for Efficient Assembly of Heme Proteins. Microb. Cell Factories 2023, 22, 59. [Google Scholar] [CrossRef] [PubMed]
- Rebello, S.; Abraham, A.; Madhavan, A.; Sindhu, R.; Binod, P.; Bahuleyan, A.K.; Aneesh, E.M.; Pandey, A. Non-Conventional Yeast Cell Factories for Sustainable Bioprocesses. FEMS Microbiol. Lett 2018, 365, fny222. [Google Scholar] [CrossRef] [PubMed]
- Emalfarb, M.A.; Punt, P.J.; Van Zeijl, C.; van den Hondel, C.; Verdoes, J.C.; Burlingame, R.P. Expression and High-Throughput Screening of Complex Expressed DNA Libraries in Filamentous Fungi. U.S. Patent 8,680,252, 25 March 2014. [Google Scholar]
- Liu, X.; Yang, Y.; Zhang, W.; Sun, Y.; Peng, F.; Jeffrey, L.; Bai, Z.; Liu, X.; Yang, Y.; Zhang, W.; et al. Expression of Recombinant Protein Using Corynebacterium Glutamicum: Progress, Challenges and Applications. Crit. Rev. Biotechnol. 2015, 36, 652–664. [Google Scholar] [CrossRef]
- Vanier, G.; Stelter, S.; Vanier, J.; Hempel, F.; Maier, U.G.; Lerouge, P.; Ma, J.; Bardor, M. Alga-Made Anti-Hepatitis B Antibody Binds to Human FCΓ Receptors. Biotechnol. J. 2017, 13, 1700496. [Google Scholar] [CrossRef]
- Ramos-Martinez, E.M.; Fimognari, L.; Sakuragi, Y. High-yield Secretion of Recombinant Proteins from the Microalga Chlamydomonas Reinhardtii. Plant Biotechnol. J. 2017, 15, 1214–1224. [Google Scholar] [CrossRef]
- Palme, J.; Wang, J.; Springer, M. Variation in the Modality of a Yeast Signaling Pathway Is Mediated by a Single Regulator. eLife 2021, 10, e69974. [Google Scholar] [CrossRef]
- Peng, B.; Williams, T.; Henry, M.; Nielsen, L.K.; Vickers, C.E. Controlling Heterologous Gene Expression in Yeast Cell Factories on Different Carbon Substrates and across the Diauxic Shift: A Comparison of Yeast Promoter Activities. Microb. Cell Factories 2015, 14, 91. [Google Scholar] [CrossRef] [PubMed]
- Solow, S.P.; Sengbusch, J.; Laird, M.W. Heterologous Protein Production from the Inducible MET25 Promoter in Saccharomyces Cerevisiae. Biotechnol. Prog. 2008, 21, 617–620. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, J.R.M.; De Moraes, L.M.P.; Torres, F.A.G. Molecular Characterization of the 3-Phosphoglycerate Kinase Gene (PGK1) from the Methylotrophic yeast Pichia Pastoris. Yeast 2005, 22, 725–737. [Google Scholar] [CrossRef] [PubMed]
- De Brabander, P.; Uitterhaegen, E.; Delmulle, T.; De Winter, K.; Soetaert, W. Challenges and progress towards industrial recombinant protein production in yeasts: A review. Biotechnol. Adv. 2023, 64, 108121. [Google Scholar] [CrossRef] [PubMed]
- Langer, E.S.; Rader, R. Single-use technologies in biopharmaceutical manufacturing: A 10-year review of trends and the future. Eng. Life Sci. 2014, 14, 238–243. [Google Scholar] [CrossRef]
- Sanchez-Garcia, L.M.; Martín, L.; Mangues, R.; Ferrer-Miralles, N.; Vázquez, E.; Villaverde, A. Recombinant pharmaceuticals from microbial cells: A 2015 update. Microb. Cell Factories 2016, 15, 33. [Google Scholar] [CrossRef] [PubMed]
- Mićić, M.; Whyte, J.J.; Karsten, V. High performance bead beating based lysing, homogenization and grinding for DNA, RNA and proteins extraction with FastPrep® systems. In Sample Preparation Techniques for Soil, Plant, and Animal Samples; Springer Protocols, Handbooks; Mimic, M., Ed.; Humana Press: New York, NY, USA, 2016; pp. 99–116. [Google Scholar]
- Lee, C.-H. A Simple outline of methods for protein isolation and purification. Endocrinol. Metab. 2017, 32, 18. [Google Scholar] [CrossRef]
- Harrison, S.T.L. Cell disruption. In Comprehensive Biotechnology, 2nd ed.; Elsevier: Oxford, UK, 2011; Volume 2, pp. 619–639. [Google Scholar]
- Crapisi, A.; Lante, A.; Pasini, G.; Spettoli, P. Enhanced Microbial Cell Lysis by the Use of Lysozyme Immobilized on Different Carriers. Process Biochem. 1993, 28, 17–21. [Google Scholar] [CrossRef]
- Scott, J.H.; Schekman, R. Lyticase: Endoglucanase and Protease Activities That Act Together in Yeast Cell Lysis. J. Bacteriol. 1980, 142, 414–423. [Google Scholar] [CrossRef]
- Bărbieru, O.-G.; Tritean, N.; Trica, B.; Constantinescu-Aruxandei, D.; Oancea, F. Optimization of yeast protein extraction through a combined enzymatic and high-pressure homogenization method. Proceedings 2020, 57, 75. [Google Scholar]
- Wingfield, P.T. Overview of the Purification of Recombinant Proteins. Curr. Protoc. Protein Sci. 2015, 80, 6.1.1–6.1.35. [Google Scholar] [CrossRef] [PubMed]
- Scopes, R.K. Protein Purification: Principles and Practice; Springer Science & Business Media: Berlin/Heidelberg, Germany, 1982. [Google Scholar]
- Walter, H.; Johansson, G. Partitioning in Aqueous Two-Phase Systems: An Overview. Anal. Biochem. 1986, 155, 215–242. [Google Scholar] [CrossRef] [PubMed]
- Van Reis, R.; Zydney, A.L. Membrane Separations in Biotechnology. Curr. Opin. Biotechnol. 2001, 12, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Rathore, A.S.; Kumar, D.; Kateja, N. Recent developments in chromatographic purification of biopharmaceuticals. Biotechnol. Lett. 2018, 40, 895–905. [Google Scholar] [CrossRef]
- Gronemeyer, P.; Ditz, R.; Strube, J. Trends in upstream and downstream process development for antibody manufacturing. Bioengineering 2014, 1, 188–212. [Google Scholar] [CrossRef] [PubMed]
- Challener, C.A. A look at the affinity chromatography landscape. Biopharm. Int. 2019, 32, 24–28. [Google Scholar]
- Zhao, X.; Li, G.; Liang, S. Several affinity tags commonly used in chromatographic purification. J. Anal. Methods Chem. 2013, 2013, 581093. [Google Scholar] [CrossRef]
- Sifniotis, V.; Cruz, E.; Eroglu, B.; Kayser, V. Current advancements in addressing key challenges of therapeutic antibody design, manufacture, and formulation. Antibodies 2019, 8, 36. [Google Scholar] [CrossRef]
- Tarrant, R.N.; Velez-Suberbie, M.L.; Tait, A.; Smales, C.M.; Bracewell, D.G. Host cell protein adsorption characteristics during protein A chromatography. Biotechnol. Prog. 2012, 28, 1037–1044. [Google Scholar] [CrossRef]
- Kimia, Z.; Hosseini, S.A.; Talesh, S.S.A.; Khatami, M.; Kavianpour, A.; Javidanbardan, A. A novel application of ion exchange chromatography in recombinant Hepatitis B vaccine downstream processing: Improving recombinant HBsAg homogeneity by removing associated aggregates. J. Chromatogr. B 2019, 1113, 20–29. [Google Scholar] [CrossRef]
- Fekete, S.; Veuthey, J.-L.; Beck, A.; Guillarme, D. Hydrophobic interaction chromatography for the characterization of monoclonal antibodies and related products. J. Pharm. Biomed. Anal. 2016, 130, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Levin, M.; Otten, H.; Von Wachenfeldt, C.; Ohlin, M. A folded and immunogenic IgE-hporeactive variant of the major allergen Phl p 1 produced in Escherichia coli. BMC Biotechnol. 2015, 15, 52. [Google Scholar] [CrossRef] [PubMed]
- Burgess, R.R. A brief practical review of size exclusion chromatography: Rules of thumb, limitations, and troubleshooting. Protein Expr. Purif. 2018, 150, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Ghose, S.; Jin, M.S.; Liu, J.; Hickey, J.L.S.; Lee, S. Integrated polishing steps for monoclonal antibody purification. In Process Scale Purification Antibodies; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2017; pp. 303–323. [Google Scholar] [CrossRef]
- Rajendran, A.; Paredes, G.; Mazzotti, M. Simulated Moving Bed Chromatography for the Separation of Enantiomers. J. Chromatogr. A 2009, 1216, 709–738. [Google Scholar] [CrossRef]
- Zang, Y.; Kammerer, B.; Eisenkolb, M.; Lohr, K.; Kiefer, H. Towards Protein Crystallization as a Process Step in Downstream Processing of Therapeutic Antibodies: Screening and Optimization at Microbatch Scale. PLoS ONE 2011, 6, e25282. [Google Scholar] [CrossRef]
- Jungbauer, A.; Kaar, W. Current Status of Technical Protein Refolding. J. Biotechnol. 2007, 128, 587–596. [Google Scholar] [CrossRef]
- Peeva, L.; Da Silva Burgal, J.; Valtcheva, I.B.; Livingston, A.G. Continuous Purification of Active Pharmaceutical Ingredients Using Multistage Organic Solvent Nanofiltration Membrane Cascade. Chem. Eng. Sci. 2014, 116, 183–194. [Google Scholar] [CrossRef]
- Boi, C.; Dimartino, S. Advances in membrane chromatography for the capture step of monoclonal antibodies. Curr. Org. Chem. 2017, 21, 1753–1759. [Google Scholar] [CrossRef]
- Emami, P.; Motevalian, S.P.; Pepin, E. Purification of a conjugated polysaccharide vaccine using tangential flow diafiltration. Biotechnol. Bioeng. 2019, 116, 591–597. [Google Scholar] [CrossRef]
- Kovács, Z. Continuous diafiltration: Cocurrent and countercurrent modes. In Encyclopedia of Membranes; Drioli, E., Giorno, L., Eds.; Springer eBooks; Springer: Berlin/Heidelberg, Germany, 2016; pp. 446–448. [Google Scholar]
- Palombarini, F.; Ghirga, F.; Boffi, A.; Bonamore, A. Application of crossflow ultrafiltration for scaling up the purification of a recombinant ferritin. Protein Expr. Purif. 2019, 163, 105451. [Google Scholar] [CrossRef]
- Carvalho, S.B.; Silva, R.J.S.; Moleirinho, M.G.; Cunha, B.; Moreira, A.S.; Xenopoulos, A.; Alves, P.M.; Carrondo, M.J.T.; Peixoto, C. Membrane-based approach for the downstream processing of influenza virus-like particles. Biotechnol. J. 2019, 14, 1800570. [Google Scholar] [CrossRef] [PubMed]
- Terol, G.L.; Gallego-Jara, J.; Martínez, R.A.S.; Díaz, M.C.; De Diego, T. Engineering protein production by rationally choosing a carbon and nitrogen source using E. coli BL21 acetate metabolism knockout strains. Microb. Cell Factories 2019, 18, 151. [Google Scholar] [CrossRef]
- Baig, F.; Fernando, L.P.; Salazar, M.A.; Powell, R.; Bruce, T.F.; Harcum, S.W. Dynamic transcriptional response of Escherichia coli to inclusion body formation. Biotechnol. Bioeng. 2014, 111, 980–999. [Google Scholar] [CrossRef]
- Larentis, A.L.; Argondizzo, A.P.C.; Esteves, G.D.S.; Jessouron, E.; Galler, R.; Medeiros, M.A. Cloning and optimization of induction conditions for mature PsaA (pneumococcal surface adhesin A) expression in Escherichia coli and recombinant protein stability during long-term storage. Protein Expr. Purif. 2011, 78, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, G.; Valax, P. Expression of correctly folded proteins in Escherichia coli. Curr. Opin. Biotechnol. 1996, 7, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Ghoshoon, M.B.; Berenjian, A.; Hemmati, S.; Dabbagh, F.; Karimi, Z.; Negahdaripour, M.; Ghasemi, Y. Extracellular production of recombinant L-asparaginase II in Escherichia coli: Medium optimization using response surface methodology. Int. J. Pept. Res. Ther. 2015, 21, 487–495. [Google Scholar] [CrossRef]
- Bao, R.-M.; Yang, H.-M.; Yu, C.-M.; Zhang, W.; Tang, J.-B. An efficient protocol to enhance the extracellular production of recombinant protein from Escherichia coli by the synergistic effects of sucrose, glycine, and triton X-100. Protein Expr. Purif. 2016, 126, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Shaw, M.; Ingraham, J.L. Synthesis of macromolecules by Escherichia coli near the minimal temperature for growth. J. Bacteriol. 1967, 94, 157–164. [Google Scholar] [CrossRef]
- Bill, R.M. Playing catch-up with Escherichia coli: Using yeast to increase success rates in recombinant protein production experiments. Front. Microbiol. 2014, 5, 85. [Google Scholar] [CrossRef]
- Vogl, T.J.; Gebbie, L.; Palfreyman, R.W.; Speight, R. Effect of plasmid design and type of integration event on recombinant protein expression in Pichia pastoris. Appl. Environ. Microbiol. 2018, 84, 02712–02717. [Google Scholar] [CrossRef]
- Xie, Y.; Han, X.; Miao, Y. An effective recombinant protein expression and purification system in Saccharomyces cerevisiae. Curr. Protoc. Mol. Biol. 2018, 123, e62. [Google Scholar] [CrossRef] [PubMed]
- Nallamsetty, S.; Waugh, D.S. A Generic protocol for the expression and purification of recombinant proteins in Escherichia coli using a combinatorial his6-maltose binding protein fusion tag. Nat. Protoc. 2007, 2, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Galloway, C.A.; Sowden, M.P.; Smith, H.C. Increasing the yield of soluble recombinant protein expressed in E. coli by induction during late log phase. Biotechniques 2003, 34, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Vasina, J.A.; Baneyx, F. Expression of aggregation-prone recombinant proteins at low temperatures: A comparative study of the Escherichia coli CspA and Tac promoter systems. Protein Expr. Purif. 1997, 9, 211–218. [Google Scholar] [CrossRef]
- Kiefhaber, T.; Rudolph, R.; Kohler, H.-H.; Buchner, J. Protein aggregation in vitro and in vivo: A quantitative model of the kinetic competition between folding and aggregation. Nat. Biotechnol. 1991, 9, 825–829. [Google Scholar] [CrossRef]
- Ramírez, O.T.; Zamora, R.; Espinosa, G.; Merino, E.; Bolívar, F.; Quintero, R. Kinetic study of penicillin acylase production by recombinant E. coli in batch cultures. Process Biochem. 1994, 29, 197–206. [Google Scholar] [CrossRef]
- Terol, G.L.; Gallego-Jara, J.; Martínez, R.A.S.; Vivancos, A.M.; Díaz, M.C.; De Diego, T. Impact of the expression system on recombinant protein production in Escherichia coli BL21. Front. Microbiol. 2021, 12, 682001. [Google Scholar]
- Wang, Z.; Jin, L.; Yuan, Z.; Węgrzyn, G.; Węgrzyn, A. Classification of plasmid vectors using replication origin, selection marker and promoter as criteria. Plasmid 2009, 61, 47–51. [Google Scholar] [CrossRef]
- Gomes, A.M.V.; Carmo, T.S.; Carvalho, L.S.; Bahia, F.M.; Parachin, N.S. Comparison of yeasts as hosts for recombinant protein production. Microorganisms 2018, 6, 38. [Google Scholar] [CrossRef]
- Scannell, J.W.; Blanckley, A.; Boldon, H.; Warrington, B.H. Diagnosing the decline in pharmaceutical R&D efficiency. Nat. Rev. Drug. Discov. 2012, 11, 191–200. [Google Scholar]
- Farid, S.S.; Baron, M.; Stamatis, C.; Nie, W.; Coffman, J. Benchmarking biopharmaceutical process development and manufacturing cost contributions to R&D. mAbs 2020, 12, 1754999. [Google Scholar] [PubMed]
- Puetz, J.; Wurm, F.M. Recombinant proteins for industrial versus pharmaceutical purposes: A review of process and pricing. Processes 2019, 7, 476. [Google Scholar] [CrossRef]
- Lexchin, J. Pharmaceutical company spending on research and development and promotion in Canada, 2013–2016: A cohort analysis. J. Pharm. Policy Pract. 2018, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Helland, E.; Lakdawalla, D.N.; Malani, A.; Seabury, S.A. Unintended consequences of products liability: Evidence from the pharmaceutical market. J. Law Econ. Organ. 2020, 36, 598–632. [Google Scholar] [CrossRef]
- Lagassé, H.A.D.; Alexaki, A.; Simhadri, V.L.; Katagiri, N.; Jankowski, W.; Sauna, Z.E.; Kimchi-Sarfaty, C. Recent advances in (therapeutic protein) drug development. F1000Research 2017, 6, 113. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, Z.; Xu, Y.; Shi, T.-Q.; Ye, C.; Sun, X.; Huang, H. CRISPR-based construction of a BL21 (DE3)-derived variant strain library to rapidly improve recombinant protein production. ACS Synth. Biol. 2021, 11, 343–352. [Google Scholar] [CrossRef]
- Öztürk, S.; Ergün, B.G.; Çalık, P. Double promoter expression systems for recombinant protein production by industrial microorganisms. Appl. Microbiol. Biotechnol. 2017, 101, 7459–7475. [Google Scholar] [CrossRef]
- De Marco, A.; Deuerling, E.; Mogk, A.; Tomoyasu, T.; Bukau, B. Chaperone-based procedure to increase yields of soluble recombinant proteins produced in E. coli. BMC Biotechnol. 2007, 7, 32. [Google Scholar] [CrossRef]
- Bui, L.M.; Geraldi, A.; Nguyen, T.H.O.; Lee, J.Y.; Lee, J.; Cho, B.-K.; Kim, S.Y. MRNA Engineering for the efficient chaperone-mediated co-translational folding of recombinant proteins in Escherichia coli. Int. J. Mol. Sci. 2019, 20, 3163. [Google Scholar] [CrossRef]
- De Macedo Robert, J.; Garcia-Ortega, X.; Montesinos-Seguí, J.L.; Freire, D.M.G.; Valero, F. Continuous operation, a realistic alternative to fed-batch fermentation for the production of recombinant lipase B from Candida antarctica under the constitutive promoter PGK in Pichia pastoris. Biochem. Eng. J. 2019, 147, 39–47. [Google Scholar] [CrossRef]
- Kopp, J.; Kolkmann, A.-M.; Veleenturf, P.G.; Spadiut, O.; Herwig, C.; Slouka, C. Boosting recombinant inclusion body production—From classical fed-batch approach to continuous cultivation. Front. Bioeng. Biotechnol. 2019, 7, 297. [Google Scholar] [CrossRef] [PubMed]
- Matanguihan, C.; Wu, P. Upstream continuous processing: Recent advances in production of biopharmaceuticals and challenges in manufacturing. Curr. Opin. Biotechnol. 2022, 78, 102828. [Google Scholar] [CrossRef] [PubMed]
- Choe, D.; Cho, S.; Kim, S.C.; Cho, B.-K. Minimal genome: Worthwhile or worthless efforts toward being smaller? Biotechnol. J. 2015, 11, 199–211. [Google Scholar] [CrossRef] [PubMed]
- Cho, B.-K.; Zengler, K.; Qiu, Y.; Park, Y.S.; Knight, E.M.; Barrett, C.; Gao, Y.; Palsson, B.O. The transcription unit architecture of the Escherichia coli genome. Nat. Biotechnol. 2009, 27, 1043–1049. [Google Scholar] [CrossRef] [PubMed]
- Nakahigashi, K.; Toya, Y.; Ishii, N.; Soga, T.; Hasegawa, M.; Watanabe, H.; Takai, Y.; Honma, M.; Mori, H.; Tomita, M. Systematic phenome analysis of Escherichia coli multiple-knockout mutants reveals hidden reactions in central carbon metabolism. Mol. Syst. Biol. 2009, 5, 306. [Google Scholar] [CrossRef]
- Gibson, D.J.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Carr, P.W.; Church, G.M. Genome engineering. Nat. Biotechnol. 2009, 27, 1151–1162. [Google Scholar] [CrossRef]
- Morimoto, T.; Kadoya, R.; Endo, K.; Tohata, M.; Sawada, K.; Liu, S.-A.; Ozawa, T.; Kodama, T.; Kakeshita, H.; Kageyama, Y.; et al. Enhanced recombinant protein productivity by genome reduction in Bacillus subtilis. DNA Res. 2008, 15, 73–81. [Google Scholar] [CrossRef]
- Bu, Q.-T.; Yu, P.; Wang, J.; Li, Z.; Chen, X.-A.; Mao, X.-M.; Li, Y.-Q. Rational construction of genome-reduced and high-efficient industrial streptomyces chassis based on multiple comparative genomic approaches. Microb. Cell Factories 2019, 18, 16. [Google Scholar] [CrossRef]
- Choi, I.Y.; Oh, J.-H.; Wang, Z.; Van Pijkeren, J.-P. Bioluminescent Monitoring of Recombinant Lactic Acid Bacteria and Their Products. MBio 2023, 14, e01197-23. [Google Scholar] [CrossRef]
- Heavey, M.K.; Anselmo, A.C. Modulating Oral Delivery and Gastrointestinal Kinetics of Recombinant Proteins via Engineered Fungi. AAPS J. 2021, 23, 76. [Google Scholar] [CrossRef] [PubMed]
- Calvanese, M.; D’Angelo, C.; Lauro, C.; Tutino, M.L. Recombinant Protein Production in Pseudoalteromonas Haloplanktis Tac125 Biofilm; Social Science Research Network: Rochester, NY, USA, 2023. [Google Scholar]
- Kabarkouhi, Z.; Arjmand, S.; Siadat, S.O.R.; Shokri, B. Cold Atmospheric Plasma Treatment Enhances Recombinant Model Protein Production in Yeast Pichia Pastoris. Sci. Rep. 2023, 13, 6797. [Google Scholar] [CrossRef] [PubMed]
- Bonanni, D.; Litrico, M.; Ahmed, W.; Morerio, P.; Cazzorla, T.; Spaccapaniccia, E.; Cattani, F.; Allegretti, M.; Beccari, A.R.; Del Bue, A.; et al. A Deep Learning Approach to Optimize Recombinant Protein Production in Escherichia coli Fermentations. Fermentation 2023, 9, 503. [Google Scholar] [CrossRef]
- Johnson, I. Human insulin from recombinant DNA technology. Science 1983, 219, 632–637. [Google Scholar] [CrossRef]
- Naik, S.K.; Sethi, M.; Verma, D.; Naliath, R.; Chaudhary, D.P. Benefits and biohazards of microbial recombinants. In Microbial Diversity, Interventions and Scope; Sharma, S., Sharma, N., Sharma, M., Eds.; Springer: Singapore, 2020; pp. 123–134. [Google Scholar]
- Ivashkiv, L.B. IFNγ: Signalling, Epigenetics and Roles in Immunity, Metabolism, Disease and Cancer Immunotherapy. Nat. Rev. Immunol. 2018, 18, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Maughan, A.; Ogbuagu, O. Pegylated Interferon Alpha 2a for the Treatment of Hepatitis C Virus Infection. Expert Opin. Drug Metab. Toxicol. 2018, 14, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Kontogeorgos, G.; Krantz, E.; Trimpou, P.; Lainé, C.; Landin-Wilhelmsen, K. Teriparatide Treatment in Severe Osteoporosis—A Controlled 10-Year Follow-up Study. BMC Musculoskelet. Disord. 2022, 23, 1011. [Google Scholar] [CrossRef] [PubMed]
- Cheung, L.S.; Fu, J.; Kumar, P.; Kumar, A.; Urbanowski, M.E.; Ihms, E.A.; Parveen, S.; Bullen, C.K.; Patrick, G.J.; Harrison, R.; et al. Second-Generation IL-2 Receptor-Targeted Diphtheria Fusion Toxin Exhibits Antitumor Activity and Synergy with Anti–PD-1 in Melanoma. Proc. Natl. Acad. Sci. USA 2019, 116, 3100–3105. [Google Scholar] [CrossRef]
- Van Trimpont, M.; Peeters, E.; De Visser, Y.; Schalk, A.M.; Mondelaers, V.; De Moerloose, B.; Lavie, A.; Lammens, T.; Goossens, S.; Van Vlierberghe, P. Novel Insights on the Use of L-Asparaginase as an Efficient and Safe Anti-Cancer Therapy. Cancers 2022, 14, 902. [Google Scholar] [CrossRef]
- De Pinho Favaro, M.T.; Atienza-Garriga, J.; Martínez-Torró, C.; Parladé, E.; Vázquez, E.; Corchero, J.L.; Ferrer-Miralles, N.; Villaverde, A. Recombinant vaccines in 2022: A perspective from the cell factory. Microb. Cell Factories 2022, 21, 203. [Google Scholar] [CrossRef]
- Home, P.; Bartley, P.C.; Russell-Jones, D.; Hanaire-Broutin, H.; Heeg, J.-E.; Abrams, P.; Landin-Olsson, M.; Hylleberg, B.; Lang, H.; Draeger, E. Insulin Detemir Offers Improved Glycemic Control Compared with NPH Insulin in People with Type 1 Diabetes. Diabetes Care 2004, 27, 1081–1087. [Google Scholar] [CrossRef] [PubMed]
- Tran, K.; Grobelna, A. Somatropin for Growth Hormone Deficiency. Can. J. Health Technol. 2023, 3. [Google Scholar] [CrossRef]
- Wilding, J.; Batterham, R.L.; Calanna, S.; Davies, M.J.; Van Gaal, L.F.; Lingvay, I.; McGowan, B.; Rosenstock, J.; Tran, M.T.D.; Wadden, T.A.; et al. Once-Weekly Semaglutide in Adults with Overweight or Obesity. N. Engl. J. Med. 2021, 384, 989–1002. [Google Scholar] [CrossRef] [PubMed]
- Savarirayan, R.; Tofts, L.; Irving, M.; Wilcox, W.R.; Bacino, C.A.; Hoover-Fong, J.; Font, R.U.; Harmatz, P.; Rutsch, F.; Bober, M.B.; et al. Once-daily, subcutaneous vosoritide therapy in children with achondroplasia: A randomised, double-blind, phase 3, placebo-controlled, multicentre trial. Lancet 2020, 396, 684–692. [Google Scholar] [CrossRef] [PubMed]
- Pi-Sunyer, X.; Astrup, A.; Fujioka, K.; Greenway, F.L.; Halpern, A.; Krempf, M.; Lau, D.C.W.; Roux, C.W.L.; Ortíz, R.V.; Jensen, C.B.; et al. A Randomized, Controlled Trial of 3.0 Mg of Liraglutide in Weight Management. N. Engl. J. Med. 2015, 373, 11–22. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, C.M.; Starling, R.C.; Hernandez, A.F.; Armstrong, P.W.; Dickstein, K.; Hasselblad, V.; Heizer, G.; Komajda, M.; Massie, B.M.; McMurray, J.J.V.; et al. Effect of Nesiritide in Patients with Acute Decompensated Heart Failure. N. Engl. J. Med. 2011, 365, 32–43. [Google Scholar] [CrossRef]
- Yamanaka, G.; Ishida, Y.; Kanou, K.; Suzuki, S.; Watanabe, Y.; Takamatsu, T.; Morichi, S.; Go, S.; Oana, S.; Yamazaki, T.; et al. Towards a Treatment for Neuroinflammation in Epilepsy: Interleukin-1 Receptor Antagonist, Anakinra, as a Potential Treatment in Intractable Epilepsy. Int. J. Mol. Sci. 2021, 22, 6282. [Google Scholar] [CrossRef]
- Wieman, T.J.; Smiell, J.M.; Su, Y. Efficacy and Safely of a Topical Gel Formulation of Recombinant Human Platelet-Derived Growth Factor-BB (Becaplermin) in Patients with Chronic Neuropathic Diabetic Ulcers: A Phase III Randomized Placebo-Controlled Double-Blind Study. Diabetes Care 1998, 21, 822–827. [Google Scholar] [CrossRef]
- Kim, H.-A.; Yoo, S.J.; Kang, H.A. Yeast Synthetic Biology for the Production of Recombinant Therapeutic Proteins. FEMS Yeast Res. 2014, 15, 1–16. [Google Scholar] [CrossRef]
| rDNA | Trade Name | Cell Factory | Application | Company | Reference |
|---|---|---|---|---|---|
| Insulin | Humulin | E. coli | Diabetes | Eli Lilly & Co., Indianapolis, IN, USA | [39] |
| Growth hormone | Protropin | E. coli | Pituitary dwarfism | Genentech, San Francisco, CA, USA | [39] |
| Interferon | Intron A | E. coli | Hairy cell leukemia | Schering-Plough Corporation, Kenilworth, NJ, USA | [39] |
| Hepatitis B Vaccine | Recombinax | S. cerevisiae | Hepatatis | Merck & Co., Rahway, NJ, USA | [39] |
| Interferon alpha 2a | Roferon A | E. coli | Chronic myeloid leukemia | Roche, Basel, Switzerland | [39] |
| Granulocyte Colony Stimulating Factor | rG-CSF | E. coli | Chemotherapy-induced neutropenia | Amgen, Fairfield, NJ, USA | [93] |
| Granulocyte Macrophage Colony Stimulating Factor | rGM-CSF | S. cerevisiae | Chemotherapy-induced neutropenia | Sanofi, Paris, France | [93] |
| IFN-gamma protein | IFN-gamma protein | E. coli | Chronic granulomatous disease and Severe malignant osteopetrosis | Genentech, San Francisco, CA, USA | [115] |
| Pegylated interferon alpha-2a | Pegasys | E. coli | Hepatitis C | Genentech, San Francisco, CA, USA | [116] |
| Teriparatide | Forteo | E. coli | Osteoporosis | Eli Lilly & Co., Indianapolis, IN, USA | [117] |
| Somatotropin | Humatrope | E. coli | Growth hormone | Eli Lilly & Co., Indianapolis, IN, USA | [39] |
| Filgrastim | Neupogen | E. coli | Neutropenia | Amgen Inc., Fairfield, NJ, USA | [39] |
| PEG-Filgrastim | Neulasta | E. coli | Neutropenia | Amgen Inc., Fairfield, NJ, USA | [39] |
| rIL-2-diptheria toxin | Ontak | E. coli | Cancer | Eisai Inc., Tokyo, Japan | [118] |
| Asparginase | Erwinaze | E. coli | Cancer | EUSA Pharma, Hertford, UK | [119] |
| Glucarpidase | Voraxase | E. coli | Nephrology, Kidney failure | BTG International Inc., Conshohocken, PA, USA | [93] |
| Metreleptin | Myalept | E. coli | Lipodystrophy | Amylin Pharmaceutical, San Diego, CA, USA | [93] |
| Albiglutide | Tanzeum | S. cerevisiae | Diabetes | GlaxoSmithKline, GlaxoSmithKline, Middlesex, UK | [93] |
| Insulin glargine | Lantus | E. coli | Insulin receptor agonist | Sanofi-aventis, Paris, France | [39] |
| Endostatin | Endostar | E. coli | Lung cancer, Colorectal cancer | Simcere, Nanjing, China | [39] |
| Aldesleukin | Proleukin | E. coli | Metastatic renal cell carcinoma, metastatic melanoma | Novartis, Basel, Switzerland | [39] |
| Interferon alfa-2b | Intron-A | E. coli | Chronic hepatitis C, hairy cell leukemia, Behcet’s disease etc. | Schering-Plough Corporation, Kenilworth, NJ, USA | [39] |
| Interferon gamma-1a | Actimmune | E. coli | Kidney cancer, sezary syndrome, mycosis fungoides | Genentech, San Francisco, CA, USA | [39] |
| Tasonermin | Beromun | E. coli | Soft tissue sarcoma | Boehringer Ingelheim, Ingelheim am Rhein, Shanghai, China | [39] |
| Granulocyte macrophage colony-stimulating factor | Molgramostim | E. coli | Myelodysplastic syndrome | Swiss Pharma Pvt Ltd., Gujarat, India | [39] |
| Nartograstim | Neu-up | E. coli | Solid tumor | Yakult Honsha, Tokyo, Japan | [39] |
| Palifermin | Kepivance | E. coli | Metastatic renal cell carcinoma, metastatic melanoma | Biovitrum, Stockholm, Sweden | [39] |
| Sargramostim | Leukine | S. cerevisiae | Acute myelocyticleukaemia | Bayer HealthCare Pharmaceuticals, Stockholm, Sweden | [39] |
| L1 capsid protein | Gardasil | S. cerevisiae | Papillomavirus vaccine | Merck & Co., Stockholm, Sweden | [120] |
| ORF2 protein | Hecolin | E. coli | Hepatitis E virus vaccine | Xiamen Innovax Biotech, Xiamen, China | [120] |
| 2 fHbp variants | Trumenba | E. coli | Neisseria meningitidis serogrup B vaccine | Wyeth Pharmaceuticals, Inc., Collegeville, PA, USA | [120] |
| HBsAg + RTS chimera | Mosquirix | S. cerevisiae | Malaria vaccine | GlaxoSmithKline, Middlesex, UK | [120] |
| OspA & C chimeric | Vanguard crLyme | E. coli | Borrelia burgdorferi vaccine | Zoetis, Parsippany-Troy Hills, NJ, USA | [120] |
| Chimeric Protein Q | Letifend | E. coli | Leishmania vaccine | Zoetis, Parsippany-Troy Hills, NJ, USA | [120] |
| P45 env. antigen | Leucogen | E. coli | Feline leukemia virus vaccine | Kalbio Global Medika, Kalbio Global Medika, Bekasi Regency, Indonesia | [120] |
| CCE, mEq84, IdeE | Strangvac | E. coli | Streptococcus equi vaccine | Dechra Veterinary Products, Northwich, UK | [120] |
| Detemir Insulin | Levemir | S. cerevisiae | Diabetes | Novo Nordisk, Bagsvarerd, Denmark | [121] |
| Glucarpidase | Voraxaze | E. coli | Treatment of patients at risk of methotrexate toxicity | BTG Specialty Pharmaceuticals, Conshohocken, PA, USA | [93] |
| Parathyroid hormone | Natpara | E. coli | Treatment of hypoparathyroidism | Takeda Pharmaceuticals, Tokyo, Japan | [93] |
| Somatropin | Omnitrope | E. coli | Treatment of growth hormone deficiency | Sandoz, Basel, Switzerland | [122] |
| Semaglutide | Wegovy | S. cerevisiae | Weight loss and weight control | Novo Nordisk Novo Nordisk, Bagsvarerd, Denmark | [123] |
| Vosoritide | Voxzogo | E. coli | Achondroplasia | BioMarin Pharmaceutical, Novato, CA, USA | [124] |
| Liraglutide | Saxenda | S. cerevisiae | Obesity | Novo Nordisk, Bagsvarerd, Denmark | [125] |
| Nesiritide | Natrecor | E. coli | Acutely decompensated congestive heart failure | Johnson & Johnson, New Brunswick, NJ, USA | [126] |
| Anakinra | Kineret | E. coli | Rheumatoid arthritis | Swedish Orphan Biovitrum, Biovitrum, Stockholm, Sweden | [127] |
| Becaplermin | Regranex | S. cerevisiae | Lower-extremity diabetic neuropathic ulcers | Johnson & Johnson, New Brunswick, NJ, USA | [128] |
| Insulin | Insugen | P. pastoris | Diabetes therapy | Biocon, Karnataka, India | [129] |
| Ecallantide | Kalbitor | P. pastoris | Hereditary angioedema | Dyax, Lexington, MA, USA | [129] |
| Human serum albumin | Medway | P. pastoris | Blood volume expansion | Mitsubishi Tanabe Pharma, Osaka, Japan | [129] |
| IFN-α 2b | Shanferon | P. pastoris | Hepatitis C | Sanofi, Paris, France | [129] |
| Ocriplasmin | Jetrea | P. pastoris | Vitreomacular adhesion | ThromboGenics, Iselin, NJ, USA | [129] |
| HBV vaccine | Hepavax-Gene | H. polymorpha | Hepatitis B | Rhein Biotech, Maastricht, Netherlands | [129] |
| Pancrelipase | Creon | Y. lipolytica | Exocrine pancreatic insufficiency | Aptalis Pharma, Bridgewater, NJ, USA | [129] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jayakrishnan, A.; Wan Rosli, W.R.; Tahir, A.R.M.; Razak, F.S.A.; Kee, P.E.; Ng, H.S.; Chew, Y.-L.; Lee, S.-K.; Ramasamy, M.; Tan, C.S.; et al. Evolving Paradigms of Recombinant Protein Production in Pharmaceutical Industry: A Rigorous Review. Sci 2024, 6, 9. https://doi.org/10.3390/sci6010009
Jayakrishnan A, Wan Rosli WR, Tahir ARM, Razak FSA, Kee PE, Ng HS, Chew Y-L, Lee S-K, Ramasamy M, Tan CS, et al. Evolving Paradigms of Recombinant Protein Production in Pharmaceutical Industry: A Rigorous Review. Sci. 2024; 6(1):9. https://doi.org/10.3390/sci6010009
Chicago/Turabian StyleJayakrishnan, Achuth, Wan Rosalina Wan Rosli, Ahmad Rashidi Mohd Tahir, Fashli Syafiq Abd Razak, Phei Er Kee, Hui Suan Ng, Yik-Ling Chew, Siew-Keah Lee, Mahenthiran Ramasamy, Ching Siang Tan, and et al. 2024. "Evolving Paradigms of Recombinant Protein Production in Pharmaceutical Industry: A Rigorous Review" Sci 6, no. 1: 9. https://doi.org/10.3390/sci6010009
APA StyleJayakrishnan, A., Wan Rosli, W. R., Tahir, A. R. M., Razak, F. S. A., Kee, P. E., Ng, H. S., Chew, Y.-L., Lee, S.-K., Ramasamy, M., Tan, C. S., & Liew, K. B. (2024). Evolving Paradigms of Recombinant Protein Production in Pharmaceutical Industry: A Rigorous Review. Sci, 6(1), 9. https://doi.org/10.3390/sci6010009






