Identification of Anti-Tuberculosis Drugs Targeting DNA Gyrase A and Serine/Threonine Protein Kinase PknB: A Machine Learning-Assisted Drug-Repurposing Approach
Abstract
1. Introduction
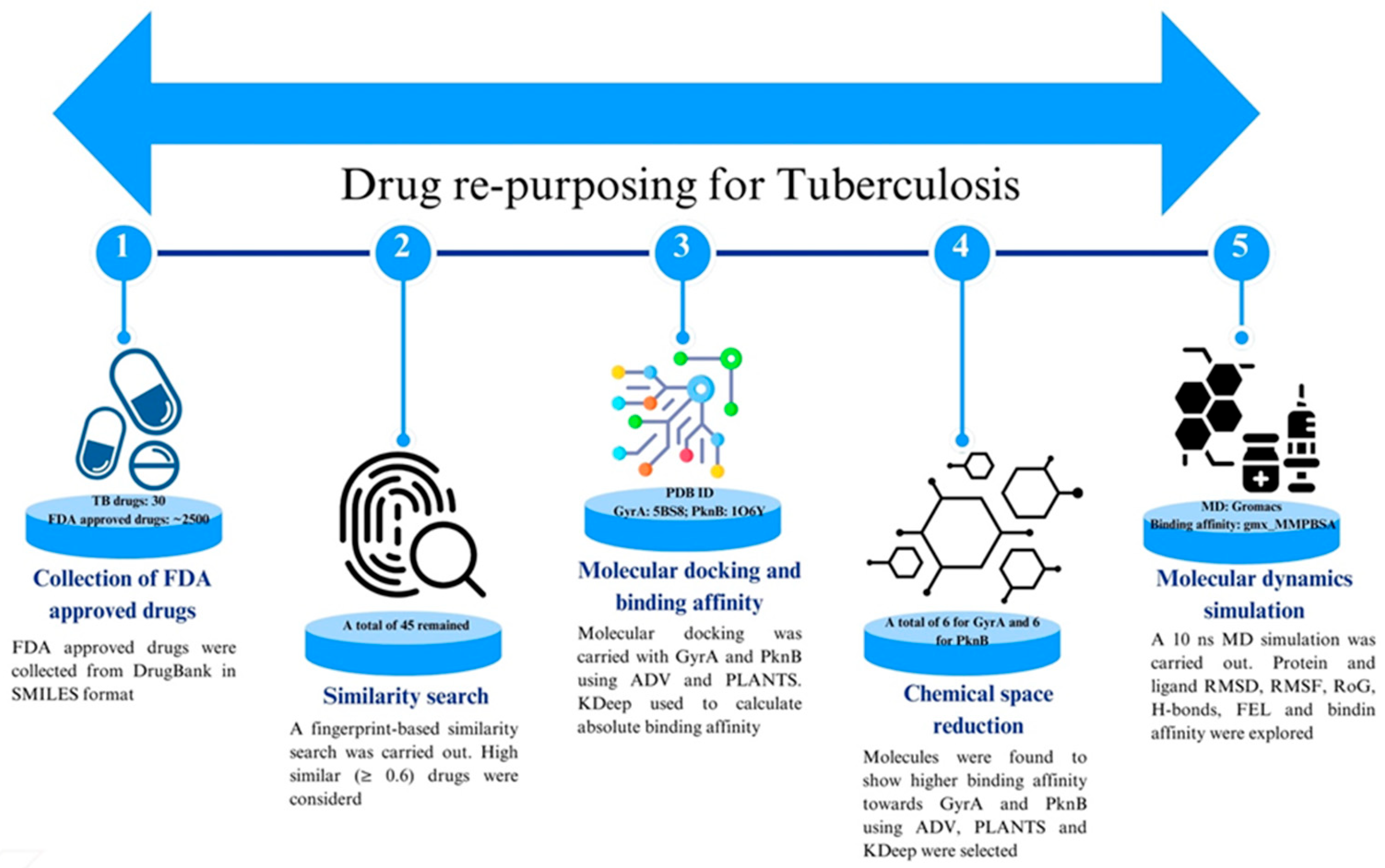
2. Materials and Methods
2.1. Selection of GyrA and PknB Structure and Preparation
2.2. Collection and Preparation of FDA-Approved Drugs
2.3. Fingerprint Based Similarity Search
2.4. Molecular Docking Using Autodock Vina and PLANTS
2.5. Absolute Binding Affinity Using KDeep
2.6. Molecular Dynamics Simulation
2.7. Binding Free Energy Calculation Using MM-GBSA Approach
3. Results and Discussion
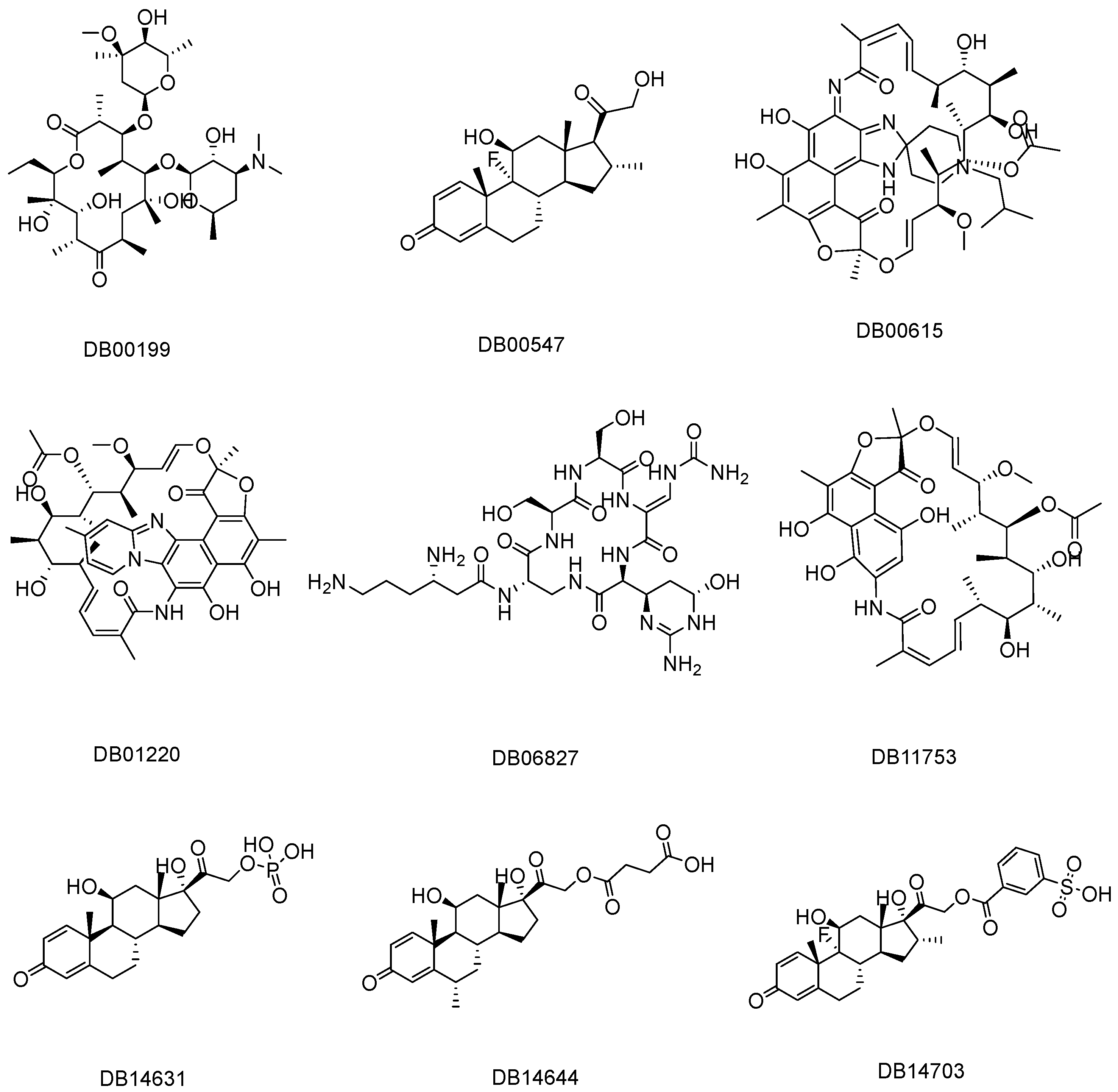
3.1. Similarity Search
3.2. Screening Through Molecular Docking and Absolute Binding Affinity
3.3. Binding Interactions Profile
3.3.1. Binding Interaction with DNA GyrA
3.3.2. Binding Interactions with PknB
3.4. Molecular Dynamics Simulation
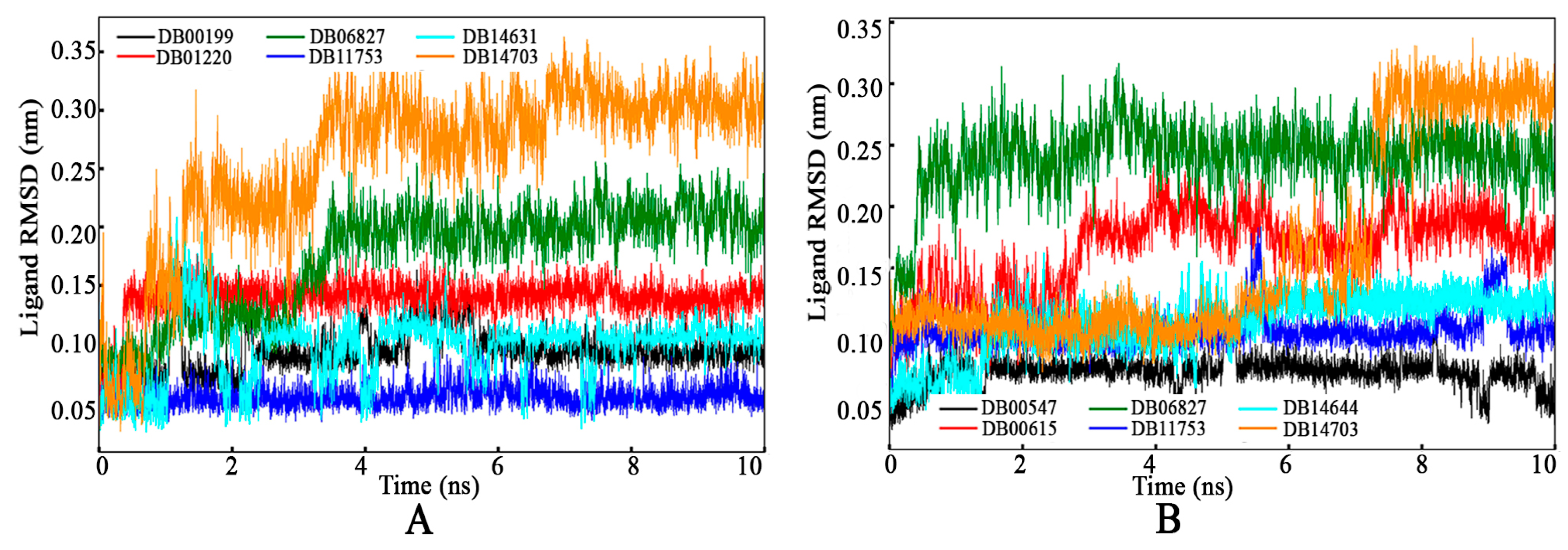
3.4.1. Root-Mean-Square Deviation
Protein Backbone RMSD
Ligand RMSD
3.4.2. Root-Means-Square Fluctuation
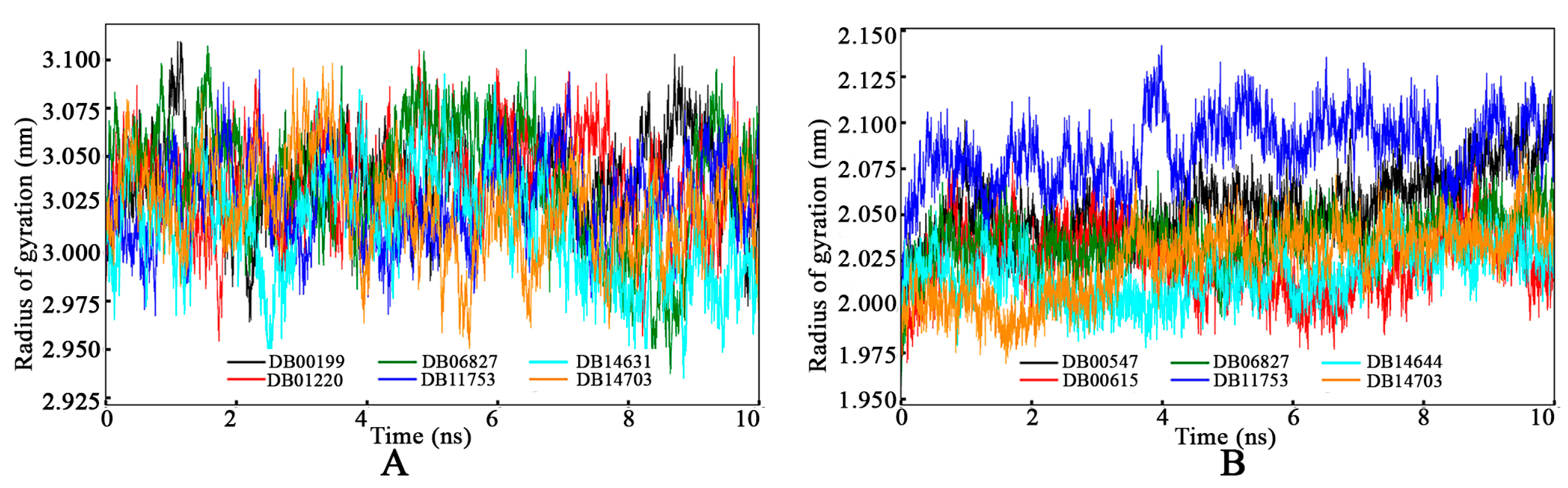
3.4.3. Radius of Gyration
3.4.4. Intermolecular Hydrogen Bond
3.4.5. Binding Free Energy Calculation Using MM-GBSA Approach
3.4.6. Free Energy Landscape
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alsayed, S.S.R.; Gunosewoyo, H. Tuberculosis: Pathogenesis, Current Treatment Regimens and New Drug Targets. Int. J. Mol. Sci. 2023, 24, 5202. [Google Scholar] [CrossRef] [PubMed]
- Bourzac, K. Infectious disease: Beating the big three. Nature 2014, 507, S4–S7. [Google Scholar] [CrossRef] [PubMed]
- Udoakang, A.J.; Djomkam Zune, A.L.; Tapela, K.; Nganyewo, N.N.; Olisaka, F.N.; Anyigba, C.A.; Tawiah-Eshun, S.; Owusu, I.A.; Paemka, L.; Awandare, G.A.; et al. The COVID-19, tuberculosis and HIV/AIDS: Ménage à Trois. Front. Immunol. 2023, 14, 1104828. [Google Scholar] [CrossRef] [PubMed]
- Harries, A.D.; Lin, Y.; Kumar, A.M.V.; Satyanarayana, S.; Takarinda, K.C.; Dlodlo, R.A.; Zachariah, R.; Olliaro, P. What can National TB Control Programmes in low- and middle-income countries do to end tuberculosis by 2030? F1000Research 2018, 7. [Google Scholar] [CrossRef]
- Monedero-Recuero, I.; Gegia, M.; Wares, D.F.; Chadha, S.S.; Mirzayev, F. Situational analysis of 10 countries with a high burden of drug-resistant tuberculosis 2 years post-UNHLM declaration: Progress and setbacks in a changing landscape. Int. J. Infect. Dis. 2021, 108, 557–567. [Google Scholar] [CrossRef]
- Smith, I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 2003, 16, 463–496. [Google Scholar] [CrossRef]
- Cozzarelli, N.R. DNA Gyrase and the supercoiling of DNA. Science 1980, 207, 953–960. [Google Scholar] [CrossRef]
- Collin, F.; Karkare, S.; Maxwell, A. Exploiting bacterial DNA gyrase as a drug target: Current state and perspectives. Appl. Microbiol. Biotechnol. 2011, 92, 479–497. [Google Scholar] [CrossRef]
- Khan, T.; Sankhe, K.; Suvarna, V.; Sherje, A.; Patel, K.; Dravyakar, B. DNA gyrase inhibitors: Progress and synthesis of potent compounds as antibacterial agents. Biomed. Pharmacother. 2018, 103, 923–938. [Google Scholar] [CrossRef]
- Dwyer, D.J.; Kohanski, M.A.; Hayete, B.; Collins, J.J. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol. Syst. Biol. 2007, 3, 91. [Google Scholar] [CrossRef]
- Prisic, S.; Husson, R.N. Mycobacterium tuberculosis Serine/Threonine Protein Kinases. Microbiol. Spectr. 2014, 2, 681–708. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.Z.; Kaur, P.; Nandicoori, V.K. Targeting the messengers: Serine/threonine protein kinases as potential targets for antimycobacterial drug development. IUBMB Life 2018, 70, 889–904. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.M.; Abbott, D.W.; Sang, T.P.; Dascher, C.C.; Cantley, L.C.; Husson, R.N. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: Substrate identification and regulation of cell shape. Genes Dev. 2005, 19, 1692–1704. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Sammartino, J.C.; Costantino, L.; Gelain, A.; Meneghetti, F.; Villa, S.; Chiarelli, L.R. An Overview on the Potential Antimycobacterial Agents Targeting Serine/Threonine Protein Kinases from Mycobacterium tuberculosis. Curr. Top. Med. Chem. 2019, 19, 646–661. [Google Scholar] [CrossRef]
- Wehenkel, A.; Fernandez, P.; Bellinzoni, M.; Catherinot, V.; Barilone, N.; Labesse, G.; Jackson, M.; Alzari, P.M. The structure of PknB in complex with mitoxantrone, an ATP-competitive inhibitor, suggests a mode of protein kinase regulation in mycobacteria. FEBS Lett. 2006, 580, 3018–3022. [Google Scholar] [CrossRef]
- Fernandez, P.; Saint-Joanis, B.; Barilone, N.; Jackson, M.; Gicquel, B.; Cole, S.T.; Alzari, P.M. The Ser/Thr protein kinase PknB is essential for sustaining mycobacterial growth. J. Bacteriol. 2006, 188, 7778–7784. [Google Scholar] [CrossRef]
- Mieczkowski, C.; Iavarone, A.T.; Alber, T. Auto-activation mechanism of the Mycobacterium tuberculosis PknB receptor Ser/Thr kinase. EMBO J. 2008, 27, 3186–3197. [Google Scholar] [CrossRef]
- Chapman, T.M.; Bouloc, N.; Buxton, R.S.; Chugh, J.; Lougheed, K.E.A.; Osborne, S.A.; Saxty, B.; Smerdon, S.J.; Taylor, D.L.; Whalley, D. Substituted aminopyrimidine protein kinase B (PknB) inhibitors show activity against Mycobacterium tuberculosis. Bioorganic Med. Chem. Lett. 2012, 22, 3349–3353. [Google Scholar] [CrossRef]
- Lougheed, K.E.A.; Osborne, S.A.; Saxty, B.; Whalley, D.; Chapman, T.; Bouloc, N.; Chugh, J.; Nott, T.J.; Patel, D.; Spivey, V.L.; et al. Effective inhibitors of the essential kinase PknB and their potential as anti-mycobacterial agents. Tuberculosis 2011, 91, 277–286. [Google Scholar] [CrossRef]
- Székely, R.; Wáczek, F.; Szabadkai, I.; Németh, G.; Hegymegi-Barakonyi, B.; Eros, D.; Szokol, B.; Pató, J.; Hafenbradl, D.; Satchell, J.; et al. A novel drug discovery concept for tuberculosis: Inhibition of bacterial and host cell signalling. Immunol. Lett. 2008, 116, 225–231. [Google Scholar] [CrossRef]
- Wang, T.; Bemis, G.; Hanzelka, B.; Zuccola, H.; Wynn, M.; Moody, C.S.; Green, J.; Locher, C.; Liu, A.; Gao, H.; et al. Mtb PKNA/PKNB Dual Inhibition Provides Selectivity Advantages for Inhibitor Design to Minimize Host Kinase Interactions. ACS Med. Chem. Lett. 2017, 8, 1224–1229. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, J.X.; Zhou, J.M.; Xu, C.L.; Huang, B.; Xing, Y.; Wang, B.; Luo, R.; Wang, Y.C.; You, X.F.; et al. A novel protein kinase inhibitor IMB-YH-8 with anti-tuberculosis activity. Sci. Rep. 2017, 7, 5093. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Q.; Pang, J.; Li, G.; Li, C.; Yang, X.; Yu, L.; Wang, Y.; Li, J.; You, X. Validated LC–MS/MS method for determination of YH-8, a novel PKnB inhibitor, in rat plasma and its application to pharmacokinetic study. Acta Pharm. Sin. B 2015, 5, 467–472. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhai, Q.Q.; Pang, J.; Li, G.Q.; Li, C.R.; Wang, Y.C.; Yu, L.Y.; Li, J.; You, X.F. Preclinical pharmacokinetic analysis of (E)-Methyl-4-aryl-4-oxabut-2-enoate, a novel Ser/Thr protein kinase B inhibitor, in rats. Acta Pol. Pharm.-Drug Res. 2017, 74, 299–307. [Google Scholar]
- Ren, J.; Xu, J.; Zhang, G.; Xu, C.; Zhao, L.L.; You, X.F.; Wang, Y.; Lu, Y.; Yu, L.; Wang, J. Design, synthesis, and bioevaluation of a novel class of (E)-4-oxo-crotonamide derivatives as potent antituberculosis agents. Bioorganic Med. Chem. Lett. 2019, 29, 539–543. [Google Scholar] [CrossRef]
- Xu, C.; Bai, X.; Xu, J.; Ren, J.; Xing, Y.; Li, Z.; Wang, J.; Shi, J.; Yu, L.; Wang, Y. Substituted 4-oxo-crotonic acid derivatives as a new class of protein kinase B (PknB) inhibitors: Synthesis and SAR study. RSC Adv. 2017, 7, 4763–4775. [Google Scholar] [CrossRef]
- Hegymegi-Barakonyi, B.; Szekely, R.; Varga, Z.; Kiss, R.; Borbely, G.; Nemeth, G.; Banhegyi, P.; Pato, J.; Greff, Z.; Horvath, Z.; et al. Signalling Inhibitors Against Mycobacterium tuberculosis—Early Days of a New Therapeutic Concept in Tuberculosis. Curr. Med. Chem. 2008, 15, 2760–2770. [Google Scholar] [CrossRef]
- Sipos, A.; Pató, J.; Székely, R.; Hartkoorn, R.C.; Kékesi, L.; Orfi, L.; Szántai-Kis, C.; Mikušová, K.; Svetlíková, Z.; Korduláková, J.; et al. Lead selection and characterization of antitubercular compounds using the Nested Chemical Library. Tuberculosis 2015, 95, S200–S206. [Google Scholar] [CrossRef]
- Bais, V.S.; Mohapatra, B.; Ahamad, N.; Boggaram, S.; Verma, S.; Prakash, B. Investigating the inhibitory potential of 2-Aminopurine metal complexes against serine/threonine protein kinases from Mycobacterium tuberculosis. Tuberculosis 2018, 108, 47–55. [Google Scholar] [CrossRef]
- Coluccia, A.; La Regina, G.; Barilone, N.; Lisa, M.-N.; Brancale, A.; André-Leroux, G.; Alzari, P.M.; Silvestri, R. Structure-based Virtual Screening to Get New Scaffold Inhibitors of the Ser/Thr Protein Kinase PknB from Mycobacterium tuberculosis. Lett. Drug Des. Discov. 2016, 13, 1012–1018. [Google Scholar] [CrossRef]
- Wlodarchak, N.; Teachout, N.; Beczkiewicz, J.; Procknow, R.; Schaenzer, A.J.; Satyshur, K.; Pavelka, M.; Zuercher, W.; Drewry, D.; Sauer, J.D.; et al. In Silico Screen and Structural Analysis Identifies Bacterial Kinase Inhibitors which Act with β-Lactams to Inhibit Mycobacterial Growth. Mol. Pharm. 2018, 15, 5410–5426. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Gao, W.; Hu, H.; Zhou, S. Why 90% of clinical drug development fails and how to improve it? Acta Pharm. Sin. B 2022, 12, 3049–3062. [Google Scholar] [CrossRef] [PubMed]
- Hinkson, V.I.; Madej, B.; Stahlberg, E.A. Accelerating Therapeutics for Opportunities in Medicine: A Paradigm Shift in Drug Discovery. Front. Pharmacol. 2020, 11, 770. [Google Scholar] [CrossRef]
- Sadybekov, V.A.; Katritch, V. Computational approaches streamlining drug discovery. Nature 2023, 616, 673–685. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2018, 18, 41–58. [Google Scholar] [CrossRef]
- Ashburn, T.T.; Thor, K.B. Drug repositioning: Identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004, 3, 673–683. [Google Scholar] [CrossRef]
- Breckenridge, A.; Jacob, R. Overcoming the legal and regulatory barriers to drug repurposing. Nat. Rev. Drug Discov. 2018, 18, 1–2. [Google Scholar] [CrossRef]
- Blower, T.R.; Williamson, B.H.; Kerns, R.J.; Berger, J.M. Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2016, 113, 1706–1713. [Google Scholar] [CrossRef]
- Ortiz-Lombardía, M.; Pompeo, F.; Boitel, B.; Alzari, P.M. Crystal structure of the catalytic domain of the PknB serine/threonine kinase from Mycobacterium tuberculosis. J. Biol. Chem. 2003, 278, 13094–13100. [Google Scholar] [CrossRef]
- Knox, C.; Wilson, M.; Klinger, C.M.; Franklin, M.; Oler, E.; Wilson, A.; Pon, A.; Cox, J.; Chin, N.E.L.; Strawbridge, S.A.; et al. DrugBank 6.0: The DrugBank Knowledgebase for 2024. Nucleic Acids Res. 2024, 52, D1265–D1275. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open chemical toolbox. J. Cheminform. 2011, 3, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, J.; Marsili, M. A new model for calculating atomic charges in molecules. Tetrahedron Lett. 1978, 19, 3181–3184. [Google Scholar] [CrossRef]
- Cai, L.; Chu, J.; Xu, J.; Meng, Y.; Lu, C.; Tang, X.; Wang, G.; Tian, G.; Yang, J. Machine learning for drug repositioning: Recent advances and challenges. Curr. Res. Chem. Biol. 2023, 3, 100042. [Google Scholar] [CrossRef]
- Rogers, D.; Hahn, M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. [Google Scholar] [CrossRef]
- Hu, Y.; Lounkine, E.; Bajorath, J. Improving the search performance of extended connectivity fingerprints through activity-oriented feature filtering and application of a bit-density-dependent similarity function. ChemMedChem 2009, 4, 540–548. [Google Scholar] [CrossRef]
- RDKit: Open-Source Cheminformatics Software. 2016. Available online: http://www.rdkit.org/ (accessed on 19 November 2024).
- Bajusz, D.; Rácz, A.; Héberger, K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J. Cheminform. 2015, 7, 1–13. [Google Scholar] [CrossRef]
- Pinzi, L.; Rastelli, G. Molecular docking: Shifting paradigms in drug discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Korb, O.; Stützle, T.; Exner, T.E. An ant colony optimization approach to flexible protein–ligand docking. Swarm Intell. 2007, 1, 115–134. [Google Scholar] [CrossRef]
- Kastritis, P.L.; Bonvin, A.M.J.J. On the binding affinity of macromolecular interactions: Daring to ask why proteins interact. J. R. Soc. Interface 2013, 10, 20120835. [Google Scholar] [CrossRef]
- Jimenez, J.; Skalic, M.; Martinez-Rosell, G.; De Fabritiis, G. K(DEEP): Protein-Ligand Absolute Binding Affinity Prediction via 3D-Convolutional Neural Networks. J. Chem. Inf. Model. 2018, 58, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Lindahl, E.; Abraham, M.J.; Hess, B.; van der Spoel, D. GROMACS 2021.3; Zenodo, 2021. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; De Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2016, 14, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Zoete, V.; Cuendet, M.A.; Grosdidier, A.; Michielin, O. SwissParam: A fast force field generation tool for small organic molecules. J. Comput. Chem. 2011, 32, 2359–2368. [Google Scholar] [CrossRef]
- Mark, P.; Nilsson, L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Simmonett, A.C.; Brooks, B.R. A compression strategy for particle mesh Ewald theory. J. Chem. Phys. 2021, 154, 5. [Google Scholar] [CrossRef]
- Maity, D.; Pal, D. MD DaVis: Interactive data visualization of protein molecular dynamics. Bioinformatics 2022, 38, 3299–3301. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. Gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef]







| DrugBank ID | Binding Energy (kcal/mol) | |||||
|---|---|---|---|---|---|---|
| Autodock Vina | PLANTS | KDeep | ||||
| GyrA | PknB | GyrA | PknB | GyrA | PknB | |
| DB00199 | −8.000 | −6.600 | −81.060 | −67.163 | −9.650 | −9.869 |
| DB00547 | −8.000 | −8.000 | −72.456 | −78.860 | −8.435 | −9.961 |
| DB00615 | −8.400 | −7.600 | −69.230 | −75.846 | −11.475 | −11.719 |
| DB01220 | −8.600 | −7.200 | −74.562 | −70.409 | −9.928 | −10.194 |
| DB06827 | −8.400 | −7.500 | −85.054 | −93.591 | −11.251 | −12.061 |
| DB11753 | −9.100 | −8.500 | −77.073 | −79.248 | −9.339 | −9.995 |
| DB14631 | −8.300 | −7.600 | −77.792 | −95.175 | −9.003 | −8.090 |
| DB14644 | −7.800 | −8.100 | −87.791 | −96.450 | −9.254 | −10.889 |
| DB14703 | −9.100 | −8.900 | −81.491 | −102.005 | −10.626 | −11.840 |
| DrugBank IDs | Hydrogen Bonds | Hydrophobic Interactions | Salt Bridge |
|---|---|---|---|
| DNA GyrA | |||
| DB00199 | Asp94, Arg98 | Ala40, Lys49, Val51, Ile92, Asp94, Ile181 | Lys49, His52, Arg98 |
| DB01220 | Asp94, Arg98, Gln277, Asn279 | Asp94, Arg98, Pro119 | - |
| DB06827 | Asp94, Arg98, Gln101, Trp103, Ser118, Pro119, Gly120, Pro124, Asn279 | - | - |
| DB11753 | Arg98, Gln277 | Val97, Arg98, Pro124, Asn279 | - |
| DB14631 | Asp94, Ser95, Arg98, Arg98, Gln101, Gln101, Gln277, Val278 | Gln277 | - |
| DB14703 | Asn115, Gly117, Ser307, Asp308, Gly311 | Trp103, Pro119, Asp308, Leu312 | - |
| Serine/threonine-protein kinase PknB | |||
| DB00547 | Val95, Ala142 | Leu17, Val25, Thr99 | |
| DB00615 | Gly21, Asp138, Lys140, Asn143, Asp156, Ala180 | Phe19 | - |
| DB06827 | Phe19, Gly21, Ser23, Glu59, Arg101, Asp102, Asp156 | Met155 | Asp102 |
| DB11753 | Leu17, Phe19, Arg101 | Val25, Ala38, Val95, Val98, Asp156 | - |
| DB14644 | Ser23, Val95, Thr99, Ala142 | Leu17, Thr99 | Lys40 |
| DB14703 | Glu59, Asp138, Ala142, Gly158, Ile159 | Leu17, Val25, Thr99 | Lys140 |
| DNA GyrA | Serine/Threonine-Protein Kinase PknB | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | G5 | G6 | S1 | S2 | G3 | G4 | S3 | G6 | ||
| Backbone RMSD (nm) | Max. | 0.396 | 0.464 | 0.451 | 0.396 | 0.398 | 0.352 | 0.429 | 0.320 | 0.364 | 0.446 | 0.342 | 0.316 |
| Min. | 0.000 | 0.001 | 0.000 | 0.000 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| Avg. | 0.254 | 0.328 | 0.283 | 0.262 | 0.277 | 0.267 | 0.289 | 0.228 | 0.275 | 0.343 | 0.238 | 0.218 | |
| Ligand RMSD (nm) | Max. | 0.172 | 0.178 | 0.256 | 0.113 | 0.208 | 0.363 | 0.110 | 0.245 | 0.317 | 0.184 | 0.162 | 0.337 |
| Min. | 0.001 | 0.000 | 0.001 | 0.000 | 0.000 | 0.000 | 0.001 | 0.000 | 0.001 | 0.000 | 0.001 | 0.000 | |
| Avg. | 0.093 | 0.139 | 0.173 | 0.054 | 0.097 | 0.257 | 0.064 | 0.164 | 0.239 | 0.098 | 0.104 | 0.163 | |
| RMSF (nm) | Max. | 0.957 | 1.198 | 1.352 | 0.987 | 0.880 | 0.790 | 0.695 | 0.495 | 0.373 | 0.368 | 0.324 | 0.313 |
| Min. | 0.063 | 0.067 | 0.060 | 0.061 | 0.059 | 0.057 | 0.050 | 0.051 | 0.043 | 0.067 | 0.047 | 0.047 | |
| Avg. | 0.146 | 0.163 | 0.171 | 0.146 | 0.149 | 0.142 | 0.142 | 0.134 | 0.113 | 0.142 | 0.131 | 0.124 | |
| RoG (nm) | Max. | 3.110 | 3.105 | 3.107 | 3.095 | 3.093 | 3.098 | 2.115 | 2.082 | 2.078 | 2.142 | 2.064 | 2.084 |
| Min. | 2.964 | 2.954 | 2.937 | 2.967 | 2.930 | 2.949 | 1.938 | 1.936 | 1.937 | 1.938 | 1.937 | 1.937 | |
| Avg. | 3.038 | 3.038 | 3.038 | 3.027 | 3.013 | 3.022 | 2.054 | 2.025 | 2.037 | 2.085 | 2.018 | 2.025 | |
| Molecule | ΔGbind (kcal/mol) | Standard Deviation | |
|---|---|---|---|
| GyrA | DB00199 | −21.450 | ±4.050 |
| DB01220 | −9.650 | ±4.630 | |
| DB06827 | −20.130 | ±5.050 | |
| DB11753 | −19.540 | ±3.870 | |
| DB14631 | −20.070 | ±4.800 | |
| DB14703 | −27.750 | ±4.760 | |
| PknB | DB00547 | −25.720 | ±3.980 |
| DB00615 | −8.580 | ±3.770 | |
| DB06827 | −19.570 | ±5.880 | |
| DB11753 | −24.750 | ±3.850 | |
| DB14644 | −36.460 | ±6.270 | |
| DB14703 | −51.510 | ±14.150 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, D.; Islam, M.A.; Natarajan, S.; Dudekula, D.B.; Chung, H.; Park, J.; Oh, B. Identification of Anti-Tuberculosis Drugs Targeting DNA Gyrase A and Serine/Threonine Protein Kinase PknB: A Machine Learning-Assisted Drug-Repurposing Approach. Trop. Med. Infect. Dis. 2024, 9, 288. https://doi.org/10.3390/tropicalmed9120288
Lee D, Islam MA, Natarajan S, Dudekula DB, Chung H, Park J, Oh B. Identification of Anti-Tuberculosis Drugs Targeting DNA Gyrase A and Serine/Threonine Protein Kinase PknB: A Machine Learning-Assisted Drug-Repurposing Approach. Tropical Medicine and Infectious Disease. 2024; 9(12):288. https://doi.org/10.3390/tropicalmed9120288
Chicago/Turabian StyleLee, Dongwoo, Md Ataul Islam, Sathishkumar Natarajan, Dawood Babu Dudekula, Hoyong Chung, Junhyung Park, and Bermseok Oh. 2024. "Identification of Anti-Tuberculosis Drugs Targeting DNA Gyrase A and Serine/Threonine Protein Kinase PknB: A Machine Learning-Assisted Drug-Repurposing Approach" Tropical Medicine and Infectious Disease 9, no. 12: 288. https://doi.org/10.3390/tropicalmed9120288
APA StyleLee, D., Islam, M. A., Natarajan, S., Dudekula, D. B., Chung, H., Park, J., & Oh, B. (2024). Identification of Anti-Tuberculosis Drugs Targeting DNA Gyrase A and Serine/Threonine Protein Kinase PknB: A Machine Learning-Assisted Drug-Repurposing Approach. Tropical Medicine and Infectious Disease, 9(12), 288. https://doi.org/10.3390/tropicalmed9120288








