Multiple-Drug Resistant Shiga Toxin-Producing E. coli in Raw Milk of Dairy Bovine
Abstract
1. Introduction
2. Materials and Methods
2.1. Source of Samples
2.2. Isolation and Characterization of E. coli from Raw Milk Sample
2.3. Biochemical Characterization
2.4. Serotyping of the E. coli Isolates
2.5. Phenotypic Detection of ESBL-Producing STEC
2.6. Detection of Shiga Toxin-Producing and ESBL Virulence Genes through Polymerase Chain Reaction (PCR)
Bacterial Culture
2.7. Antibiotic Susceptibility of STEC
2.8. Detection of Multidrug Resistance (MDR) among STEC Isolates
2.9. Statistical Analysis
3. Results
3.1. Prevalence of E. coli in Raw Milk
3.2. Multiplex PCR for Virulence Genes
3.3. Detection of ESBL Genes
3.4. Antimicrobial Resistance among STEC
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paton, A.W.; Paton, J.C. Detection and characterization of Shigatoxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 1998, 36, 598–602. [Google Scholar] [CrossRef] [PubMed]
- Riley, L.W.; Remis, R.S.; Helgerson, S.D.; McGee, H.B.; Wells, J.G.; Davis, B.R.; Hebert, R.J.; Olcott, E.S.; Johnson, L.M.; Hargrett, N.T.; et al. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 1983, 308, 681–685. [Google Scholar] [CrossRef] [PubMed]
- Vidal, M.; Kruger, E.; Durán, C.; Lagos, R.; Levine, M.; Prado, V.; Toro, C.; Vidal, R. Single multiplex PCR assay to identify simultaneously the six categories of diarrheagenic Escherichia coli associated with entericinfections. J. Clin. Microbiol. 2005, 43, 5362–5365. [Google Scholar] [CrossRef] [PubMed]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Burk, C.; Braumiller, I.; Becker, H.; Martlbauer, E. Nuclease fluorescence assay for the detection of verotoxin genes in raw milk. Lett. Appl. Microbiol. 2002, 35, 153–156. [Google Scholar] [CrossRef]
- Crump, J.A.; Sulka, A.C.; Langer, A.J.; Schaben, C.; Crielly, A.S.; Gage, R.; Baysinger, M.; Moll, M.; Withers, G.; Toney, D.M.; et al. Anout break of Escherichia coli O157:H7 infections among visitors to a dairyfarm. N. Engl. J. Med. 2002, 347, 555–560. [Google Scholar] [CrossRef]
- Zhao, S.; White, D.G.; Ge, B.; Ayers, S.; Friedman, S.; English, L.; Wagner, D.; Gaines, S.; Meng, J. Identification and characterization of integron-mediated antibiotic resistance among Shigatoxin-producing Escherichia coli isolates. Appl. Environ. Microbiol. 2001, 67, 1558–1564. [Google Scholar] [CrossRef] [PubMed]
- Keene, W.E.; Hedberg, K.; Herriott, D.E.; Hancock, D.D.; McKay, R.W.; Barrett, T.J.; Fleming, D.W. A Prolonged Outbreakof Escherichia coli O157:H7 infections caused by commercially distributed raw milk. J. Infect. Dis. 1997, 176, 815–818. [Google Scholar] [CrossRef]
- Wilson, J.B.; Clarke, R.C.; Renwick, S.A.; Rahn, V.; Johnson, R.P.; Karmali, M.A.; Lior, H.; Alves, D.; Gyles, C.L.; Sandhu, K.S.; et al. Verocytotoxigenic Escherichia coli infection in dairy farm families. J. Infect. Dis. 1996, 174, 1021–1027. [Google Scholar] [CrossRef]
- Lahti, E. Cattleand Reindeer as Possible Sources of Escherichia coli O157 Infection in Humans. Ph.D. Thesis, Food Hygiene and Environmental Health, University of Helsinki, Helsinki, Finland, 2003. [Google Scholar]
- Morgan, D.; Newman, C.; Hutchinson, D.; Walker, A.M.; Rowe, B.; Majid, F. Verotox in producing Escherichia coli O157 infections as sociated with the consumption of yoghurt. Epidemiol. Infect. 1993, 111, 181–188. [Google Scholar] [CrossRef]
- Chiueh, L.-C.; Liu, F.-M.; Shih, D.-C. Prevalence of Shigatoxin-producing Escherichia coli in feces and raw milk of domestic cattle and sheep. J. Food Drug Anal. 2002, 10, 39–46. [Google Scholar] [CrossRef]
- Pradel, N.; Boukhors, K.; Bertin, Y.; Forestier, C.; Martin, C.; Livrelli, V. Heterogeneity of Shigatoxin-producing Escherichia coli strains isolated from hemolytic-uremic syndrome patients, cattle, and food samples in central France. Appl. Environ. Microbiol. 2001, 67, 2460–2468. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dehkdi, F.S.; Yazdani, F.; Mozafari, J.; Valizadeh, Y. Virulence factors, serogroups and antimicrobial resistance properties of Escherichia coli strains in fermented dairy products. BMC Res. Notes 2014, 7, 217. [Google Scholar] [CrossRef]
- Ali, T.; ur Rahman, S.; Zhang, L.; Shahid, M.; Zhang, S.; Liu, G.; Gao, J.; Han, B. ESBL-producing Escherichia coli from cows suffering mastitis in China contain clinical class 1 integrons with CTX-M linked to ISCR1. Front. Microbiol. 2016, 7, 1931. [Google Scholar] [CrossRef] [PubMed]
- Vlat, C.; Auvray, F.; Forest, K.; Métayer, V.; Gay, E.; de Garam, C.P.; Madec, J.-Y.; Haenni, M. Phylogenetic grouping and virulence potential of extended-spectrum-β-lactamase-producing Escherichia coli strains in cattle. Appl. Environ. Microbiol. 2012, 78, 4677–4682. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, P.; Abiri, R.; Rezaei, M.; Salmanzadeh-Ahrabi, S. Isolation of Shigatoxin-producing Escherichia coli from raw milk in Kermanshah, Iran. Iran. J. Microbiol. 2013, 5, 233–238. [Google Scholar]
- Ptel, A.A.; Zhang, Y.; Fullerton, J.N.; Boelen, L.; Rongvaux, A.; Maini, A.A.; Bigley, V.; Flavell, R.A.; Gilroy, D.W.; Asquith, B.; et al. The fate and life span of human monocyte subsets in stady state and systemic inflammation. J. Exp. Med. 2017, 214, 1913–1923. [Google Scholar] [CrossRef]
- Abbey, T.C.; Deak, E. What’s new from the CLSI subcommittee on antimicrobial susceptibility testing M100. Clin. Microbiol. Newsl. 2019, 41, 203–209. [Google Scholar] [CrossRef]
- Irshad, H.; Cookson, A.; Hotter, G.; Besser, T.; On, S.; French, N. Epidemiology of Shigatox in producing Escherichia coli O157 in very young calves in the North Island of New Zealand. N. Z. Vet J. 2012, 60, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Shma, J.; Sharma, M.; Ray, P. Detection of TEM & SHV genes in Escherichia coli & Klebsiella pneumonia isolates in atertiary care hospital from India. Indian J. Med. Res. 2010, 132, 332–336. [Google Scholar]
- Weintin, M.P.; Limbago, B.; Patel, J.; Mathers, A.; Campeau, S.; Mazzulli, T. 100 performance standards for antimicrobial susceptibility testing. Clin. Lab. Stand. Inst. 2018, 27, 210–214. [Google Scholar]
- Hamida, A.; Javed, A.; Mohammad, W.; Yasir, A.; Javed, U. Microbial quality assessment study of branded and unbranded milk sold in Peshawar City, Pakistan. Pak. J. Nutr. 2009, 8, 704–709. [Google Scholar]
- Soomro, A.; Arain, M.; Khaskheli, M.; Bhutto, B. Isolation of Escherichia coli from raw milk and milk products in relation to public health sold under market conditions at Tandojam. Pak. J. Nutr. 2002, 1, 151–152. [Google Scholar]
- Ahmad, I.; Khattak, S.; Ali, R.; Nawaz, N.; Ullah, K.; Khan, S.B.; Ali, M.; Patching, S.G.; Mustafa, M.Z. Prevalence and molecular characterization of multidrug-resistant Escherichia coli O157:H7 from dairy milk in the Peshawar region of Pakistan. J. Food Saf. 2021, 41, e12941. [Google Scholar] [CrossRef]
- Shahzad, K.; Muhammad, K.; Sheikh, A.; Yaqub, T.; Rabbani, M.; Hussain, T.; Anjum, A.A.; Anees, M. Isolation and molecular characterization of shigatox in producing E. coli O157. J. Anim. Plant Sci. 2013, 23, 1618–1621. [Google Scholar]
- Samad, A.; Abbas, F.; Ahmad, Z.; Tanveer, Z.; Ahmad, I.; Patching, S.G.; Nawaz, N.; Asmat, M.T.; Raziq, A.; Naeem, M.; et al. Multiplex polymerase chain reaction detection of Shigatox in genes and antibiotic sensitivity of Escherichia coli O157:H7 isolated from beef meat in Quetta, Pakistan. J. Food Saf. 2018, 38, e12540. [Google Scholar] [CrossRef]
- Haile, A.F.; Alonso, S.; Berhe, N.; Atoma, T.B.; Boyaka, P.N.; Grace, D. Prevalence, antibiogram, and multidrug-resistant profile of E. coli O157:H7 in retail raw beef in Addis Ababa, Ethiopia. Front. Vet. Sci. 2022, 9, 734896. [Google Scholar] [CrossRef] [PubMed]
- Karmali, M.A. Factors in the emergence of serious human infections associated with highly pathogenics trains of shigatoxin-producing Escherichia coli. Int. J. Med. Microbiol. 2018, 308, 1067–1072. [Google Scholar] [CrossRef]
- Tarr, C.L.; Nelson, A.M.; Beutin, L.; Olsen, K.E.P.; Whittam, T.S. Molecular characterization reveals similar virulence gene content in unrelated clonal groups of Escherichia coli of serogroup O174(OX3). J. Bacteriol. 2008, 190, 1344–1349. [Google Scholar] [CrossRef][Green Version]
- Tahra, B.; Ullah, K.; Samad, A.; Ali, S.; Nabi, S.; Naeem, M.; Hussain, H.; Mohammad, A. Isolation and molecular characterization of Shigatox in producing E. coli O157:h7 in raw milk using mPCR. Int. J. Pharm. Sci. Res. 2017, 8, 3107–3112. [Google Scholar]
- Abdullah, S.; Rahman, S.; Muhammad, F.; Mohsin, M. Association between antimicrobial consumption and resistantce rate of Escherichia coli in hospital settings. J. Appl. Microbiol. 2023, 134, lxac003. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Wang, J.; Ho, H.; Wang, Y.; Huang, S.; Han, R. Prevalence and antimicrobial-resistance phenotypes and genotypes of Escherichia coli isolated from raw milk samples from mastitis cases in four regions of China. J. Glob. Antimicrob. Resist. 2019, 22, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Pakbin, B.; Allahyari, S.; Amani, Z.; Brück, W.M.; Mahmoudi, R.; Peymani, A. Prevalence, phylogroups and antimicrobial susceptibility of Escherichia coli isolates from food products. Antibiotics 2021, 10, 1291. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.; Kotwal, S.; Malik, M.; Singh, M. Prevalence, genetic profile of virulence determinants and multi drug resistance of Escherichia coli isolates from foods of animal origin. Vet. World 2013, 6, 139–142. [Google Scholar] [CrossRef]

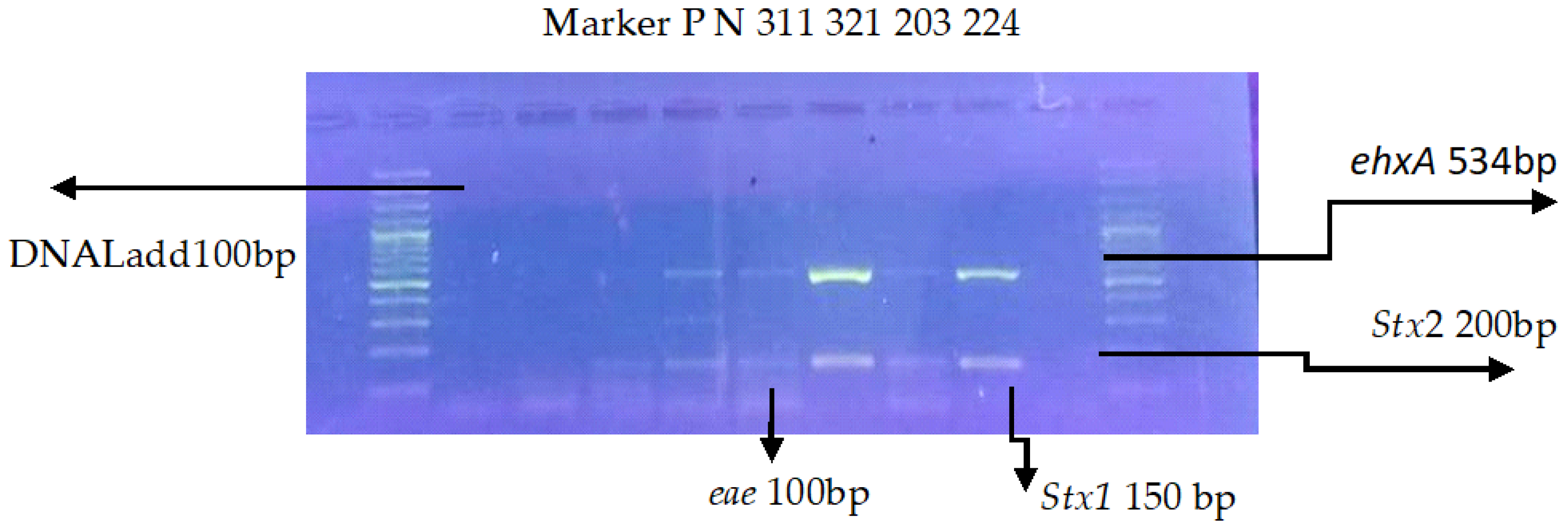
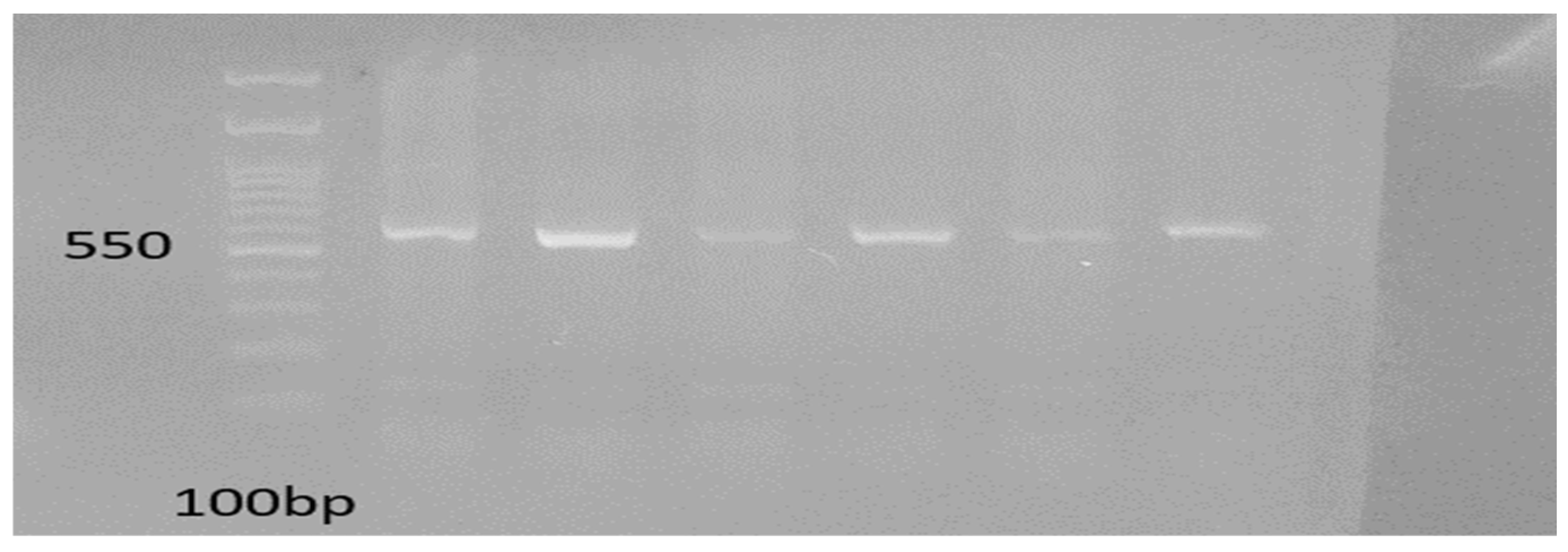
| Genes | PCR Conditions | PCR Reaction Volume |
|---|---|---|
| Stx1, Stx2, eae, ehxA | 1cycle | 2.5 μL of 10× PCR buffer |
| 96 °C-10 min | 0.15 mM MgCl2 | |
| 35 cycles | 0.1 mM of each dNTP | |
| 95 °C-45 s | 0.5 μL of each primer | |
| 60 °C-45 s | One unit of Taq DNA polymerase | |
| 72 °C-45 s | 3 μL of DNA | |
| 1 Cycle final volume of 25 μL with sterile water 72 °C-8 min | ||
| blaCTX-M, blaTEM, blaSHV | 1cycle 2.5 μL of | 2.5 μL of 10× PCR buffer 10× PCR |
| 96 °C-5 min | 0.15 mM MgCl2 buffer | |
| 25 cycles | 0.1 mM of each dNTP (Thermoscientific, Waltham, MA, USA) | |
| 95 °C-1 min | 1.0 μLmM of blaCTX-M, TEM, SHV primers | |
| 56 °C, 58 °C-1 min | One unit of Taq DNA polymerase | |
| 72 °C-1 min | 5 μL of DNA | |
| 1 Cycle, 72 °C-10 min held 4 °C forever the final volume of 20 μL with sterile water | ||
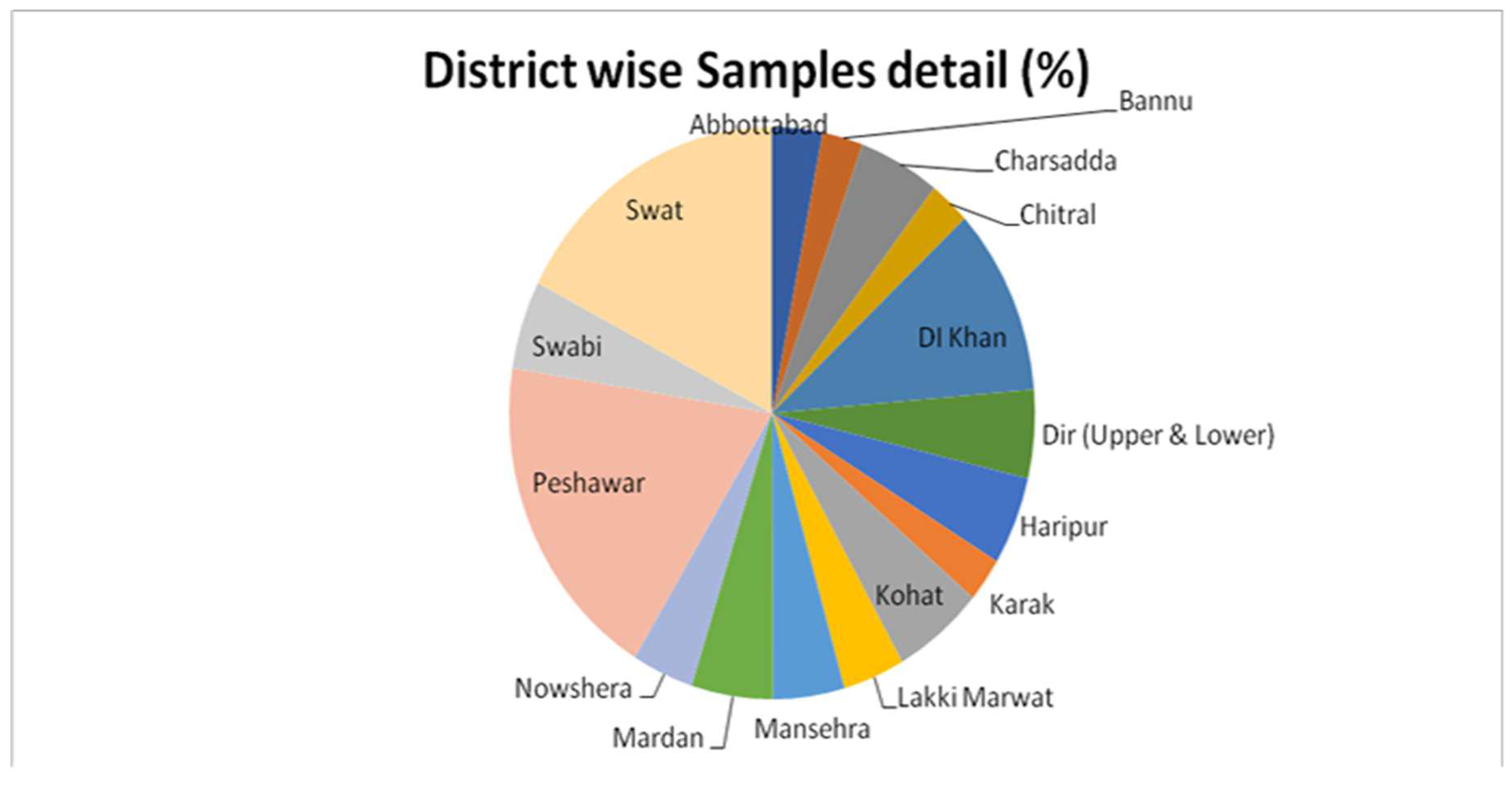
| Districts | E. coli | E. coli non-O157:H7 | Total | ||
|---|---|---|---|---|---|
| Positive | Not Detected | Positive | Not Detected | ||
| Abbottabad | 07 | 18 | 0 | 25 | 25 |
| Bannu | 10 | 10 | 1 | 19 | 20 |
| Charsadda | 21 | 19 | 2 | 38 | 40 |
| Chitral | 9 | 11 | 0 | 20 | 20 |
| Dera Ismail Khan | 32 | 53 | 6 | 79 | 85 |
| Dir (Upper and Lower) | 15 | 25 | 0 | 40 | 40 |
| Haripur | 12 | 28 | 0 | 40 | 40 |
| Karak | 8 | 12 | 0 | 20 | 20 |
| Kohat | 21 | 24 | 4 | 41 | 45 |
| Lakki Marwat | 12 | 18 | 1 | 29 | 30 |
| Mansehra | 7 | 28 | 0 | 35 | 35 |
| Mardan | 11 | 29 | 2 | 38 | 40 |
| Nowshera | 12 | 18 | 2 | 28 | 30 |
| Peshawar | 78 | 72 | 20 | 130 | 150 |
| Swabi | 19 | 21 | 2 | 38 | 40 |
| Swat | 47 | 93 | 0 | 140 | 140 |
| Total | 321 | 479 | 40 | 760 | 800 |
| Source Detail | E. coli | STEC | ESBL | Total | |||
|---|---|---|---|---|---|---|---|
| Positive | Not Detected | Positive | Not Detected | Positive | Not Detected | ||
| Dairy Farms and Individual Farmers | 073 | 127 | 08 | 192 | 02 | 198 | 200 |
| Milk Collection Centre | 013 | 013 | 01 | 025 | 00 | 026 | 026 |
| Milk Shops | 207 | 293 | 30 | 470 | 08 | 492 | 500 |
| Milk Vendors | 028 | 046 | 01 | 073 | 01 | 073 | 074 |
| Total | 321 | 479 | 40 | 760 | 11 | 789 | 800 |
| Probability Statistics | p-Value = 0.449 | p-Value = 0.305 | p-Value = 0.860 | ||||
| STEC Virulence Genes | N (%) |
|---|---|
| Stx1, eae | 4 (10.0) |
| Stx1, stx2, ehxA | 5 12.5) |
| eae | 5 (12.5) |
| Stx1, ehxA | 6 (15.0) |
| ehxA | 4 (10.0) |
| Stx1, eae, ehxA | 3 (7.5) |
| Stx2, eae, ehxA | 2 (5.0) |
| Stx1, stx2 | 6 (15.0) |
| Stx2 | 3(7.5) |
| Stx1 | 2 (5.0) |
| Total | 40 (100.0%) |
| Antibiotic | Disc Concert | Resistance | % | Intermediate | % | Sensitive | % |
|---|---|---|---|---|---|---|---|
| Penicillin | P 10 IU | 11 | 50 | 7 | 31 | 4 | 18 |
| Amoxicillin | AML 30 µG | 17 | 77 | 5 | 22 | 0 | 0 |
| Amoxicillin/Clavulanic acid | AUG 30 µG | 13 | 59 | 4 | 18 | 5 | 22 |
| Sulphamathoxole | SMX 50 µG | 9 | 40 | 8 | 36 | 5 | 22 |
| Gentamicin | CN 10 µG | 7 | 31 | 7 | 31 | 8 | 36 |
| Streptomycin | S 10 µG | 11 | 50 | 6 | 27 | 5 | 22 |
| Oxytetracyline | OT 30 µG | 6 | 27 | 8 | 36 | 8 | 36 |
| Ceftriaxone | CRO 30 µG | 12 | 54 | 6 | 27 | 4 | 18 |
| Norfloxacin | NOR 10 µG | 5 | 22 | 4 | 18 | 13 | 59 |
| Enrofloxacin | ENR 5 µG | 4 | 18 | 6 | 27 | 12 | 54 |
| Florefencial | FFC 30 µG | 6 | 27 | 5 | 22 | 11 | 50 |
| Cefotaxime/Clavulnic acid | CTL 40 µG | 8 | 36 | 9 | 40 | 5 | 22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ullah, S.; Khan, S.U.H.; Khan, M.J.; Khattak, B.; Fozia, F.; Ahmad, I.; Wadaan, M.A.; Khan, M.F.; Baabbad, A.; Goyal, S.M. Multiple-Drug Resistant Shiga Toxin-Producing E. coli in Raw Milk of Dairy Bovine. Trop. Med. Infect. Dis. 2024, 9, 64. https://doi.org/10.3390/tropicalmed9030064
Ullah S, Khan SUH, Khan MJ, Khattak B, Fozia F, Ahmad I, Wadaan MA, Khan MF, Baabbad A, Goyal SM. Multiple-Drug Resistant Shiga Toxin-Producing E. coli in Raw Milk of Dairy Bovine. Tropical Medicine and Infectious Disease. 2024; 9(3):64. https://doi.org/10.3390/tropicalmed9030064
Chicago/Turabian StyleUllah, Safir, Saeed Ul Hassan Khan, Muhammad Jamil Khan, Baharullah Khattak, Fozia Fozia, Ijaz Ahmad, Mohammad Ahmad Wadaan, Muhammad Farooq Khan, Almohannad Baabbad, and Sagar M. Goyal. 2024. "Multiple-Drug Resistant Shiga Toxin-Producing E. coli in Raw Milk of Dairy Bovine" Tropical Medicine and Infectious Disease 9, no. 3: 64. https://doi.org/10.3390/tropicalmed9030064
APA StyleUllah, S., Khan, S. U. H., Khan, M. J., Khattak, B., Fozia, F., Ahmad, I., Wadaan, M. A., Khan, M. F., Baabbad, A., & Goyal, S. M. (2024). Multiple-Drug Resistant Shiga Toxin-Producing E. coli in Raw Milk of Dairy Bovine. Tropical Medicine and Infectious Disease, 9(3), 64. https://doi.org/10.3390/tropicalmed9030064






