Molecular Characterization of Leptospira Species among Patients with Acute Undifferentiated Febrile Illness from the Municipality of Villeta, Colombia
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Consideration
2.2. Study Area
2.3. Febrile Patient Recruitment
2.4. DNA Extraction
2.5. Detection of Leptospira spp.
2.6. Identification of Leptospira spp.
2.7. Phylogenetic Analyses
3. Results
3.1. Detection of Leptospira spp. in Febrile Patients
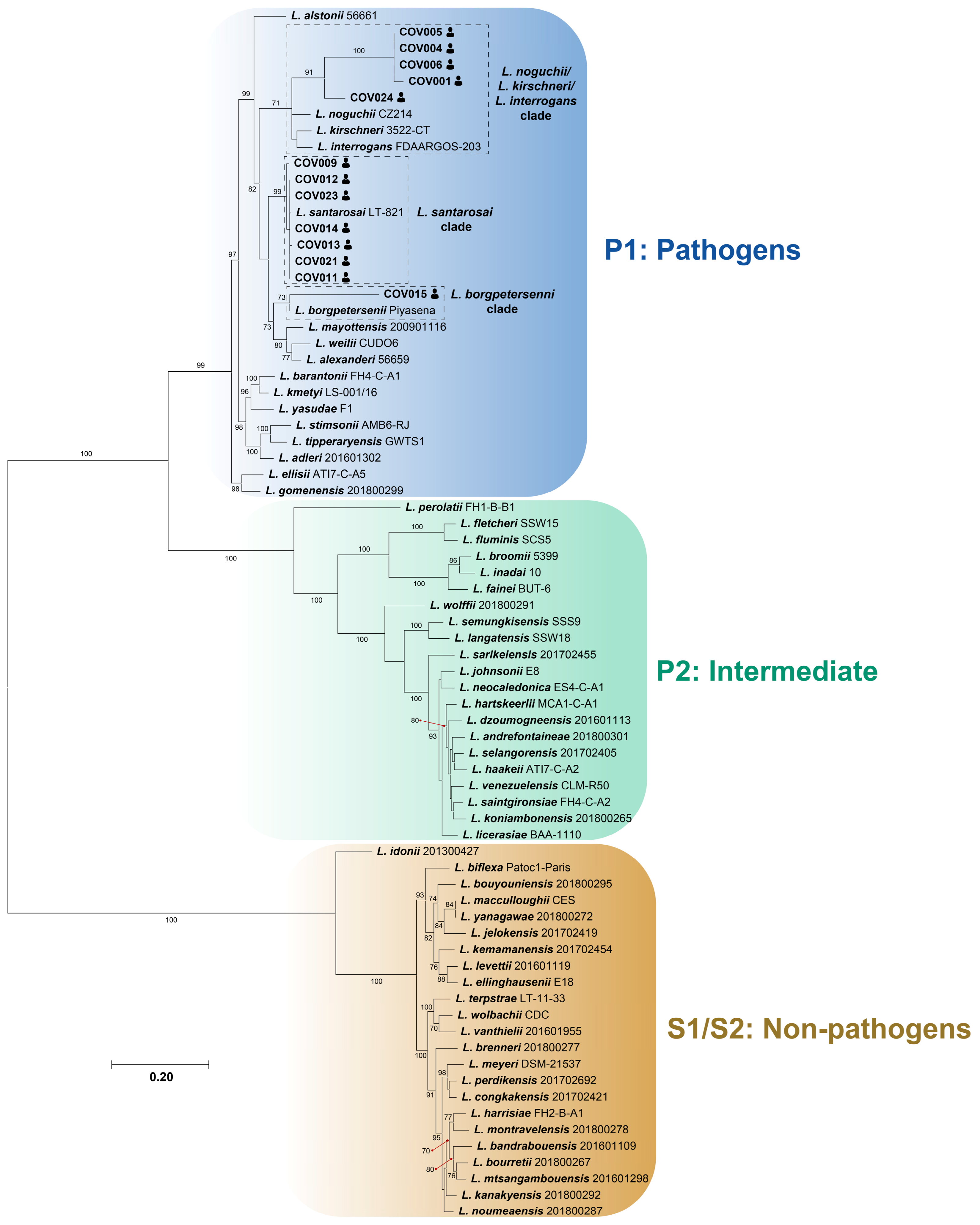
3.2. Molecular Identification of Leptospira spp. in Febrile Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adler, B.; de la Peña Moctezuma, A. Leptospira and leptospirosis. Vet. Microbiol. 2010, 140, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Picardeau, M. Virulence of the zoonotic agent of leptospirosis: Still terra incognita? Nat. Rev. Microbiol. 2017, 15, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.T.; Schiettekatte, O.; Goarant, C.; Neela, V.K.; Bernet, E.; Thibeaux, R.; Ismail, N.; Khalid, M.K.N.M.; Amran, F.; Masuzawa, T.; et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 2019, 13, e0007270. [Google Scholar] [CrossRef] [PubMed]
- Ellis, W.A. Animal Leptospirosis. In Leptospira and Leptospirosis; Adler, B., Ed.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 99–137. [Google Scholar] [CrossRef]
- Cilia, G.; Bertelloni, F.; Fratini, F. Leptospira Infections in Domestic and Wild Animals. Pathogens 2020, 9, 573. [Google Scholar] [CrossRef] [PubMed]
- Cilia, G.; Bertelloni, F.; Albini, S.; Fratini, F. Insight into the Epidemiology of Leptospirosis: A Review of Leptospira Isolations from “Unconventional” Hosts. Animals 2021, 11, 191. [Google Scholar] [CrossRef] [PubMed]
- Stimson, A.M. Note on an organism found in yellow-fever tissue. Public Health Rep. 1907, 22, 541. [Google Scholar] [CrossRef]
- Perolat, P.; Chappel, R.J.; Adler, B.; Baranton, G.; Bulach, D.M.; Billinghurst, M.L.; Letocart, M.; Merien, F.; Serrano, M.S. Leptospira fainei sp. nov., isolated from pigs in Australia. Int. J. Syst. Bacteriol. 1998, 48, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Costa, F.; Hagan, J.E.; Calcagno, J.; Kane, M.; Torgerson, P.; Martinez-Silveira, M.S.; Stein, C.; Abela-Ridder, B.; Ko, A.I. Global Morbidity and Mortality of Leptospirosis: A Systematic Review. PLoS Negl. Trop. Dis. 2015, 9, e0003898. [Google Scholar] [CrossRef] [PubMed]
- Karpagam, K.B.; Ganesh, B. Leptospirosis: A neglected tropical zoonotic infection of public health importance-an updated review. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Halliday, J.E.; Carugati, M.; Snavely, M.E.; Allan, K.J.; Beamesderfer, J.; Ladbury, G.A.; Hoyle, D.V.; Holland, P.; Crump, J.A.; Cleaveland, S.; et al. Zoonotic causes of febrile illness in malaria endemic countries: A systematic review. Lancet Infect Dis. 2020, 20, e27–e37. [Google Scholar] [CrossRef]
- Browne, E.S.; Pereira, M.; Barreto, A.; Zeppelini, C.G.; de Oliveira, D.; Costa, F. Prevalence of human leptospirosis in the Americas: A systematic review and meta-analysis. Rev. Panam. Salud Publica 2023, 47, e126. [Google Scholar] [CrossRef]
- Viroj, J.; Claude, J.; Lajaunie, C.; Cappelle, J.; Kritiyakan, A.; Thuainan, P.; Chewnarupai, W.; Morand, S. Agro-Environmental Determinants of Leptospirosis: A Retrospective Spatiotemporal Analysis (2004-2014) in Mahasarakham Province (Thailand). Trop. Med. Infect. Dis. 2021, 6, 115. [Google Scholar] [CrossRef]
- Yupiana, Y.; Wilson, P.R.; Weston, J.F.; Vallée, E.; Collins-Emerson, J.M.; Benschop, J.; Scotland, T.; Heuer, C. Epidemiological investigation of Leptospira spp. in a dairy farming enterprise after the occurrence of three human leptospirosis cases. Zoonoses Public Health 2019, 66, 470–479. [Google Scholar] [CrossRef]
- Hassell, J.M.; Begon, M.; Ward, M.J.; Fèvre, E.M. Urbanization and Disease Emergence: Dynamics at the Wildlife-Livestock-Human Interface. Trends Ecol. Evol. 2017, 32, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Rajapakse, S. Leptospirosis: Clinical aspects. Clin. Med. 2022, 22, 14–17. [Google Scholar] [CrossRef]
- Sykes, J.E.; Reagan, K.L.; Nally, J.E.; Galloway, R.L.; Haake, D.A. Role of Diagnostics in Epidemiology, Management, Surveillance, and Control of Leptospirosis. Pathogens 2022, 11, 395. [Google Scholar] [CrossRef]
- Villegas Vélez, Á.A.; Castrillón Gallego, C. Territorio, enfermedad y población en la producción de la geografía tropical colombiana, 1872–1934. Hist. Crit. 2006, 32, 94–117. [Google Scholar] [CrossRef]
- Silva-Ramos, C.R.; Faccini-Martínez, Á.A.; Serna-Rivera, C.C.; Mattar, S.; Hidalgo, M. Etiologies of Zoonotic Tropical Febrile Illnesses That Are Not Part of the Notifiable Diseases in Colombia. Microorganisms 2023, 11, 2154. [Google Scholar] [CrossRef] [PubMed]
- Barrera, E.L.P.; Reales-González, J.; Salas, D.; Santamaría, E.R.; Bello, S.; Rico, A.; Pardo, L.; Parra, E.; Rodriguez, K.; Alarcon, Z.; et al. Fatal acute undifferentiated febrile illness among clinically suspected leptospirosis cases in Colombia, 2016–2019. PLoS Negl. Trop. Dis. 2023, 17, e0011683. [Google Scholar] [CrossRef]
- Calderón, A.; Rodríguez, V.; Máttar, S.; Arrieta, G. Leptospirosis in pigs, dogs, rodents, humans, and water in an area of the Colombian tropics. Trop. Anim. Health Prod. 2014, 46, 427–432. [Google Scholar] [CrossRef]
- Ensuncho-Hoyos, C.; Rodríguez-Rodríguez, V.; Pérez-Doria, A.; Vergara, O.; Calderón-Rangel, A. Epidemiology behavior of leptospirosis in Ciénaga de Oro, Córdoba (Colombia). Trop. Anim. Health Prod. 2017, 49, 1345–1351. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Vélez, J.C.; Rodas, J.D.; Rojas, C.A.; Ko, A.I.; Wunder, E.A. Leptospira Infection in Rural Areas of Urabá Region, Colombia: A Prospective Study. Am. J. Trop. Med. Hyg. 2022, 107, 1267–1277. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rodríguez, V.; Castro-Cordero, A.; Calderón-Rangel, A.; Martínez-Ibarra, E.; Yasnot, M.; Agudelo-Flórez, P.; Monroy, F.P. Acute human leptospirosis in a Caribbean region of Colombia: From classic to emerging risk factors. Zoonoses Public Health 2024, 71, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Faccini-Martínez, Á.A.; Ramírez-Hernández, A.; Barreto, C.; Forero-Becerra, E.; Millán, D.; Valbuena, E.; Sánchez-Alfonso, A.C.; Imbacuán-Pantoja, W.O.; Cortés-Vecino, J.A.; Polo-Terán, L.J.; et al. Epidemiology of Spotted Fever Group Rickettsioses and Acute Undifferentiated Febrile Illness in Villeta, Colombia. Am. J. Trop. Med. Hyg. 2017, 97, 782–788. [Google Scholar] [CrossRef] [PubMed]
- Silva-Ramos, C.R.; Gil-Mora, J.; Serna-Rivera, C.C.; Martínez Díaz, H.C.; Restrepo-López, N.; Agudelo-Flórez, P.; Arboleda, M.; Díaz, F.J.; Faccini-Martínez, Á.A.; Hidalgo, M.; et al. Etiological characterization of acute undifferentiated febrile illness in Apartadó and Villeta municipalities, Colombia, during COVID-19 pandemic. Infez. Med. 2023, 31, 517–532. [Google Scholar] [CrossRef] [PubMed]
- du Breuil, R.M.; Patel, J.M.; Mendelow, B.V. Quantitation of beta-actin-specific mRNA transcripts using xeno-competitive PCR. PCR Methods Appl. 1993, 3, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Bessa, T.A.; Spichler, A.; Chapola, E.G.; Husch, A.C.; de Almeida, M.F.; Sodré, M.M.; Mouriz Savani, E.S.M.; Veiga Sacramento, D.R.; Vinetz, J.M. The contribution of bats to leptospirosis transmission in Sao Paulo City, Brazil. Am. J. Trop. Med. Hyg. 2010, 82, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Devi, S.M.; De los Á Valverde, L.; Vijayachari, P.; Machang’u, R.S.; Ellis, W.A.; Hartskeerl, R.A. Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann. Clin. Microbiol. Antimicrob. 2006, 5, 28. [Google Scholar] [CrossRef][Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J.; Clustal, W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Patino, L.; Afanador, A.; Paul, J.H. A spotted fever in Tobia, Colombia. Preliminary report. Am. J. Trop. Med. Hyg. 1937, 17, 639–653. [Google Scholar] [CrossRef]
- Hidalgo, M.; Orejuela, L.; Fuya, P.; Carrillo, P.; Hernandez, J.; Parra, E.; Keng, C.; Small, M.; Olano, J.P.; Bouyer, D.; et al. Rocky Mountain spotted fever, Colombia. Emerg. Infect. Dis. 2007, 13, 1058–1060. [Google Scholar] [CrossRef] [PubMed]
- Bharti, A.R.; Nally, J.E.; Ricaldi, J.N.; Matthias, M.A.; Diaz, M.M.; Lovett, M.A.; Levett, P.N.; Gilman, R.H.; Willig, M.R.; Gotuzzo, E.; et al. Leptospirosis: A zoonotic disease of global importance. Lancet Infect. Dis. 2003, 3, 757–771. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.D.; Martínez-Vega, R.A. Spatiotemporal dynamics of human leptospirosis and its relationship with rainfall anomalies in Colombia. Trans. R. Soc. Trop. Med. Hyg. 2018, 112, 115–123. [Google Scholar] [CrossRef]
- Gutiérrez, J.D.; Martínez-Vega, R.A.; Botello, H.; Ruiz-Herrera, F.J.; Arenas-López, L.C.; Hernandez-Tellez, K.D. Environmental and socioeconomic determinants of leptospirosis incidence in Colombia. Cad. Saude Publica 2019, 35, e00118417. [Google Scholar] [CrossRef]
- Astudillo Hernández, M.; González Rodríguez, A.; Batista Santiesteban, N.; Mirabal Sosa, M.; Menéndez Hernández, J. Estudio seroepidemiológico de la leptospirosis humana en el departamento del Valle del Cauca, Colombia. Rev. Cubana Med. Trop. 2009, 61, 1–10. [Google Scholar]
- Arroyave, E.; Londoño, A.F.; Quintero, J.C.; Agudelo-Flórez, P.; Arboleda, M.; Díaz, F.J.; Rodas, J.D. Etiología y caracterización epidemiológica del síndrome febril no palúdico en tres municipios del Urabá antioqueño, Colombia. Biomedica 2013, 33, 99–107. [Google Scholar] [PubMed]
- Mattar, S.; Tique, V.; Miranda, J.; Montes, E.; Garzon, D. Undifferentiated tropical febrile illness in Cordoba, Colombia: Not everything is dengue. J. Infect. Public Health 2017, 10, 507–512. [Google Scholar] [CrossRef]
- Arboleda, M.; Mejía-Torres, M.; Posada, M.; Restrepo, N.; Ríos-Tapias, P.; Rivera-Pedroza, L.A.; Calle, D.; Sánchez-Jiménez, M.M.; Marín, K.; Agudelo-Flórez, P. Molecular Diagnosis as an Alternative for Public Health Surveillance of Leptospirosis in Colombia. Microorganisms 2023, 11, 2759. [Google Scholar] [CrossRef]
- Waggoner, J.J.; Pinsky, B.A. Molecular diagnostics for human leptospirosis. Curr. Opin. Infect. Dis. 2016, 29, 440–445. [Google Scholar] [CrossRef] [PubMed]
- Guglielmini, J.; Bourhy, P.; Schiettekatte, O.; Zinini, F.; Brisse, S.; Picardeau, M. Genus-wide Leptospira core genome multilocus sequence typing for strain taxonomy and global surveillance. PLoS Negl. Trop. Dis. 2019, 13, e0007374. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, P.H.; Steigerwalt, A.G.; Sulzer, K.R.; Kaufmann, A.F.; Rogers, F.; Brenner, D.J. Deoxyribonucleic acid relatedness between serogroups and serovars in the family Leptospiraceae with proposals for seven new Leptospira species. Int. J. Syst. Evol. Microbiol. 1987, 37, 407–415. [Google Scholar] [CrossRef]
- Kallel, H.; Bourhy, P.; Mayence, C.; Houcke, S.; Hommel, D.; Picardeau, M.; Caro, V.; Matheus, S. First report of human Leptospira santarosai infection in French Guiana. J. Infect. Public Health 2020, 13, 1181–1183. [Google Scholar] [CrossRef] [PubMed]
- Aymée, L.; Nogueira Di Azevedo, M.I.; de Souza Pedrosa, J.; Loria de Melo, J.D.S.; Carvalho-Costa, F.A.; Lilenbaum, W. The role of Leptospira santarosai serovar Guaricura as agent of Bovine Genital Leptospirosis. Vet. Microbiol. 2022, 268, 109413. [Google Scholar] [CrossRef] [PubMed]
- Chinchilla, D.; Nieves, C.; Gutiérrez, R.; Sordoillet, V.; Veyrier, F.J.; Picardeau, M. Phylogenomics of Leptospira santarosai, a prevalent pathogenic species in the Americas. PLoS Negl. Trop. Dis. 2023, 17, e0011733. [Google Scholar] [CrossRef] [PubMed]
- Naotunna, C.; Agampodi, S.B.; Agampodi, T.C. Etiological agents causing leptospirosis in Sri Lanka: A review. Asian Pac. J. Trop. Med. 2016, 9, 390–394. [Google Scholar] [CrossRef]
- Lata, K.S.; Vaghasia, V.; Bhairappanavar, S.B.; Kumar, S.; Ayachit, G.; Patel, S.; Das, J. Whole genome sequencing and de novo assembly of three virulent Indian isolates of Leptospira. Infect. Genet. Evol. 2020, 85, 104579. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.K.; Lee, M.H.; Chen, Y.C.; Hsueh, P.R.; Chang, S.C. Factors associated with severity and mortality in patients with confirmed leptospirosis at a regional hospital in northern Taiwan. J. Microbiol. Immunol. Infect. 2020, 53, 307–314. [Google Scholar] [CrossRef]
- Miotto, B.A.; Moreno, L.Z.; Guilloux, A.G.A.; de Sousa, G.O.; Loureiro, A.P.; Moreno, A.M.; Lilenbaum, W.; Vasconcellos, S.A.; Heinemann, M.B.; Hagiwara, M.K. Molecular and serological characterization of the first Leptospira santarosai strain isolated from a dog. Acta Trop. 2016, 162, 1–4. [Google Scholar] [CrossRef]
- Moreno, L.Z.; Miraglia, F.; Marvulo, M.F.V.; Silva, J.C.R.; Paula, C.D.; Costa, B.L.P.; Morais, Z.M.; Ferreira, F.; Neto, J.S.F.; Dellagostin, O.A.; et al. Characterization of Leptospira santarosai Serogroup Grippotyphosa Serovar Bananal Isolated from Capybara (Hydrochaeris hydrochaeris) in Brazil. J. Wildl. Dis. 2016, 52, 688–693. [Google Scholar] [CrossRef]
- Kremer, F.S.; Eslabão, M.R.; Provisor, M.; Woloski, R.D.S.; Ramires, O.V.; Moreno, L.Z.; Moreno, A.M.; Hamond, C.; Lilenbaum, W.; Dellagostin, O.A. Draft Genome Sequences of Leptospira santarosai Strains U160, U164, and U233, Isolated from Asymptomatic Cattle. Genome Announc. 2015, 3, e00910-15. [Google Scholar] [CrossRef]
- Lilenbaum, W.; Kremer, F.; Ristow, P.; Dellagostin, O.; Bourhy, P.; Hartskeerl, R.; Vasconcellos, S. Molecular characterization of the first leptospires isolated from goats in Brazil. Braz. J. Microbiol. 2015, 45, 1527–1530. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hamond, C.; Dirsmith, K.L.; LeCount, K.; Soltero, F.V.; Rivera-Garcia, S.; Camp, P.; Anderson, T.; Hicks, J.A.; Galloway, R.; Sutherland, G.; et al. Leptospira borgpetersenii serovar Hardjo and Leptospira santarosai serogroup Pyrogenes isolated from bovine dairy herds in Puerto Rico. Front. Vet. Sci. 2022, 9, 1025282. [Google Scholar] [CrossRef]
- Peláez Sanchez, R.G.; Lopez, J.Á.; Pereira, M.M.; Arboleda Naranjo, M.; Agudelo-Flórez, P. Genetic diversity of Leptospira in northwestern Colombia: First report of Leptospira santarosai as a recognised leptospirosis agent. Mem. Inst. Oswaldo Cruz. 2016, 111, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Perez-Garcia, J.; Monroy, F.P.; Agudelo-Florez, P. Canine Leptospirosis in a Northwestern Region of Colombia: Serological, Molecular and Epidemiological Factors. Pathogens 2022, 11, 1040. [Google Scholar] [CrossRef]
- Silva, E.F.; Cerqueira, G.M.; Seyffert, N.; Seixas, F.K.; Hartwig, D.D.; Athanazio, D.A.; Pinto, L.S.; Queiroz, A.; Ko, A.I.; Brod, C.S.; et al. Leptospira noguchii and human and animal leptospirosis, Southern Brazil. Emerg. Infect. Dis. 2009, 15, 621–623. [Google Scholar] [CrossRef]
- Philip, N.; Affendy, N.B.; Ramli, S.N.A.; Arif, M.; Raja, P.; Nagandran, E.; Renganathan, P.; Taib, N.M.; Masri, S.N.; Yuhana, M.Y.; et al. Leptospira interrogans and Leptospira kirschneri are the dominant Leptospira species causing human leptospirosis in Central Malaysia. PLoS Negl. Trop. Dis. 2020, 14, e0008197. [Google Scholar] [CrossRef] [PubMed]
- Soares, P.M.; Gomes, D.O.; Macedo, F.P.; Soares, M.M.; Lemes, K.R.; Jaeger, L.H.; Lilenbaum, W.; Lima, A.M. Serological and molecular characterization of Leptospira kirschneri serogroup Grippotyphosa isolated from bovine in Brazil. Microb. Pathog. 2020, 138, 103803. [Google Scholar] [CrossRef]
- Bandara, A.G.N.M.K.; Kalaivarny, G.; Perera, N.; Indrakumar, J. Aseptic meningitis as the initial presentation of Leptospira borgpetersenii serovar Tarassovi: Two case reports and a literature review. BMC Infect. Dis. 2021, 21, 488. [Google Scholar] [CrossRef]
- Moinet, M.; Wilkinson, D.A.; Aberdein, D.; Russell, J.C.; Vallée, E.; Collins-Emerson, J.M.; Heuer, C.; Benschop, J. Of Mice, Cattle, and Men: A Review of the Eco-Epidemiology of Leptospira borgpetersenii Serovar Ballum. Trop. Med. Infect. Dis. 2021, 6, 189. [Google Scholar] [CrossRef] [PubMed]
- Giraud-Gatineau, A.; Nieves, C.; Harrison, L.B.; Benaroudj, N.; Veyrier, F.J.; Picardeau, M. Evolutionary insights into the emergence of virulent Leptospira spirochetes. bioRxiv 2024. [Google Scholar] [CrossRef] [PubMed]
- Barragan, V.; Chiriboga, J.; Miller, E.; Olivas, S.; Birdsell, D.; Hepp, C.; Hornstra, H.; Schupp, J.M.; Morales, M.; Gonzalez, M.; et al. High Leptospira Diversity in Animals and Humans Complicates the Search for Common Reservoirs of Human Disease in Rural Ecuador. PLoS Negl. Trop. Dis. 2016, 10, e0004990. [Google Scholar] [CrossRef] [PubMed]
| Demographic Characteristic | n (%) | Leptospira spp. Detection (%) |
|---|---|---|
| No. of patients | 56 (100) | 14 (25) |
| Gender | ||
| Male | 30 (53.6) | 10 (33.3) |
| Female | 26 (46.4) | 4 (15.3) |
| Age groups | ||
| Children (3–12) | 6 (10.7) | 5 (83.3) |
| Adolescents (13–16) | 4 (7.1) | 1 (25) |
| Young adults (17–29) | 21 (37.5) | 4 (19) |
| Middle-aged adults (30–44) | 12 (21.5) | 2 (16.7) |
| Older adults (above 45) | 13 (23.2) | 2 (15.4) |
| Origin | ||
| Urban area | 47 (83.9) | 10 (21.3) |
| Rural area | 9 (16.1) | 4 (44.4) |
| Sampling month | ||
| September | 6 (10.7) | 4 (66.7) |
| October | 7 (12.5) | 5 (71.4) |
| November | 10 (17.9) | 4 (40) |
| December | 33 (58.9) | 1 (3) |
| Sample ID | Gene | BLASTn Results | Serological Results from Silva-Ramos et al. 2023 [26] | ||||
|---|---|---|---|---|---|---|---|
| Organism | Identity (%) | Coverage (%) | e-Value | GenBank ID | |||
| COV001 | rrs | L. noguchii | 96.3% | 100.0% | 1 × 10−91 | CP091936.1 | Positive IgM in acute and convalescent samples Seroconversion to serovar Bratislava Seroconversion to serovar Hardjo |
| COV004 | rrs | L. noguchii | 96.7% | 100.0% | 4 × 10−91 | CP091936.1 | |
| COV005 | rrs | L. noguchii | 96.4% | 100.0% | 4 × 10−96 | CP091936.1 | |
| COV006 | rrs | L. noguchii | 96.4% | 100.0% | 3 × 10−82 | CP091967.1 | |
| COV009 | rrs | L. santarosai | 100.0% | 100.0% | 2 × 10−88 | MH801931.1 | Positive IgM in convalescent sample |
| L. interrogans * | 100.0% | 100.0% | 2 × 10−88 | MH686123.1 | |||
| secY | L. santarosai | 100.0% | 100.0% | 6 × 10−169 | MK315145.1 | ||
| icdA | L. santarosai | 99.7% | 99.0% | 4 × 10−175 | KC492816.1 | ||
| COV011 | rrs | L. santarosai | 100.0% | 100.0% | 9 × 10−103 | MH801931.1 | |
| L. interrogans * | 100.0% | 100.0% | 9 × 10−103 | MH686123.1 | |||
| COV012 | rrs | L. santarosai | 100.0% | 100.0% | 9 × 10−103 | MH801931.1 | |
| L. interrogans * | 100.0% | 100.0% | 9 × 10−103 | MH686123.1 | |||
| COV013 | rrs | L. santarosai | 100.0% | 100.0% | 9 × 10−103 | MH801931.1 | Positive IgM in convalescent sample |
| L. interrogans * | 100.0% | 100.0% | 9 × 10−103 | MH686123.1 | |||
| secY | L. santarosai | 99.7% | 100.0% | 0 | EU358050.1 | ||
| LipL32 | L. santarosai | 100.0% | 100.0% | 9 × 10−116 | PP554251.1 | ||
| LipL41 | L. santarosai | 99.5% | 100.0% | 0 | AY461959.1 | ||
| COV014 | rrs | L. santarosai | 100.0% | 100.0% | 5 × 10−90 | MH801931.1 | Positive IgM in convalescent sample |
| L. interrogans * | 100.0% | 100.0% | 5 × 10−90 | MH686123.1 | |||
| COV015 | rrs | L. borgpetersenii | 94.7% | 100.0% | 3 × 10−66 | CP047520.1 | |
| COV021 | rrs | L. santarosai | 100.0% | 100.0% | 9 × 10−103 | MH801931.1 | |
| L. interrogans * | 100.0% | 100.0% | 9 × 10−103 | MH686123.1 | |||
| secY | L. santarosai | 100.0% | 100.0% | 6 × 10−148 | MK315143.1 | ||
| LipL32 | L. santarosai | 100.0% | 100.0% | 7 × 10−117 | AY461928.1 | ||
| icdA | L. santarosai | 100.0% | 100.0% | 0 | CP028377.1 | ||
| COV023 | rrs | L. santarosai | 100.0% | 100.0% | 1 × 10−96 | MH801931.1 | Positive IgM in convalescent sample |
| L. interrogans * | 100.0% | 100.0% | 1 × 10−96 | MH686123.1 | |||
| secY | L. santarosai | 100.0% | 100.0% | 0 | MK315138.1 | ||
| LipL32 | L. santarosai | 100.0% | 100.0% | 7 × 10−153 | PP554251.1 | ||
| COV024 | rrs | L. kirschneri | 98.1% | 99.0% | 1 × 10−96 | CP125672.1 | |
| L. interrogans | 98.1% | 99.0% | 1 × 10−96 | KP211707.1 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva-Ramos, C.R.; Matiz-González, J.M.; Gil-Mora, J.; Martínez Díaz, H.-C.; Faccini-Martínez, Á.A.; Cuervo, C.; Melby, P.C.; Aguilar, P.V.; Cabada, M.M.; Rodas, J.D.; et al. Molecular Characterization of Leptospira Species among Patients with Acute Undifferentiated Febrile Illness from the Municipality of Villeta, Colombia. Trop. Med. Infect. Dis. 2024, 9, 168. https://doi.org/10.3390/tropicalmed9080168
Silva-Ramos CR, Matiz-González JM, Gil-Mora J, Martínez Díaz H-C, Faccini-Martínez ÁA, Cuervo C, Melby PC, Aguilar PV, Cabada MM, Rodas JD, et al. Molecular Characterization of Leptospira Species among Patients with Acute Undifferentiated Febrile Illness from the Municipality of Villeta, Colombia. Tropical Medicine and Infectious Disease. 2024; 9(8):168. https://doi.org/10.3390/tropicalmed9080168
Chicago/Turabian StyleSilva-Ramos, Carlos Ramiro, J. Manuel Matiz-González, Juliana Gil-Mora, Heidy-C. Martínez Díaz, Álvaro A. Faccini-Martínez, Claudia Cuervo, Peter C. Melby, Patricia V. Aguilar, Miguel M. Cabada, Juan David Rodas, and et al. 2024. "Molecular Characterization of Leptospira Species among Patients with Acute Undifferentiated Febrile Illness from the Municipality of Villeta, Colombia" Tropical Medicine and Infectious Disease 9, no. 8: 168. https://doi.org/10.3390/tropicalmed9080168
APA StyleSilva-Ramos, C. R., Matiz-González, J. M., Gil-Mora, J., Martínez Díaz, H.-C., Faccini-Martínez, Á. A., Cuervo, C., Melby, P. C., Aguilar, P. V., Cabada, M. M., Rodas, J. D., & Hidalgo, M. (2024). Molecular Characterization of Leptospira Species among Patients with Acute Undifferentiated Febrile Illness from the Municipality of Villeta, Colombia. Tropical Medicine and Infectious Disease, 9(8), 168. https://doi.org/10.3390/tropicalmed9080168






