Structural Studies of a Fungal Polyphenol Oxidase with Application to Bioremediation of Contaminated Water †
Abstract
:1. Introduction
2. Methods
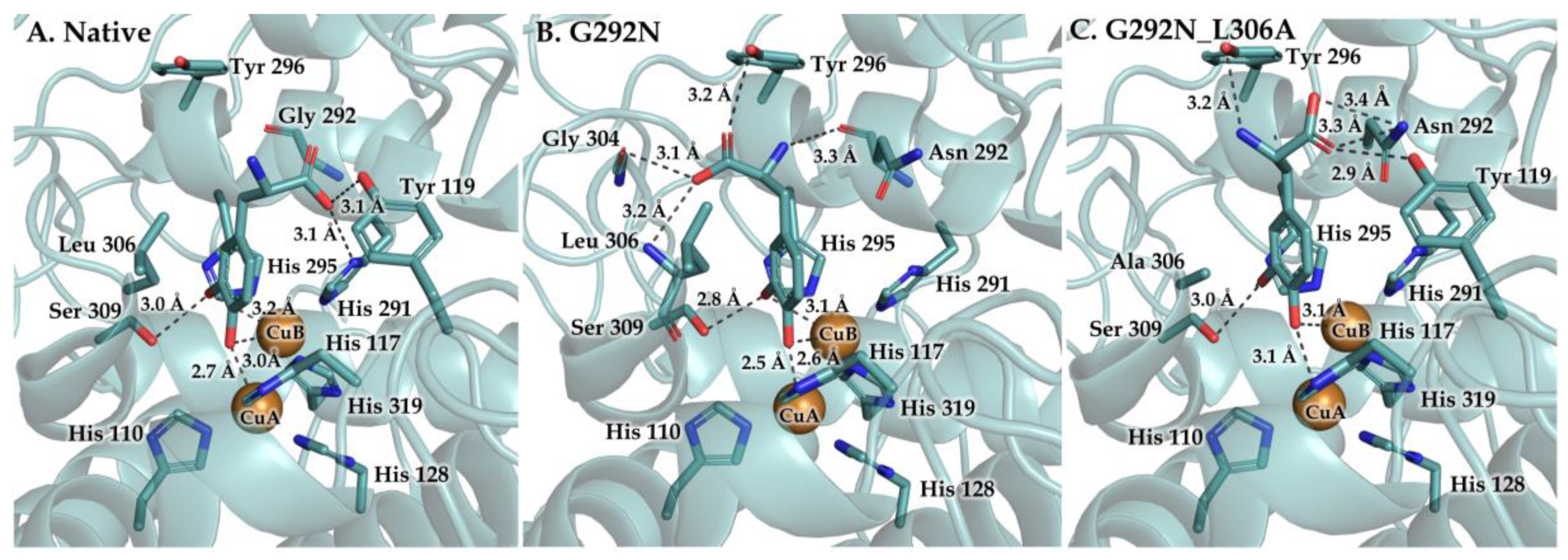
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Goldfeder, M.; Kanteev, M.; Isaschar-Ovdat, S.; Adir, N.; Fishman, A. Determination of tyrosinase substrate-binding modes reveals mechanistic differences between type-3 copper proteins. Nat. Commun. 2014, 5, 4505. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Yeon, Y.J.; Song, W.; Pack, S.P.; Choi, Y.R.; Choi, Y.S. A cold-adapted tyrosinase with an abnormally high monophenolase/diphenolase activity ratio originating from the marine archaeon Candidatus Nitrosopumilus koreensis. Biotechnol. Lett. 2016, 38, 1535–1542. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Soler-Lopez, M.; Wichers, H.J.; Dijkstra, B.W. Large-Scale Recombinant Expression and Purification of Human Tyrosinase Suitable for Structural Studies. PLoS ONE 2016, 11, e0161697. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Jiang, H.; Deng, J. Crystal Structure of Manduca Sexta Prophenoloxidase Provides Insights into the Mechanism of Type 3 Copper Enzymes. Proc. Natl. Acad. Sci. USA 2009, 106, 17002–17006. [Google Scholar] [CrossRef] [PubMed]
- Mauracher, S.G.; Molitor, C.; Michael, C.; Kragl, M.; Rizzi, A.; Rompel, A. High level protein-purification allows the unambiguous polypeptide determination of latent isoform PPO4 of mushroom tyrosinase Dedicated to Prof. Dr. H. Witzel on the occasion of his 90th birthday. Phytochemistry 2014, 99, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Kanteev, M.; Goldfeder, M.; Fishman, A. Structure-function correlations in tyrosinases. Protein Sci. 2015, 24, 1360–1369. [Google Scholar] [CrossRef] [PubMed]
- Pretzler, M.; Rompel, A. What causes the different functionality in type-III-copper enzymes? A state of the art perspective. Inorganica Chim. Acta 2018, 481, 25–31. [Google Scholar] [CrossRef]
- Solano, F. Melanins: Skin Pigments and Much More—Types, Structural Models, Biological Functions, and Formation Routes. New J. Sci. 2014, 2014, 1–28. [Google Scholar] [CrossRef]
- Gul, I.; Ahmad, M.S.; Naqvi, S.S.; Hussain, A.; Wali, R.; Farooqi, A.A.; Ahmed, I. Polyphenol oxidase (PPO) based biosensors for detection of phenolic compounds: A Review. J. Appl. Biol. Biotechnol. 2017, 5, 72–85. [Google Scholar] [CrossRef]
- Mukherjee, S.; Basak, B.; Bhunia, B.; Dey, A.; Mondal, B. Potential use of polyphenol oxidases (PPO) in the bioremediation of phenolic contaminants containing industrial wastewater. Rev. Environ. Sci. Bio/Technology 2012, 12, 61–73. [Google Scholar] [CrossRef]
- Panis, F.; Rompel, A. Identification of the amino acid position controlling the different enzymatic activities in walnut tyrosinase isoenzymes (JrPPO1 and JrPPO2). Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Nikolaivits, E.; Dimarogona, M.; Karagiannaki, I.; Chalima, A.; Fishman, A.; Topakas, E. Versatile Fungal Polyphenol Oxidase with Chlorophenol Bioremediation Potential: Characterization and Protein Engineering. Appl. Environ. Microbiol. 2018, 84, e01628-18. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Vriend, G. YASARA View—Molecular graphics for all devices—From smartphones to workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Chen, L.; Maddula, H.; Akcan, O.; Oughtred, R.; Berman, H.M.; Westbrook, J. Ligand Depot: A data warehouse for ligands bound to macromolecules. Bioinformatics 2004, 20, 2153–2155. [Google Scholar] [CrossRef] [PubMed]
- Goodsell, D.S.; Morris, G.M.; Olson, A.J. Automated docking of flexible ligands: Applications of AutoDock. JMR 1996, 9, 1–5. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Penttinen, L.; Rutanen, C.; Jänis, J.; Rouvinen, J.; Hakulinen, N. Unraveling Substrate Specificity and Catalytic Promiscuity of Aspergillus oryzae Catechol Oxidase. ChemBioChem 2018, 19, 2348–2352. [Google Scholar] [CrossRef] [PubMed]
- Solem, E.; Tuczek, F.; Decker, P.D.H. Tyrosinase versus Catechol Oxidase: One Asparagine Makes the Difference. Angew. Chem. Int. Ed. 2016, 55, 2884–2888. [Google Scholar] [CrossRef] [PubMed]
| Best Binding Energy [kcal/mol] | Dissociation Constant [μM] | |
|---|---|---|
| Native | 4.937 | 240.508 |
| G292N | 5.097 | 183.590 |
| G292N/L306A | 5.614 | 76.715 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valmas, A.; Dedes, G.; Dimarogona, M. Structural Studies of a Fungal Polyphenol Oxidase with Application to Bioremediation of Contaminated Water. Proceedings 2020, 66, 10. https://doi.org/10.3390/proceedings2020066010
Valmas A, Dedes G, Dimarogona M. Structural Studies of a Fungal Polyphenol Oxidase with Application to Bioremediation of Contaminated Water. Proceedings. 2020; 66(1):10. https://doi.org/10.3390/proceedings2020066010
Chicago/Turabian StyleValmas, Alexandros, Grigorios Dedes, and Maria Dimarogona. 2020. "Structural Studies of a Fungal Polyphenol Oxidase with Application to Bioremediation of Contaminated Water" Proceedings 66, no. 1: 10. https://doi.org/10.3390/proceedings2020066010
APA StyleValmas, A., Dedes, G., & Dimarogona, M. (2020). Structural Studies of a Fungal Polyphenol Oxidase with Application to Bioremediation of Contaminated Water. Proceedings, 66(1), 10. https://doi.org/10.3390/proceedings2020066010




