Abstract
Though therapeutic compounds can be extracted directly from medicinal plant tissues; it is now known that many phytotherapeutic compounds are actually produced by associated microorganisms or due to their interaction with the plant. Bouvardia ternifolia is a medicinal plant that can be considered a potential source of therapeutic compounds, such as the antitumoral molecule bouvardin. We characterized the endophytic, endophytic+epiphyte and soil bacterial microbiota in the flower, leaves, stems and roots in specimens of this plant through 16S rDNA sequencing. We found that Proteobacteria followed by Actinobacteria were the most abundant bacteria phyla in this plant. On the other hand, the most representative genera living endophytically were Propionibacterium, Paraccocus, Lactobacillus, Kaistobacter, Methylobacterium and Erwinia. This study provides evidence of the composition and diversity of the bacterial communities present in soil and tissues of B. ternifolia which can be considered for subsequent analysis and understanding of the therapeutic compounds that give this species its medicinal properties.
1. Introduction
Bouvardia ternifolia is a Mexican endemic plant used historically in Mexico as a general curative resource and contemporaneously as a remedy for dysentery, poison therapy, pain relief, and other afflictions [1]. The hydroalcoholic extract prepared from this plant shows acetylcholinesterase inhibition activity, as well as anti-inflammatory and antioxidant effects [2]. Bouvardin is a cyclic hexapeptide found in B. ternifolia with in vitro cytotoxicity against B16 melanoma melanotic and P388 lymphocytic leukemia cell lines [3]. These findings make this plant a potential source of therapeutic bioactive compounds. It is well known that many therapeutic secondary metabolites can be produced by plant-associated microorganisms [4]; because of this, the characterization of species that live in conjunction with the plant has become an important research activity. In this study, we report the characterization of the bacterial community associated with the Mexican medicinal plant B. ternifolia.
2. Material and Methods
2.1. Sampling
Plant specimens were collected from five geographical locations in boroughs and municipalities of Mexico City and the State of Mexico. The soil samples were taken at approximately 10 to 30 cm deep from the surface using sterile tubes. The whole plants were deposited in sterile plastic bags and with the soil samples transported to the laboratory within 1 h to 6 h.
2.2. DNA Extraction
For endophytes, stems and roots were cut in segments of approximately 3 cm. The parts were rinsed three times with autoclaved deionized water and treated with a sterilization train consisting of liquid detergent, 0.1% benzalconium chloride, 0.05% sodium nitrite, 70% ethanol, 5% sodium hypochlorite solution, 2.5–3.5% hydrogen peroxide, 0.35% ethanol, 0.05% n-phenylethanamide, ≥99.5% ethanol, and 15 min of 0.120 joules 254 nm UV treatment. Samples were crushed in aseptic conditions on a mortar with PBS pH 7.4 and liquid nitrogen. Samples were resuspended in PBS pH 7.4, filtered using sterile gauzes, and transferred to polypropylene tubes. Tubes were centrifuged at 6000 g, the supernatant discharged, and the pellet used for DNA extraction using the FavorPrep™ Stool DNA Isolation Mini Kit. For endophytes+epiphytes, specimens were rinsed once with autoclaved distilled water to remove excessive soil, crushed, and process directly for DNA extraction. DNA from soil samples was extracted using QIAGEN DNeasy PowerSoil Kit.
2.3. DNA Massive Sequencing
Barcoded amplicons (~281 bp) containing the V3 variable region of the 16S rRNA was amplified by PCR using V3-341F forward and V3-518R reverse primers [5]. For library preparation, an equal mass of amplicons was quantified by gel densitometry and pooled. The library’s size and concentration were checked using the Agilent 2100 Bioanalyzer system. High-throughput sequencing was made using Ion Torrent PGM System technology. Quality control was made using FastQC [6]; reads were trimmed to 200 nt with Trimmomatic v0.36. FASTQ files were processed with multiple QIIME v1.9.0 scripts [7]. DNA sequences were classified into operational taxonomic units (OTUs) using the Greengenes database v13.8.
2.4. Microbial Abundance and Diversity Analyses
We used QIIME pipeline v1.9.0 to determine abundance at phylum level, setting similarity at 97% with the database. Observed species, Chao1 alpha diversity indexes were calculated and plotted in R environment v3.4.4 using the phyloseq (v1.22.3) and ggplot2 (v3.3.0) packages.
2.5. Determination of Shared OTUs, Differential Abundance and Core Endophyte Bacterial Taxa
The shared bacteria among groups were determined with shared_phylotypes.py QIIME script and plotted as Venn diagram (Inkscape). The LEfSe v1.0 program was used to detect differences in the relative abundances of bacterial taxa among the endophytes, endophytes+epiphytes and soil groups at an linear discriminant analysis (LDA) cut-off score ≥ 2.5 [8]. The core microbiome shared by at least 50% was determined using compute_core_microbiome.py QIIME script, and a heatmap was generated using the gplots (v3.0.3) R package.
3. Results
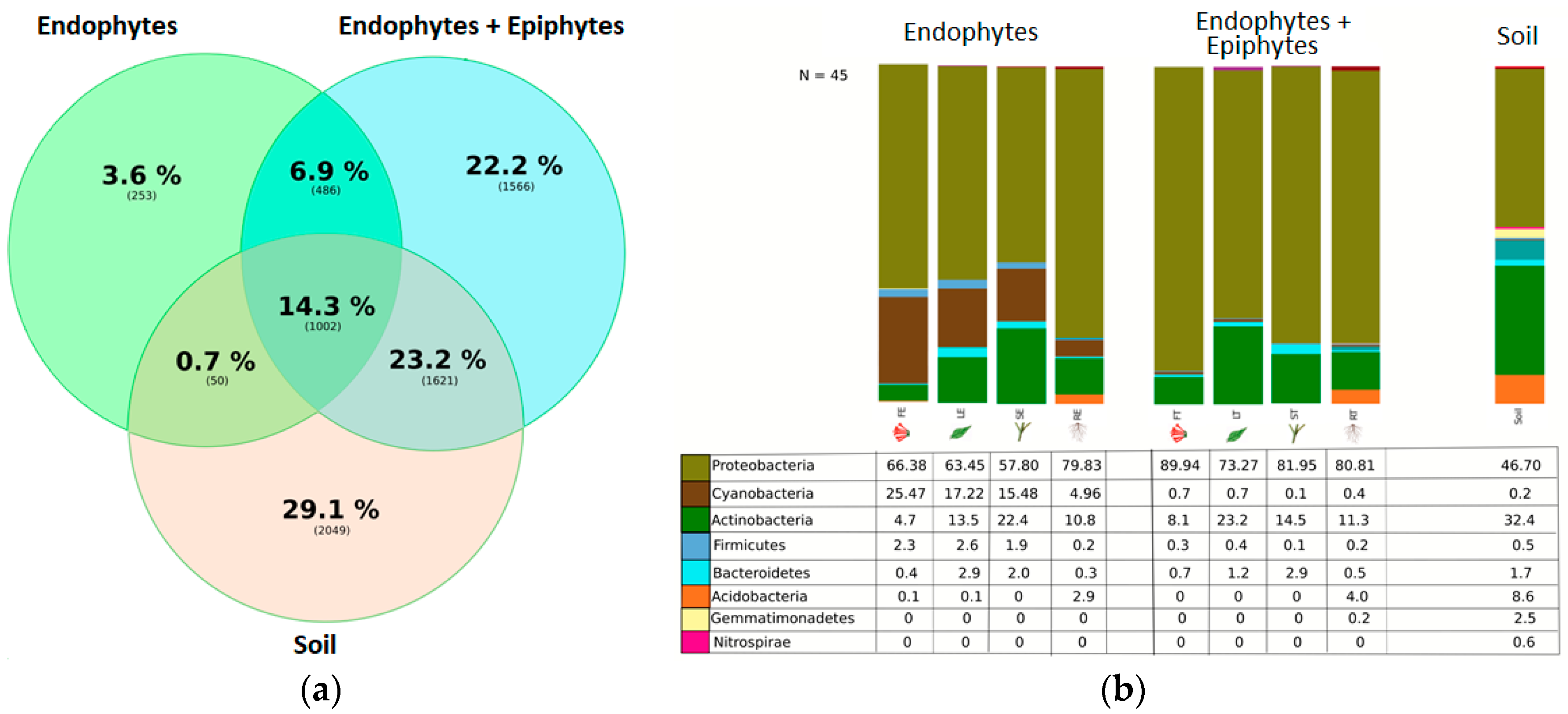
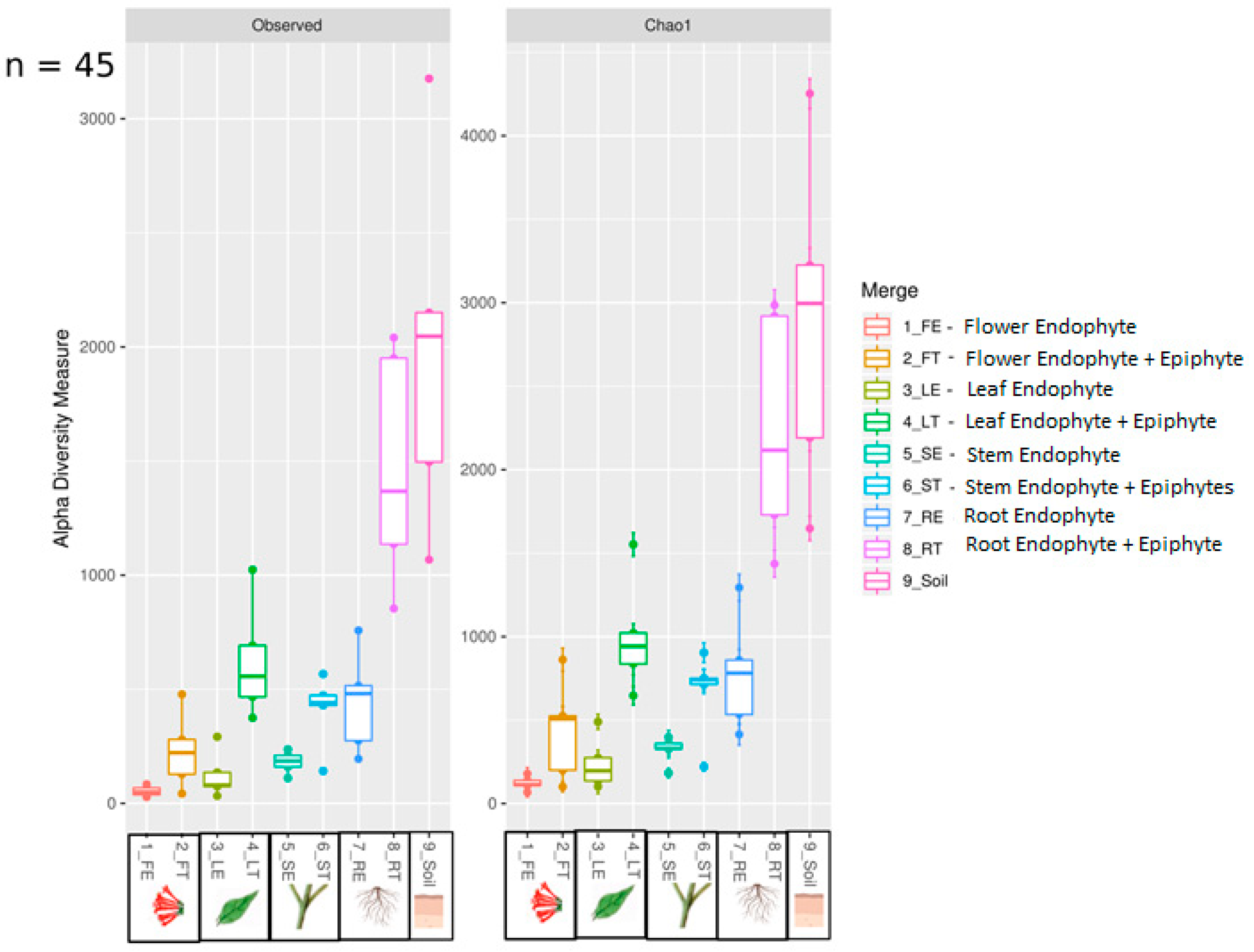
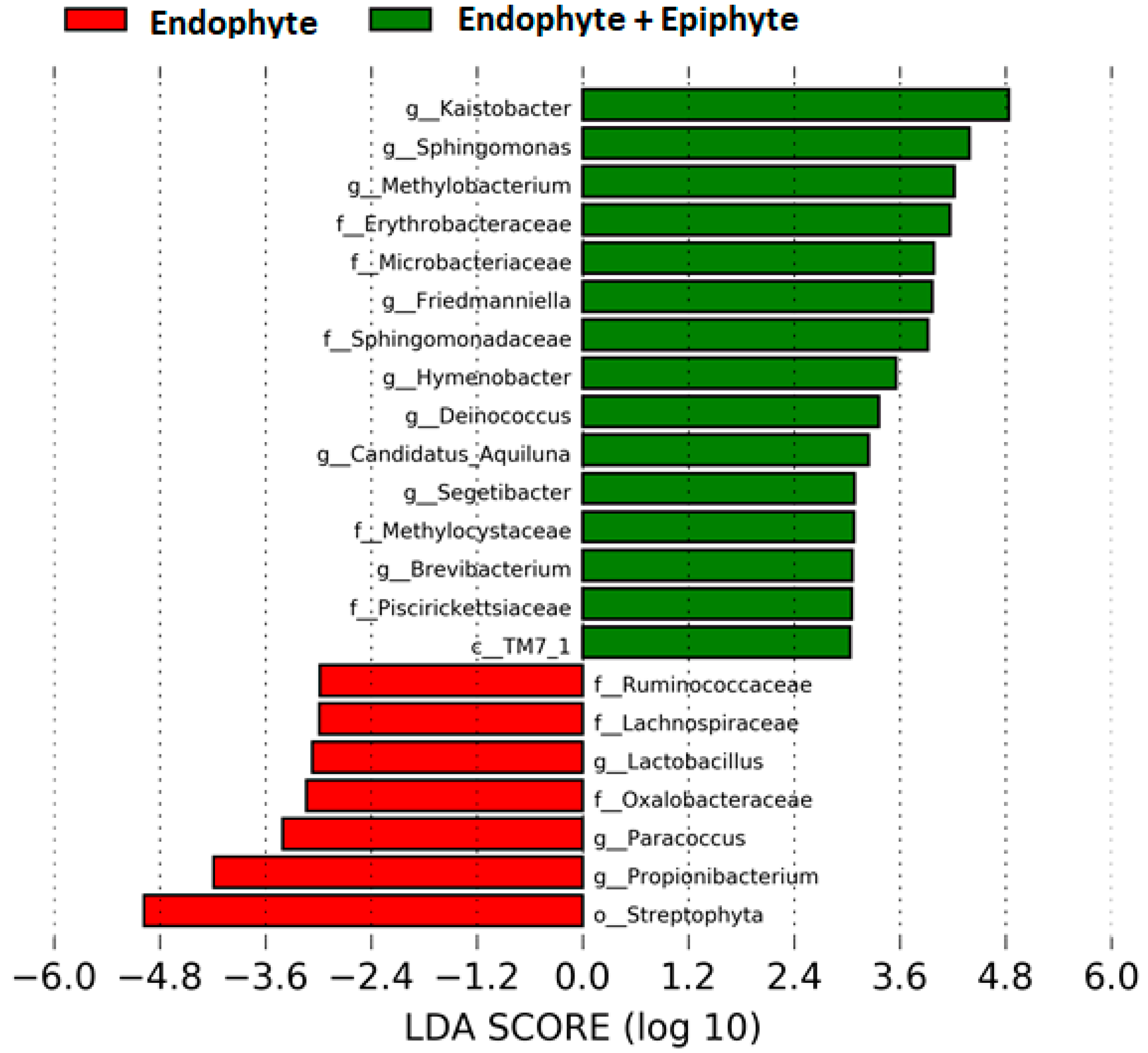
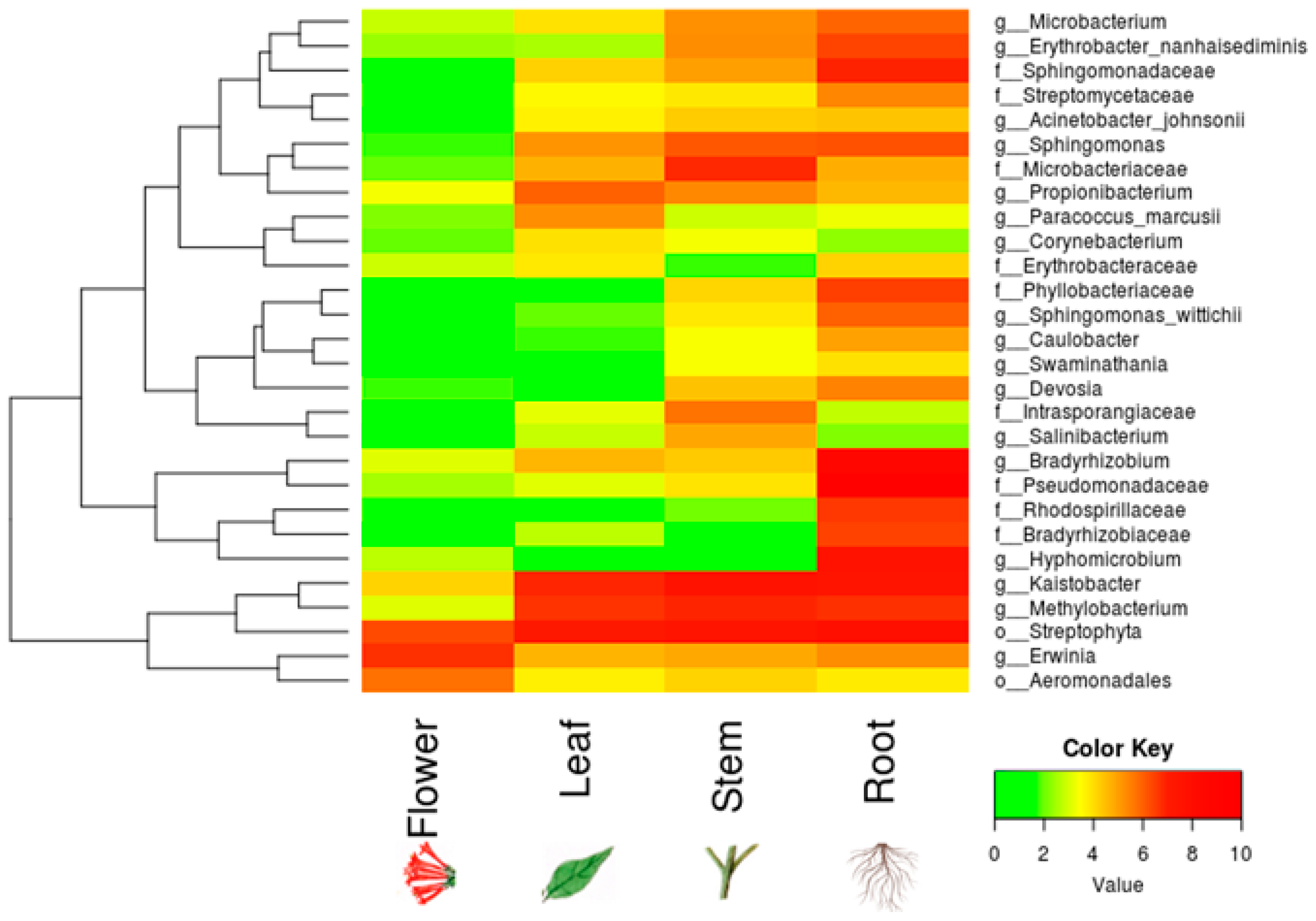
We found ~1000 OTUs shared among the total plant, endophytic, and soil samples, ~2000 OTUs exclusively for soil bacteria, ~1500 OTUs for endophyte+epiphyte plant bacteria, and ~250 OTUs for endophytic bacteria (Figure 1a). Proteobacteria (46–79%) were the most representative phylum followed by Actinobacteria and Acidobacteria. The phylum Cyanobacteria (8.5%) were more representative in endophytes samples than in total plant samples (1%) (Figure 1b). Bacterial richness and diversity by plant fractions and soil, from highest to lowest, were observed in the following order: soil > root > stem > leaf > flower (Figure 2). Linear discriminant effect size analysis showed the order: Streptophyta; the families: Oxalobacteraceae, Ruminococcaceae, Lachnospiraceae; the genera: Propionilbacterium, Paracoccus, Lactobacillus, as the most representative taxa in endophyte bacterial samples (Figure 3). Endophyte core microbiota was characterized for each part of the plant and we found 28 bacterial taxa as the most representative, including Erwinia genus predominant in flowers, Kaistobacter, Sphingomonas and Methylobacterium genera in leaf, stem, roots, and other taxa associated with roots such as Hypomicrobium and Bradyrhizobium genera (Figure 4).
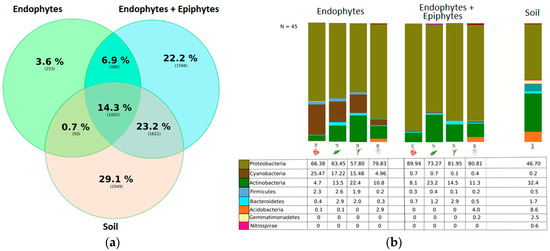
Figure 1.
(a) Ven diagram showing the operational taxonomic unit (OTU) distribution among groups. OTU phylotypes are shown in brackets and their percentage representation among endophyte, endophyte+epiphyte and soil bacterial samples. (b) Relative abundance of bacteria at phylum level. Percentage of each phylum is shown in the different plant parts and soil among the endophytes, endophytes+epiphytes and soil sample groups.
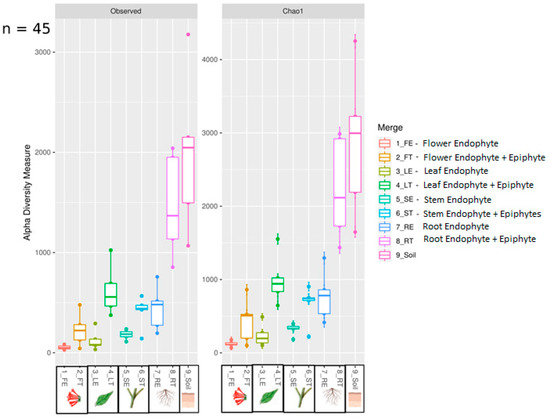
Figure 2.
Boxplot showing observed and Chao1 alpha diversity indexes. Samples were grouped by anatomical parts of the plant of endophyte and endophytes+epiphytes and soil bacterial samples.
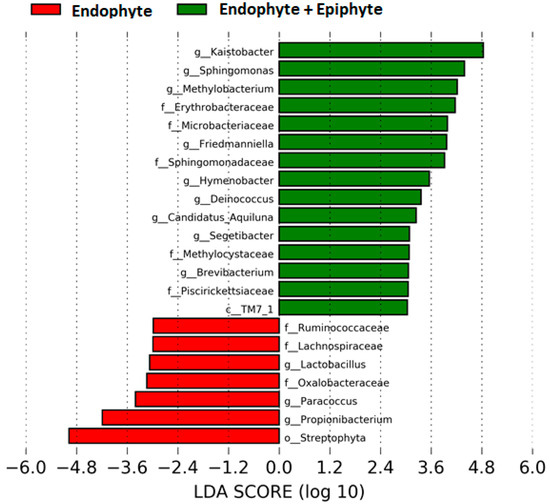
Figure 3.
Differentially abundant bacterial taxa found in the plant. Linear discriminant analysis (LDA) effect size (LEfSe) comparison of horizontal bars represent the effect size for each taxon: red color indicates taxa enriched in endophyte group while green indicates taxa enriched in endophytes+epiphytes group.
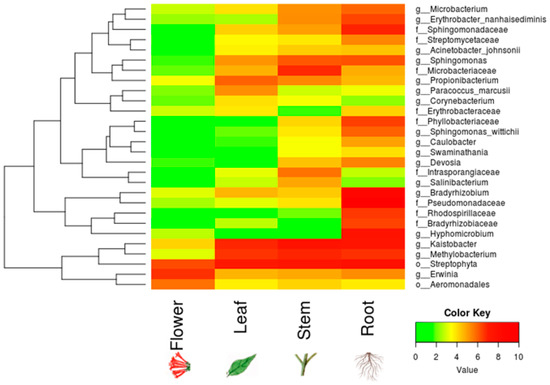
Figure 4.
Core endophyte microbiota in different anatomical parts of B. ternifolia. The heatmap shows the absolute abundance of bacterial taxa for endophytes in the flowers, leaves, stems and roots. Included counts for each taxon in the analysis were present in at least 50% of the samples. Color key from green to red indicates increasing absolute abundance in natural logarithm of counts. Green color indicates the lowest abundance while red color the highest abundance.
4. Discussion
B. ternifolia is a medicinal shrub widely distributed in Mexico and part of the plant medicinal heritage collection of this country [9]. It is important to characterize microorganisms present in this species as many bioactive compounds may be produced by them or due to its interaction with the plant. The most abundant bacterial phyla present in plants are Proteobacteria, Actinobacteria, Bacteroidetes and Firmicutes [10]. In agreement with this, we found that B. ternifolia includes Proteobacteria and Actinobacteria as the most abundant phyla. Each plant tissue provides a unique environment for endophyte and epiphyte bacterial richness above and below ground [11]. In our study, we found lower bacterial richness in flowers, leaves and stems, and higher bacterial richness in plant roots and soil samples. Abundant endophytic bacterial genera that we found in B. ternifolia, such as Kaistobater [12], Methylobacterium [13], Erwinia [10], Propionibacterium [14], Paraccocus [15] and Lactobacillus [16], have been reported to be present living endophytically in other vegetal species. Among these, some metabolites of endophytic Propionibacterium are reported to have valuable therapeutic activity [17], while members of the genus Kaistobacter are associated with plant-disease suppression [12].
5. Conclusions
This study provides evidence for the composition and diversity of the bacterial communities living in flowers, leaves, stems, roots and associated soil of the Mexican medicinal plant B. ternifolia, which can be considered for subsequent analysis to profiling therapeutic and bioactive compounds such as the antitumoral molecule bouvardin, reported to be present in this species.
Author Contributions
Conceptualization, F.V.-E., R.S., J.G.-M.; methodology, L.E.V.-F., S.D.E.-T., A.P.-E., Y.C.-N.; software, L.E.V.-F., F.H.-Q.; validation, L.E.V.-F., J.G.-M.; formal analysis, L.E.V.-F., F.H.-Q., J.G.-M.; investigation, L.E.V.-F., Y.C.-N., F.V.-E., R.S., J.G.-M.; resources, Y.C.-N., J.G.-M.; data curation, L.E.V.-F., S.D.E.-T., J.G.-M.; writing—original draft preparation, L.E.V.-F., J.G.-M.; writing—review and editing, L.E.V.-F., Y.C.-N., F.V.-E., R.S., J.G.-M.; visualization, L.E.V.-F.; supervision, J.G.-M.; project administration; J.G.-M.; funding acquisition, F.V.-E., R.S., J.G.-M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financed by Cinvestav, FONCICYT 2 267416, CONACYT-BMBF-267416, and CONACyT-163235 INFR-2011-01.
Acknowledgments
We thank CONACyT Doctoral Fellowships for LEV-F (336296) and FH-Q (291236. We are grateful to Rodrigo García-Gutiérrez for technical support, and Ms. Viridiana Rosas Ocegueda for administrative assistance. We recognize the support of the undergraduate students Isabel Montserrat Cortez de la Puente (2015) and Mariana Román Reyes (2016) for the development of this work. J.G.-M. (19815) is a Fellow from the Sistema Nacional de Investigadores, Mexico.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Jiménez-Ferrer, J.E.; Pérez-Terán, Y.Y.; Román-Ramos, R.; Tortoriello, J. Antitoxin activity of plants used in Mexican traditional medicine against scorpion poisoning. Phytomedicine 2005, 12, 116–122. [Google Scholar] [CrossRef] [PubMed]
- García-Morales, G.; Huerta-Reyes, M.; González-Cortazar, M.; Zamilpa, A.; Jiménez-Ferrer, E.; Silva-García, R.; Román-Ramos, R.; Aguilar-Rojas, A. Anti-inflammatory, antioxidant and anti-acetylcholinesterase activities of Bouvardia ternifolia: Potential implications in Alzheimer’s disease. Arch. Pharmacal Res. 2015, 38, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Jolad, S.D.; Hoffman, J.J.; Torrance, S.J.; Wiedhopf, R.M.; Cole, J.R.; Arora, S.K.; Bates, R.B.; Gargiulo, R.L.; Kriek, G.R. Bouvardin and Deoxybouvardin, Antitumor Cyclic Hexapeptides from Bouvardia ternifolia (Rubiaceae). J. Am. Chem. Soc. 1977, 99, 8040–8044. [Google Scholar] [CrossRef] [PubMed]
- Gouda, S.; Das, G.; Sen, S.K.; Shin, H.S.; Patra, J.K. Endophytes: A treasure house of bioactive compounds of medicinal importance. Front. Microbiol. 2016, 7, 1538. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Hamady, M.; Lauber, C.L.; Knight, R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 17994–17999. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 18 January 2019).
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, 1–18. [Google Scholar] [CrossRef] [PubMed]
- CONABIO. Capital Natural de México, Vol. II: Conocimiento actual de la biodiversidad. Anexo: Catalogo taxonómico de especies; Comisión Nacional para el Conocimiento y Uso de la Biodiversidad: Mexico City, Mexico, 2009; ISBN 978-607-7607-08-3. [Google Scholar]
- Afzal, I.; Shinwari, Z.K.; Sikandar, S.; Shahzad, S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 2019, 221, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, S.; Jiang, Q.; Bai, Y.; Shen, G.; Li, S.; Ding, W. Using community analysis to explore bacterial indicators for disease suppression of tobacco bacterial wilt. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Madhaiyan, M.; Alex, T.H.H.; Te Ngoh, S.; Prithiviraj, B.; Ji, L. Leaf-residing Methylobacterium species fix nitrogen and promote biomass and seed production in Jatropha curcas. Biotechnol. Biofuels 2015, 8, 222. [Google Scholar] [CrossRef] [PubMed]
- Campisano, A.; Ometto, L.; Compant, S.; Pancher, M.; Antonielli, L.; Yousaf, S.; Varotto, C.; Anfora, G.; Pertot, I.; Sessitsch, A.; et al. Interkingdom transfer of the acne-causing agent, propionibacterium acnes, from human to grapevine. Mol. Biol. Evol. 2014, 31, 1059–1065. [Google Scholar] [CrossRef]
- Sun, L.N.; Zhang, Y.F.; He, L.Y.; Chen, Z.J.; Wang, Q.Y.; Qian, M.; Sheng, X.F. Genetic diversity and characterization of heavy metal-resistant-endophytic bacteria from two copper-tolerant plant species on copper mine wasteland. Bioresour. Technol. 2010, 101, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Harrison, J.G.; Beltran, L.P.; Buerkle, C.A.; Cook, D.; Gardner, D.R.; Parchman, T.L.; Forister, M.L. A suite of rare microbes interacts with a dominant, heritable, fungal endophyte to influence plant trait expression. bioRxiv 2019, 608729. [Google Scholar] [CrossRef] [PubMed]
- Piwowarek, K.; Lipińska, E.; Hać-Szymańczuk, E.; Kieliszek, M.; Ścibisz, I. Propionibacterium spp.—Source of propionic acid, vitamin B12, and other metabolites important for the industry. Appl. Microbiol. Biotechnol. 2018, 102, 515–538. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).