Missing Plant Detection in Vineyards Using UAV Angled RGB Imagery Acquired in Dormant Period
Abstract
1. Introduction
2. Materials and Methods
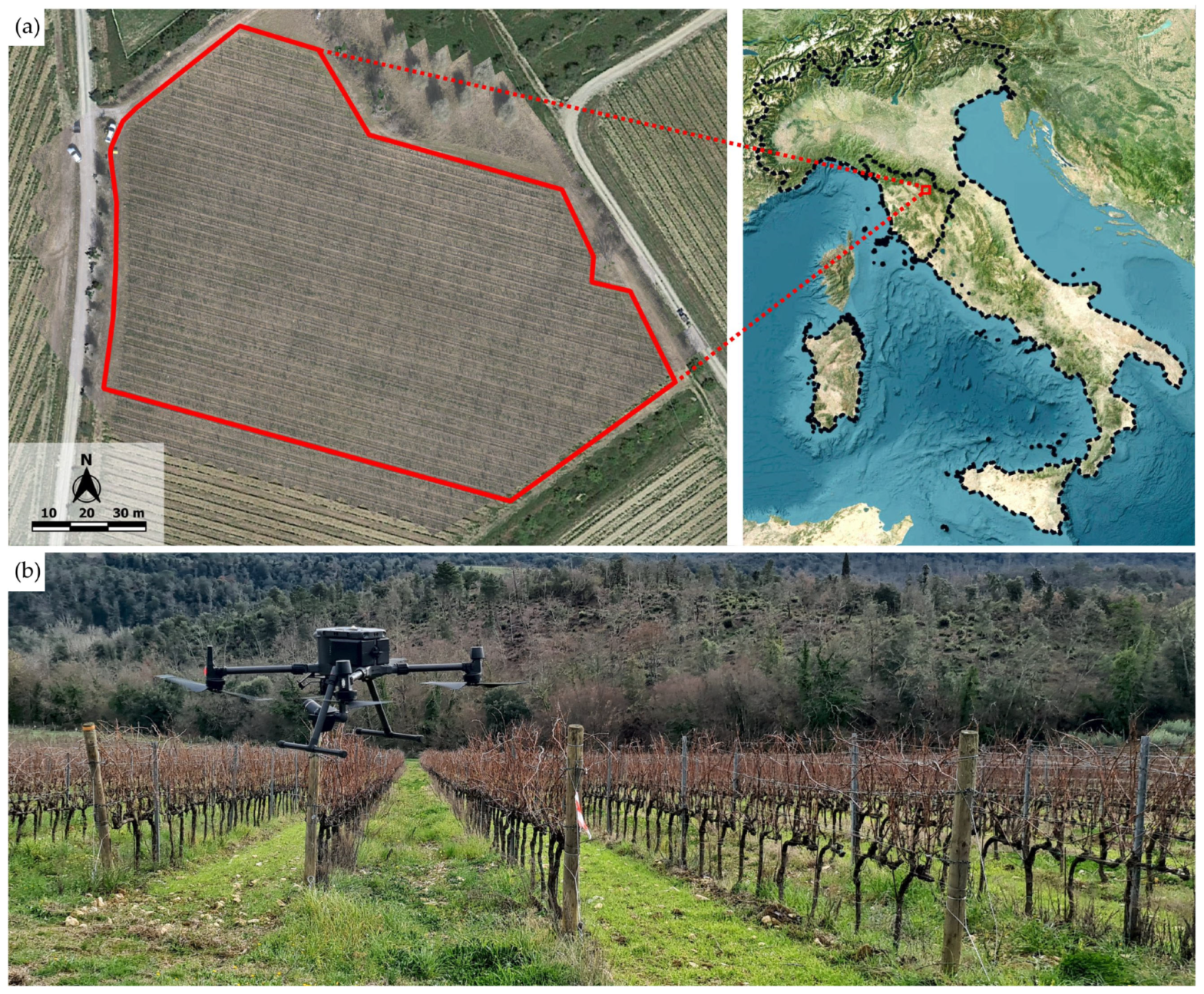
2.1. Study Site and UAV Data Acquisition
2.2. Image Processing Workflow
2.2.1. Point Cloud Classification
2.2.2. Polygons and Centroids Generation

2.2.3. Post and Trunks Extraction
2.2.4. Posts and Trunks Validation
2.2.5. Missing Vines Detection
2.2.6. Statistical Evaluation
3. Results
3.1. Posts and Vines Detection
3.2. Missing Vines Detection
3.3. Overall Evaluation of the Four Methods
4. Discussion
4.1. Posts and Vines Detection
4.2. Missing Vines Detection
4.3. Evaluation of Results vs. Other Studies in Literature
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Candiago, S.; Remondino, F.; De Giglio, M.; Dubbini, M.; Gattelli, M. Evaluating Multispectral Images and Vegetation Indices for Precision Farming Applications from UAV Images. Remote Sens. 2015, 7, 4026–4047. [Google Scholar] [CrossRef]
- Pádua, L.; Matese, A.; Di Gennaro, S.F.; Morais, R.; Peres, E.; Sousa, J.J. Vineyard Classification Using OBIA on UAV-Based RGB and Multispectral Data: A Case Study in Different Wine Regions. Comput. Electron. Agric. 2022, 196, 106905. [Google Scholar] [CrossRef]
- Sassu, A.; Gambella, F.; Ghiani, L.; Mercenaro, L.; Caria, M.; Pazzona, A.L. Advances in Unmanned Aerial System Remote Sensing for Precision Viticulture. Sensors 2021, 21, 956. [Google Scholar] [CrossRef] [PubMed]
- Andújar, D.; Moreno, H.; Bengochea-Guevara, J.M.; de Castro, A.; Ribeiro, A. Aerial Imagery or On-Ground Detection? An Economic Analysis for Vineyard Crops. Comput. Electron. Agric. 2019, 157, 351–358. [Google Scholar] [CrossRef]
- Torres-Sánchez, J.; Mesas-Carrascosa, F.J.; Santesteban, L.G.; Jiménez-Brenes, F.M.; Oneka, O.; Villa-Llop, A.; Loidi, M.; López-Granados, F. Grape Cluster Detection Using UAV Photogrammetric Point Clouds as a Low-Cost Tool for Yield Forecasting in Vineyards. Sensors 2021, 21, 3083. [Google Scholar] [CrossRef] [PubMed]
- Mesas-Carrascosa, F.J.; De Castro, A.I.; Torres-Sánchez, J.; Triviño-Tarradas, P.; Jiménez-Brenes, F.M.; García-Ferrer, A.; López-Granados, F. Classification of 3D Point Clouds Using Color Vegetation Indices for Precision Viticulture and Digitizing Applications. Remote Sens. 2020, 12, 317. [Google Scholar] [CrossRef]
- Matese, A.; Di Gennaro, S.F. Beyond the Traditional NDVI Index as a Key Factor to Mainstream the Use of UAV in Precision Viticulture. Sci. Rep. 2021, 11, 2721. [Google Scholar] [CrossRef]
- De Castro, A.I.; Jimenez-Brenes, F.M.; Torres-Sanchez, J.; Pena, J.M.; Borra-Serrano, I.; Lopez-Granados, F. 3-D Characterization of Vineyards Using a Novel UAV Imagery-Based OBIA for Precision Viticulture Applications. Remote Sens. 2018, 10, 584. [Google Scholar] [CrossRef]
- Pádua, L.; Marques, P.; Hruška, J.; Adão, T.; Bessa, J.; Sousa, A.; Peres, E.; Morais, R.; Sousa, J.J. Vineyard Properties Extraction Combining UAS-Based RGB Imagery with Elevation Data. Int. J. Remote Sens. 2018, 39, 5377–5401. [Google Scholar] [CrossRef]
- Towers, P.C.; Poblete-echeverría, C. Effect of the Illumination Angle on NDVI Data Composed of Mixed Surface Values Obtained over Vertical-Shoot-Positioned Vineyards. Remote Sens. 2021, 13, 855. [Google Scholar] [CrossRef]
- Comba, L.; Biglia, A.; Ricauda Aimonino, D.; Tortia, C.; Mania, E.; Guidoni, S.; Gay, P. Leaf Area Index Evaluation in Vineyards Using 3D Point Clouds from UAV Imagery. Precis. Agric. 2020, 21, 881–896. [Google Scholar] [CrossRef]
- Di Gennaro, S.F.; Matese, A. Evaluation of Novel Precision Viticulture Tool for Canopy Biomass Estimation and Missing Plant Detection Based on 2.5D and 3D Approaches Using RGB Images Acquired by UAV Platform. Plant Methods 2020, 16, 91. [Google Scholar] [CrossRef] [PubMed]
- Martín-Vélez, V.; Van Leeuwen, C.H.A.; Sánchez, M.I.; Hortas, F.; Shamoun-Baranes, J.; Thaxter, C.B.; Lens, L.; Camphuysen, C.J.; Green, A.J. Spatial Patterns of Weed Dispersal by Wintering Gulls within and beyond an Agricultural Landscape. J. Ecol. 2021, 109, 1947–1958. [Google Scholar] [CrossRef]
- Campos, J.; García-Ruíz, F.; Gil, E. Assessment of Vineyard Canopy Characteristics from Vigour Maps Obtained Using UAV and Satellite Imagery. Sensors 2021, 21, 2363. [Google Scholar] [CrossRef]
- Kalisperakis, I.; Stentoumis, C.; Grammatikopoulos, L.; Karantzalos, K. Leaf Area Index Estimation in Vineyards from UAV Hyperspectral Data, 2D Image Mosaics and 3D Canopy Surface Models. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2015, XL-1-W4, 299–303. [Google Scholar] [CrossRef]
- Mathews, A.J.; Jensen, J.L.R. Visualizing and Quantifying Vineyard Canopy LAI Using an Unmanned Aerial Vehicle (UAV) Collected High Density Structure from Motion Point Cloud. Remote Sens. 2013, 5, 2164–2183. [Google Scholar] [CrossRef]
- Matese, A.; Di Gennaro, S.F. Practical Applications of a Multisensor UAV Platform Based on Multispectral, Thermal and RGB High Resolution Images in Precision. Agriculture 2018, 8, 116. [Google Scholar] [CrossRef]
- Gracia-Romero, A.; Vergara-Díaz, O.; Thierfelder, C.; Cairns, J.E.; Kefauver, S.C.; Araus, J.L. Phenotyping Conservation Agriculture Management Effects on Ground and Aerial Remote Sensing Assessments of Maize Hybrids Performance in Zimbabwe. Remote Sens. 2018, 10, 349. [Google Scholar] [CrossRef]
- López-García, P.; Intrigliolo, D.S.; Moreno, M.A.; Martínez-Moreno, A.; Ortega, J.F.; Pérez-álvarez, E.P.; Ballesteros, R. Assessment of Vineyard Water Status by Multispectral and RGB Imagery Obtained from an Unmanned Aerial Vehicle. Am. J. Enol. Vitic. 2021, 72, 285–297. [Google Scholar] [CrossRef]
- Sepúlveda-Reyes, D.; Ingram, B.; Bardeen, M.; Zúñiga, M.; Ortega-Farías, S.; Poblete-Echeverría, C. Selecting Canopy Zones and Thresholding Approaches to Assess Grapevine Water Status by Using Aerial and Ground-Based Thermal Imaging. Remote Sens. 2016, 8, 822. [Google Scholar] [CrossRef]
- Bellvert, J.; Zarco-Tejada, P.J.; Girona, J.; Fereres, E. Mapping Crop Water Stress Index in a ‘Pinot-Noir’ Vineyard: Comparing Ground Measurements with Thermal Remote Sensing Imagery from an Unmanned Aerial Vehicle. Precis. Agric. 2014, 15, 361–376. [Google Scholar] [CrossRef]
- Albetis, J.; Jacquin, A.; Goulard, M.; Poilvé, H.; Rousseau, J.; Clenet, H.; Dedieu, G.; Duthoit, S. On the Potentiality of UAV Multispectral Imagery to Detect Flavescence Dorée and Grapevine Trunk Diseases. Remote Sens. 2018, 11, 23. [Google Scholar] [CrossRef]
- Bendel, N.; Kicherer, A.; Backhaus, A.; Klück, H.C.; Seiffert, U.; Fischer, M.; Voegele, R.T.; Töpfer, R. Evaluating the Suitability of Hyper- and Multispectral Imaging to Detect Foliar Symptoms of the Grapevine Trunk Disease Esca in Vineyards. Plant Methods 2020, 16, 142. [Google Scholar] [CrossRef] [PubMed]
- Vélez, S.; Ariza-Sentís, M.; Valente, J. Mapping the Spatial Variability of Botrytis Bunch Rot Risk in Vineyards Using UAV Multispectral Imagery. Eur. J. Agron. 2023, 142, 126691. [Google Scholar] [CrossRef]
- De Castro, A.I.; Peña, J.M.; Torres-Sánchez, J.; Jiménez-Brenes, F.; López-Granados, F. Mapping Cynodon Dactylon in Vineyards Using UAV Images for Site-Specific Weed Control. Adv. Anim. Biosci. 2017, 8, 267–271. [Google Scholar] [CrossRef]
- Jiménez-Brenes, F.M.; López-Granados, F.; Torres-Sánchez, J.; Peña, J.M.; Ramírez, P.; Castillejo-González, I.L.; de Castro, A.I. Automatic UAV-Based Detection of Cynodon Dactylon for Site-Specific Vineyard Management. PLoS ONE 2019, 14, e0218132. [Google Scholar] [CrossRef]
- Ballesteros, R.; Intrigliolo, D.S.; Ortega, J.F.; Ramírez-Cuesta, J.M.; Buesa, I.; Moreno, M.A. Vineyard Yield Estimation by Combining Remote Sensing, Computer Vision and Artificial Neural Network Techniques. Precis. Agric. 2020, 21, 1242–1262. [Google Scholar] [CrossRef]
- Di Gennaro, S.F.; Toscano, P.; Cinat, P.; Berton, A.; Matese, A. A Low-Cost and Unsupervised Image Recognition Methodology for Yield Estimation in a Vineyard. Front. Plant Sci. 2019, 10, 559. [Google Scholar] [CrossRef]
- López-García, P.; Ortega, J.F.; Pérez-Álvarez, E.P.; Moreno, M.A.; Ramírez, J.M.; Intrigliolo, D.S.; Ballesteros, R. Yield Estimations in a Vineyard Based on High-Resolution Spatial Imagery Acquired by a UAV. Biosyst. Eng. 2022, 224, 227–245. [Google Scholar] [CrossRef]
- Hamza, M.A.; Anderson, W.K. Soil Compaction in Cropping Systems: A Review of the Nature, Causes and Possible Solutions. Soil Tillage Res. 2005, 82, 121–145. [Google Scholar] [CrossRef]
- Pádua, L.; Marques, P.; Hruška, J.; Adão, T.; Peres, E.; Morais, R.; Sousa, J.J. Multi-Temporal Vineyard Monitoring through UAV-Based RGB Imagery. Remote Sens. 2018, 10, 1907. [Google Scholar] [CrossRef]
- Di Gennaro, S.F.; Toscano, P.; Gatti, M.; Poni, S.; Berton, A.; Matese, A. Spectral Comparison of UAV-Based Hyper and Multispectral Cameras for Precision Viticulture. Remote Sens. 2022, 14, 449. [Google Scholar] [CrossRef]
- Mielcarek, M.; Kamińska, A.; Stereńczak, K. Digital Aerial Photogrammetry (DAP) and Airborne Laser Scanning (ALS) as Sources of Information about Tree Height: Comparisons of the Accuracy of Remote Sensing Methods for Tree Height Estimation. Remote Sens. 2020, 12, 1808. [Google Scholar] [CrossRef]
- Lombardi, E.; Rodríguez-Puerta, F.; Santini, F.; Chambel, M.R.; Climent, J.; de Dios, V.R.; Voltas, J. UAV-LiDAR and RGB Imagery Reveal Large Intraspecific Variation in Tree-Level Morphometric Traits across Different Pine Species Evaluated in Common Gardens. Remote Sens. 2022, 14, 5904. [Google Scholar] [CrossRef]
- Su, B.F.; Xue, J.R.; Xie, C.Y.; Fang, Y.L.; Song, Y.Y.; Fuentes, S. Digital Surface Model Applied to Unmanned Aerial Vehicle Based Photogrammetry to Assess Potential Biotic or Abiotic Effects on Grapevine Canopies. Int. J. Agric. Biol. Eng. 2016, 9, 119–130. [Google Scholar] [CrossRef]
- Primicerio, J.; Caruso, G.; Comba, L.; Crisci, A.; Gay, P.; Guidoni, S.; Genesio, L.; Aimonino, D.R.; Vaccari, F.P. Individual Plant Definition and Missing Plant Characterization in vineyards from High-Resolution UAV Imagery. Eur. J. Remote Sens. 2017, 50, 179–186. [Google Scholar] [CrossRef]
- Comba, L.; Gay, P.; Primicerio, J.; Ricauda Aimonino, D. Vineyard Detection from Unmanned Aerial Systems Images. Comput. Electron. Agric. 2015, 114, 78–87. [Google Scholar] [CrossRef]
- Pádua, L.; Adão, T.; Sousa, A.; Peres, E.; Sousa, J.J. Individual Grapevine Analysis in a Multi-Temporal Context Using UAV-Based Multi-Sensor Imagery. Remote Sens. 2020, 12, 139. [Google Scholar] [CrossRef]
- Jurado, J.M.; Padua, L.; Feito, F.R.; Sousa, J.J. Automatic Grapevine Trunk Detection on UAV-Based Point Cloud. Remote Sens. 2020, 12, 3043. [Google Scholar] [CrossRef]
- Hajjar, C.; Ghattas, G.; Sarkis, M.K.; Chamoun, Y.G. Vine Identification and Characterization in Goblet-Trained Vineyards Using Remotely Sensed Images. Remote Sens. 2021, 13, 2992. [Google Scholar] [CrossRef]



| True Positive | False Positive | False Negative | Precision (%) | Recall (%) | F1 Score (%) | OA (%) | ||
|---|---|---|---|---|---|---|---|---|
| Correct Detection | Incorrect Detection | Not Detected | ||||||
| Posts | 797 | 4 | 9 | 99.50% | 98.88% | 99.19% | 98.40% | |
| Trunks | M1 | 4283 | 34 | 472 | 99.21% | 90.07% | 94.42% | 89.43% |
| M2 | 4462 | 118 | 293 | 97.42% | 93.84% | 95.60% | 91.57% | |
| M3 | 4612 | 316 | 143 | 93.59% | 96.99% | 95.26% | 90.95% | |
| M4 | 4665 | 214 | 90 | 95.61% | 98.11% | 96.84% | 93.88% | |
| True Positive | False Positive | False Negative | Precision (%) | Recall (%) | F1 Score (%) | OA (%) | ||
|---|---|---|---|---|---|---|---|---|
| Correct Detection | Incorrect Detection | Not Detected | ||||||
| Missing vines | M1 | 1137 | 414 | 32 | 73.31% | 97.26% | 83.60% | 71.83% |
| M2 | 1124 | 275 | 45 | 80.34% | 96.15% | 87.54% | 77.84% | |
| M3 | 1061 | 124 | 108 | 89.54% | 90.76% | 90.14% | 82.06% | |
| M4 | 1103 | 83 | 66 | 93.00% | 94.35% | 93.67% | 88.10% | |
| Global Precision | Global Recall | Global F1 Score | Global OA | ||
|---|---|---|---|---|---|
| Vines and missing vines | M1 | 92.37% | 91.49% | 91.93% | 85.06% |
| M2 | 93.43% | 94.29% | 93.86% | 88.43% | |
| M3 | 92.80% | 95.76% | 94.26% | 89.14% | |
| M4 | 95.10% | 97.37% | 96.22% | 92.72% | |
| Vines | Missing Vines | Relative Error | ||||||
|---|---|---|---|---|---|---|---|---|
| Id Row | Ground Truth | M4 | Delta | Ground Truth | M4 | Delta | Vines | Missing Vines |
| 1 | 22 | 23 | −1 | 2 | 1 | 1 | 4.55% | 4.17% |
| 2 | 26 | 26 | 0 | 5 | 7 | −2 | 0.00% | 6.45% |
| 3 | 32 | 30 | 2 | 5 | 7 | −2 | 6.25% | 5.41% |
| 4 | 37 | 38 | −1 | 6 | 6 | 0 | 2.70% | 0.00% |
| 5 | 42 | 41 | 1 | 7 | 8 | −1 | 2.38% | 2.04% |
| 6 | 42 | 41 | 1 | 12 | 16 | −4 | 2.38% | 7.41% |
| 7 | 51 | 51 | 0 | 8 | 8 | 0 | 0.00% | 0.00% |
| 8 | 107 | 105 | 2 | 20 | 22 | −2 | 1.87% | 1.57% |
| 9 | 109 | 111 | −2 | 23 | 22 | 1 | 1.83% | 0.76% |
| 10 | 102 | 101 | 1 | 32 | 35 | −3 | 0.98% | 2.24% |
| 11 | 112 | 112 | 0 | 29 | 32 | −3 | 0.00% | 2.13% |
| 12 | 122 | 122 | 0 | 24 | 25 | −1 | 0.00% | 0.68% |
| 13 | 130 | 128 | 2 | 22 | 25 | −3 | 1.54% | 1.97% |
| 14 | 139 | 136 | 3 | 18 | 22 | −4 | 2.16% | 2.55% |
| 15 | 140 | 145 | −5 | 18 | 20 | −2 | 3.57% | 1.27% |
| 16 | 132 | 131 | 1 | 25 | 28 | −3 | 0.76% | 1.91% |
| 17 | 141 | 142 | −1 | 23 | 22 | 1 | 0.71% | 0.61% |
| 18 | 152 | 150 | 2 | 18 | 21 | −3 | 1.32% | 1.76% |
| 19 | 153 | 153 | 0 | 14 | 15 | −1 | 0.00% | 0.60% |
| 20 | 155 | 160 | −5 | 19 | 19 | 0 | 3.23% | 0.00% |
| 21 | 166 | 170 | −4 | 9 | 10 | −1 | 2.41% | 0.57% |
| 22 | 156 | 159 | −3 | 20 | 20 | 0 | 1.92% | 0.00% |
| 23 | 156 | 159 | −3 | 25 | 27 | −2 | 1.92% | 1.10% |
| 24 | 172 | 171 | 1 | 11 | 16 | −5 | 0.58% | 2.73% |
| 25 | 132 | 138 | −6 | 49 | 44 | 5 | 4.55% | 2.76% |
| 26 | 135 | 139 | −4 | 43 | 45 | −2 | 2.96% | 1.12% |
| 27 | 112 | 121 | −9 | 61 | 56 | 5 | 8.04% | 2.89% |
| 28 | 137 | 140 | −3 | 35 | 41 | −6 | 2.19% | 3.49% |
| 29 | 121 | 131 | −10 | 49 | 44 | 5 | 8.26% | 2.94% |
| 30 | 141 | 148 | −7 | 26 | 25 | 1 | 4.96% | 0.60% |
| 31 | 122 | 128 | −6 | 39 | 43 | −4 | 4.92% | 2.48% |
| 32 | 114 | 121 | −7 | 47 | 45 | 2 | 6.14% | 1.24% |
| 33 | 109 | 115 | −6 | 47 | 43 | 4 | 5.50% | 2.56% |
| 34 | 105 | 109 | −4 | 48 | 47 | 1 | 3.81% | 0.65% |
| 35 | 108 | 108 | 0 | 44 | 45 | −1 | 0.00% | 0.66% |
| 36 | 115 | 121 | −6 | 34 | 35 | −1 | 5.22% | 0.67% |
| 37 | 101 | 110 | −9 | 47 | 46 | 1 | 8.91% | 0.68% |
| 38 | 92 | 96 | −4 | 51 | 48 | 3 | 4.35% | 2.10% |
| 39 | 115 | 119 | −4 | 26 | 25 | 1 | 3.48% | 0.71% |
| 40 | 101 | 110 | −9 | 35 | 30 | 5 | 8.91% | 3.68% |
| 41 | 102 | 114 | −12 | 30 | 28 | 2 | 11.76% | 1.52% |
| 42 | 94 | 97 | −3 | 35 | 34 | 1 | 3.19% | 0.78% |
| 43 | 103 | 109 | −6 | 28 | 28 | 0 | 5.83% | 0.00% |
| Total | 4755 | 4879 | −124 | 1169 | 1186 | −17 | 3.40% | 1.85% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Gennaro, S.F.; Vannini, G.L.; Berton, A.; Dainelli, R.; Toscano, P.; Matese, A. Missing Plant Detection in Vineyards Using UAV Angled RGB Imagery Acquired in Dormant Period. Drones 2023, 7, 349. https://doi.org/10.3390/drones7060349
Di Gennaro SF, Vannini GL, Berton A, Dainelli R, Toscano P, Matese A. Missing Plant Detection in Vineyards Using UAV Angled RGB Imagery Acquired in Dormant Period. Drones. 2023; 7(6):349. https://doi.org/10.3390/drones7060349
Chicago/Turabian StyleDi Gennaro, Salvatore Filippo, Gian Luca Vannini, Andrea Berton, Riccardo Dainelli, Piero Toscano, and Alessandro Matese. 2023. "Missing Plant Detection in Vineyards Using UAV Angled RGB Imagery Acquired in Dormant Period" Drones 7, no. 6: 349. https://doi.org/10.3390/drones7060349
APA StyleDi Gennaro, S. F., Vannini, G. L., Berton, A., Dainelli, R., Toscano, P., & Matese, A. (2023). Missing Plant Detection in Vineyards Using UAV Angled RGB Imagery Acquired in Dormant Period. Drones, 7(6), 349. https://doi.org/10.3390/drones7060349











