Detection and Multi-Class Classification of Invasive Knotweeds with Drones and Deep Learning Models
Abstract
1. Introduction
2. Materials and Methods
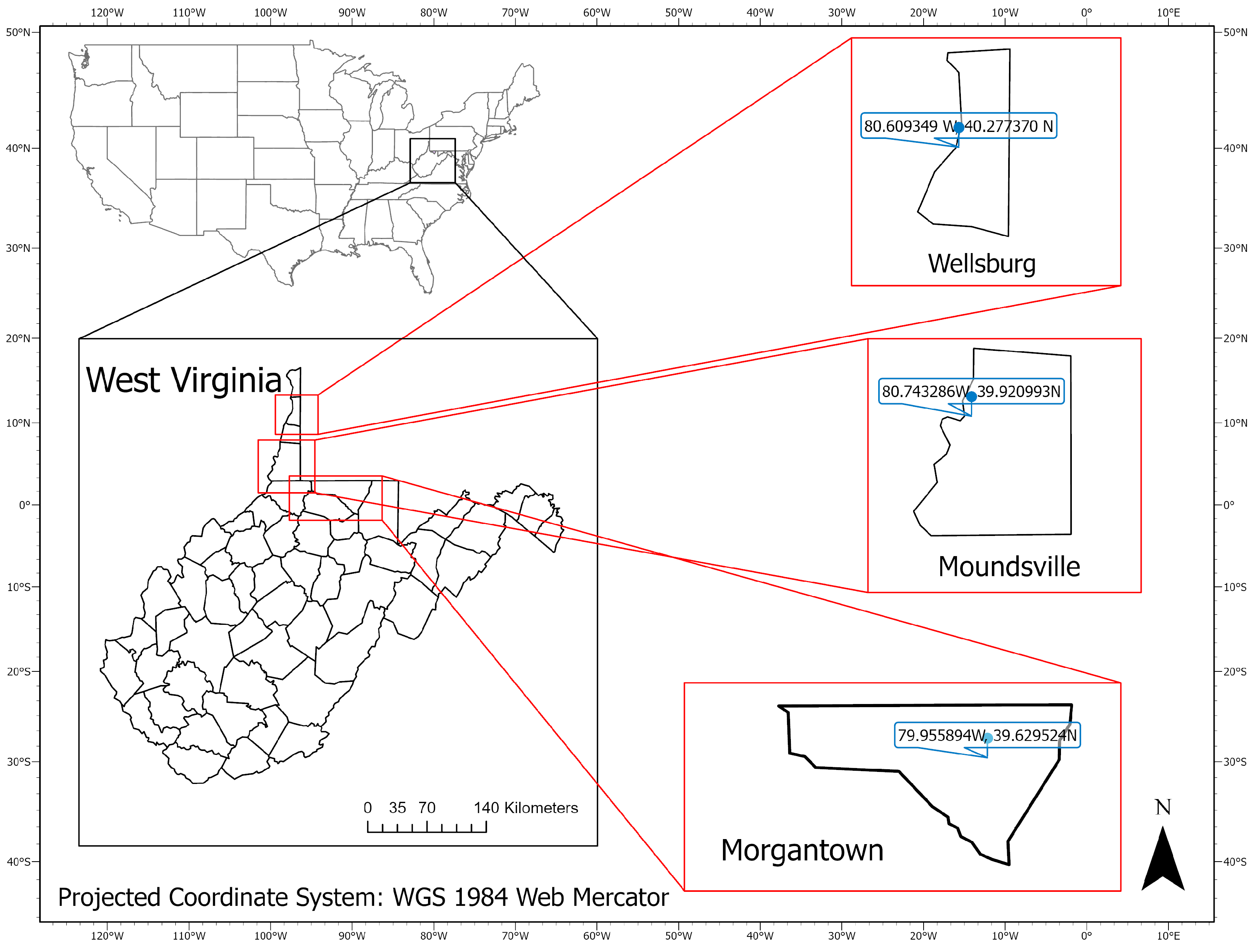
2.1. Study Sites
2.2. Aerial Surveys with Drones and Sensors
2.3. Development of Deep Learning Capability for Automated Identification
2.3.1. Aerial Surveys Using Drones
2.3.2. Data Preprocessing
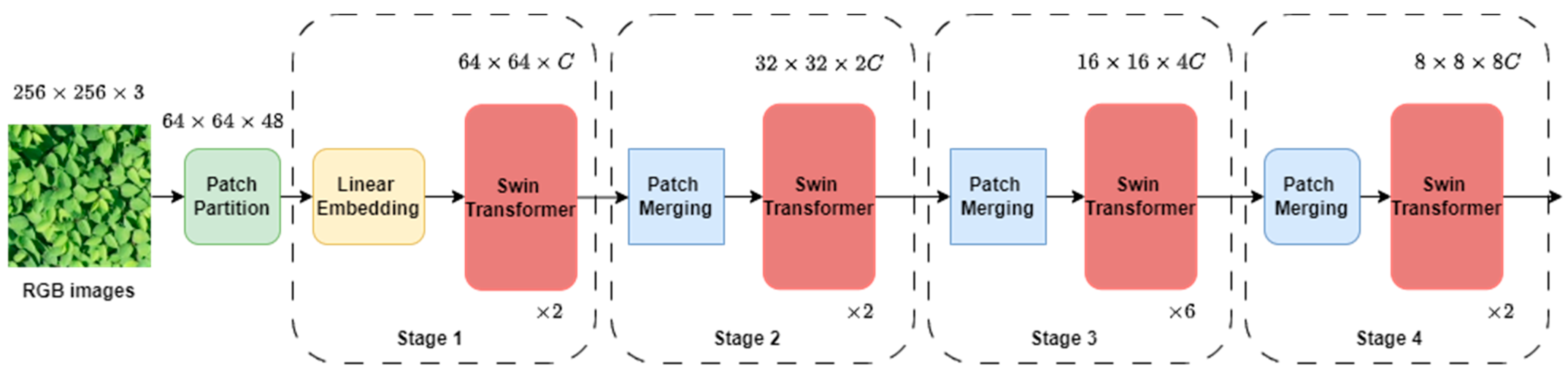
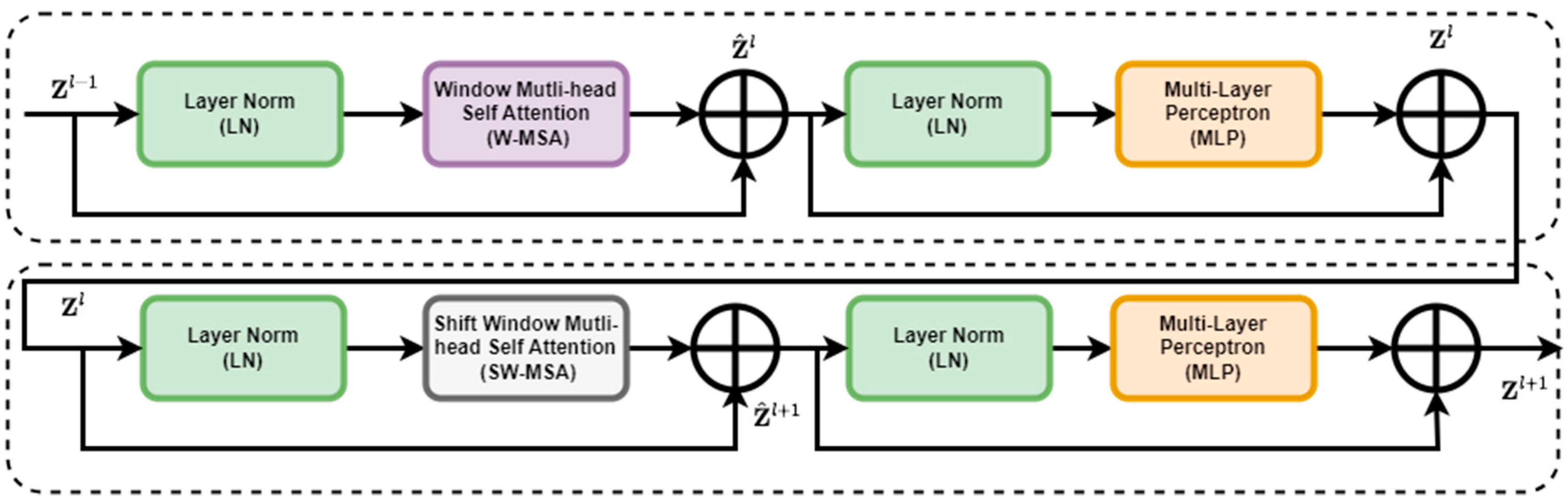
2.3.3. Overview of Swin Transformer
2.3.4. Data Augmentation and Training
3. Results
3.1. Detection of Knotweeds Using Aerial Imagery
3.2. Deep Learning for Knotweed Classification
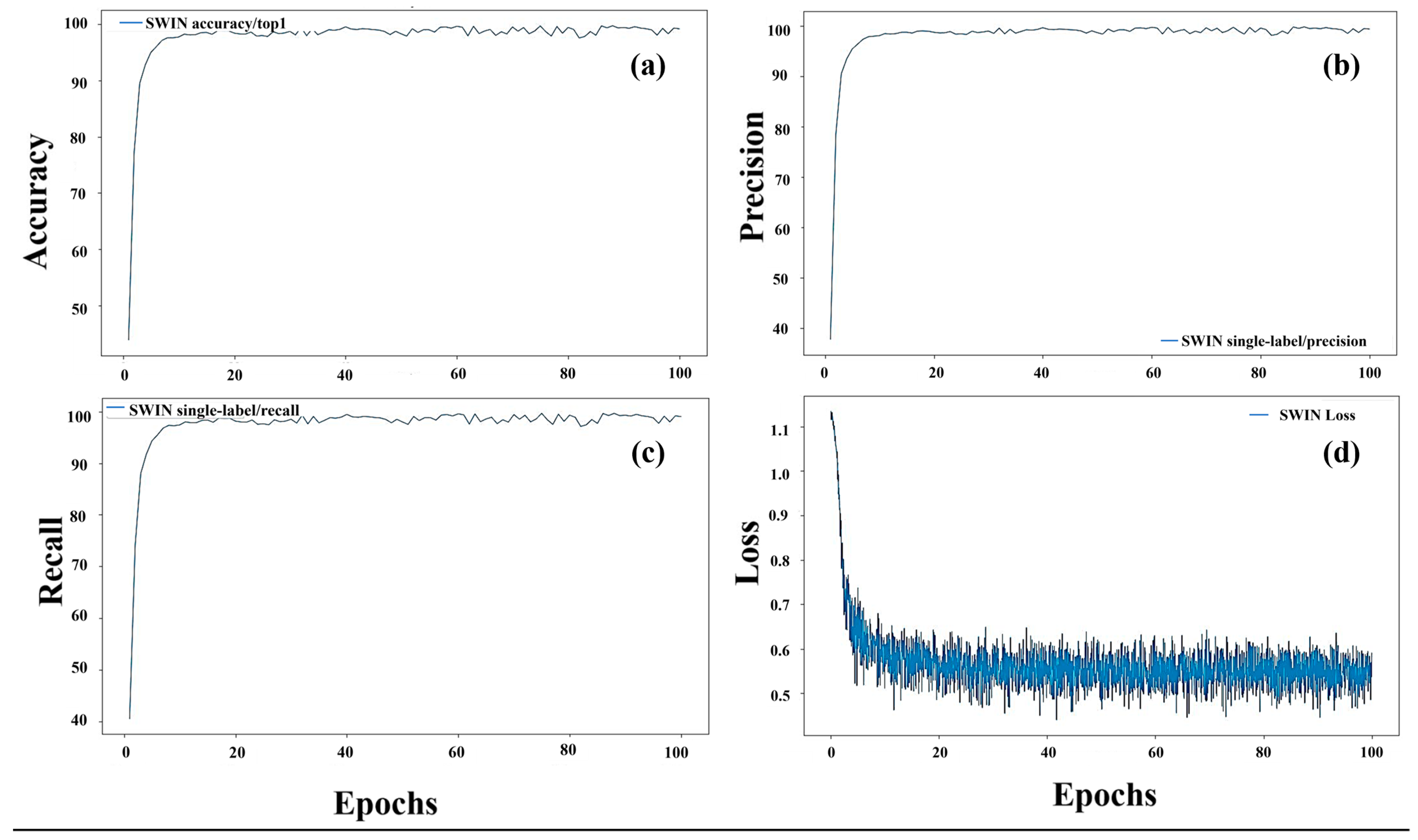
3.2.1. Evaluation of the Deep Learning Model
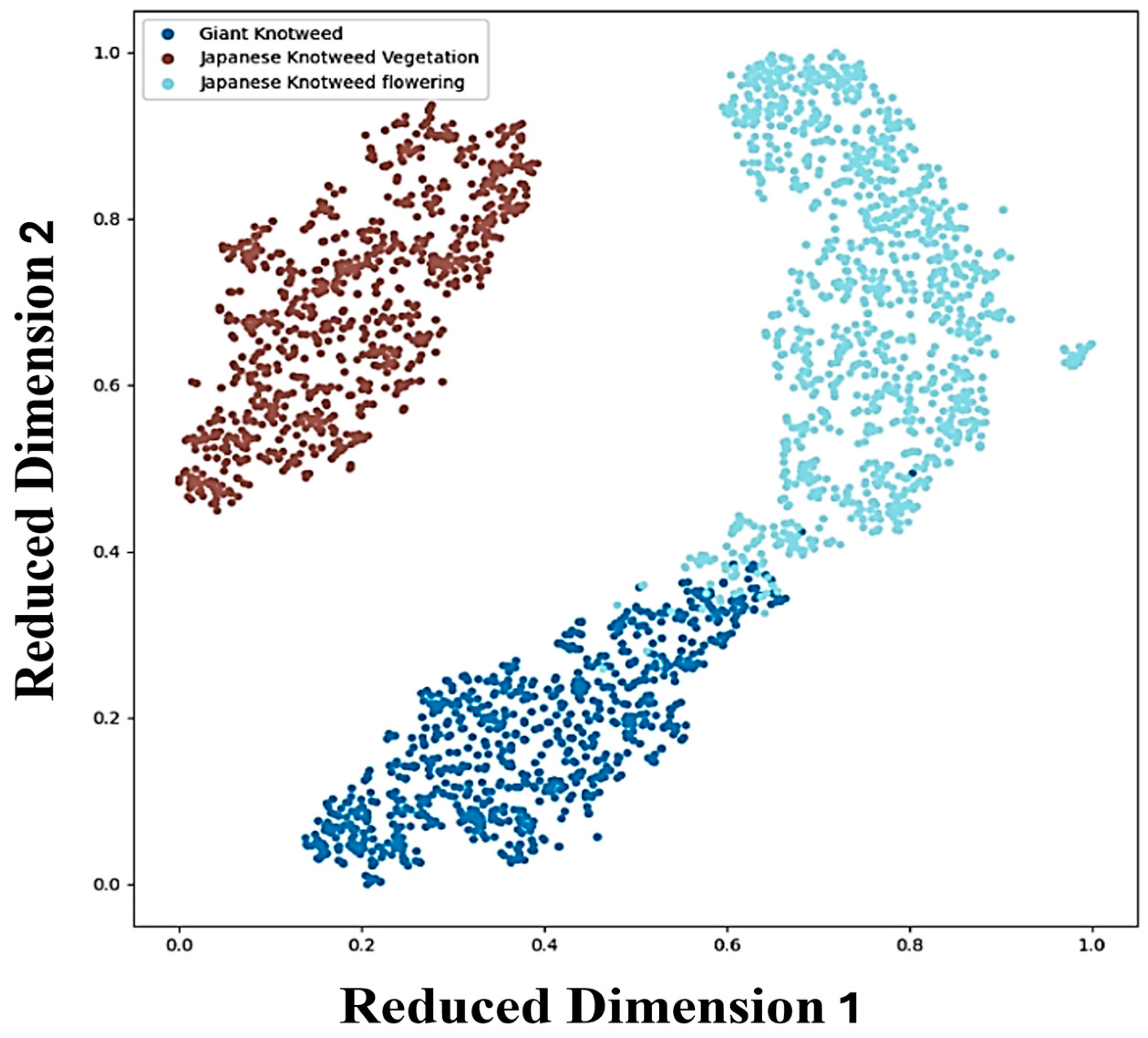
3.2.2. Confusion Matrix
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilson, L.M. Key to Identification of Invasive Knotweeds in British Columbia; Ministry of Forests and Range, Forest Practices Branch: Kamloops, BC, Canada, 2007. [Google Scholar]
- Parkinson, H.; Mangold, J. Biology, Ecology and Management of the Knotweed Complex; Montana State University Extension: Bozeman, MT, USA, 2010. [Google Scholar]
- Brousseau, P.-M.; Chauvat, M.; De Almeida, T.; Forey, E. Invasive knotweed modifies predator–prey interactions in the soil food web. Biol. Invasions 2021, 23, 1987–2002. [Google Scholar] [CrossRef]
- Kato-Noguchi, H. Allelopathy of knotweeds as invasive plants. Plants 2021, 11, 3. [Google Scholar] [CrossRef] [PubMed]
- Colleran, B.; Lacy, S.N.; Retamal, M.R. Invasive Japanese knotweed (Reynoutria japonica Houtt.) and related knotweeds as catalysts for streambank erosion. River Res. Appl. 2020, 36, 1962–1969. [Google Scholar] [CrossRef]
- Payne, T.; Hoxley, M. Identifying and eradicating Japanese knotweed in the UK built environment. Struct. Surv. 2012, 30, 24–42. [Google Scholar] [CrossRef]
- Dusz, M.-A.; Martin, F.-M.; Dommanget, F.; Petit, A.; Dechaume-Moncharmont, C.; Evette, A. Review of existing knowledge and practices of tarping for the control of invasive knotweeds. Plants 2021, 10, 2152. [Google Scholar] [CrossRef]
- Rejmánek, M.; Pitcairn, M.J. When is eradication of exotic plant pests a realistic goal? In Turning the Tide: The Eradication of Invasive Species, Proceedings of the International Conference on Eradication of Island Invasives, Auckland, New Zealand, 19–23 February 2001; Veitch, C.R., Clout, M.N., Eds.; IUCN SSC Invasive Species Specialist Group: Gland, Switzerland; Cambridge, UK, 2002; 414p. [Google Scholar]
- Hocking, S.; Toop, T.; Jones, D.; Graham, I.; Eastwood, D. Assessing the relative impacts and economic costs of Japanese knotweed management methods. Sci. Rep. 2023, 13, 3872. [Google Scholar] [CrossRef] [PubMed]
- Shahi, T.B.; Dahal, S.; Sitaula, C.; Neupane, A.; Guo, W. Deep learning-based weed detection using UAV images: A comparative study. Drones 2023, 7, 624. [Google Scholar] [CrossRef]
- López-Granados, F. Weed detection for site-specific weed management: Mapping and real-time approaches. Weed Res. 2011, 51, 1–11. [Google Scholar] [CrossRef]
- Hasan, A.S.M.M.; Sohel, F.; Diepeveen, D.; Laga, H.; Jones, M.G. A survey of deep learning techniques for weed detection from images. Comput. Electron. Agric. 2021, 184, 106067. [Google Scholar]
- Lambert, J.P.T.; Hicks, H.L.; Childs, D.Z.; Freckleton, R.P. Evaluating the potential of unmanned aerial systems for mapping weeds at field scales: A case study with Alopecurus myosuroides. Weed Res. 2018, 58, 35–45. [Google Scholar] [CrossRef]
- Singh, V.; Rana, A.; Bishop, M.; Filippi, A.M.; Cope, D.; Rajan, N.; Bagavathiannan, M. Unmanned aircraft systems for precision weed detection and management: Prospects and challenges. Adv. Agron. 2020, 159, 93–134. [Google Scholar]
- Barbizan Sühs, R.; Ziller, S.R.; Dechoum, M. Is the use of drones cost-effective and efficient in detecting invasive alien trees? A case study from a subtropical coastal ecosystem. Biol. Invasions 2024, 26, 357–363. [Google Scholar] [CrossRef]
- Bradley, B.A. Remote detection of invasive plants: A review of spectral, textural and phenological approaches. Biol. Invasions 2014, 16, 1411–1425. [Google Scholar] [CrossRef]
- Lass, L.W.; Prather, T.S.; Glenn, N.F.; Weber, K.T.; Mundt, J.T.; Pettingill, J. A review of remote sensing of invasive weeds and example of the early detection of spotted knapweed (Centaurea maculosa) and baby’s breath (Gypsophila paniculata) with a hyperspectral sensor. Weed Sci. 2005, 53, 242–251. [Google Scholar] [CrossRef]
- Miotto, R.; Wang, F.; Wang, S.; Jiang, X.; Dudley, J.T. Deep learning for healthcare: Review, opportunities and challenges. Brief. Bioinform. 2018, 19, 1236–1246. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.X.; Tuia, D.; Mou, L.; Xia, G.S.; Zhang, L.; Xu, F.; Fraundorfer, F. Deep learning in remote sensing: A comprehensive review and list of resources. IEEE Geosci. Remote Sens. Mag. 2017, 5, 8–36. [Google Scholar] [CrossRef]
- Zhang, Y.; Yan, J.; Chen, S.; Gong, M.; Gao, D.; Zhu, M.; Gan, W. Review of the applications of deep learning in bioinformatics. Curr. Bioinform. 2020, 15, 898–911. [Google Scholar] [CrossRef]
- Otter, D.W.; Medina, J.R.; Kalita, J.K. A survey of the usages of deep learning for natural language processing. IEEE Trans. Neural Netw. Learn. Syst. 2020, 32, 604–624. [Google Scholar] [CrossRef] [PubMed]
- Simonyan, K. Very deep convolutional networks for large-scale image recognition. arXiv 2015. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 4700–4708. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, PMLR 97, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the Advances in Neural Information Processing Systems 2017, Long Beach, CA, USA, 4–9 December 2017; Volume 30. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 × 16 words: Transformers for image recognition at scale. In Proceedings of the 9th International Conference on Learning Representations, ICLR 2021, Virtual, 3–7 May 2021. [Google Scholar]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jegou, H. Training data-efficient image transformers & distillation through attention. In Proceedings of the 38th International Conference on Machine Learning, PLMR 139, Virtual, 18–24 July 2021; pp. 10347–10357. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Nashville, TN, USA, 20–25 June 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 10012–10022. [Google Scholar]
- Naharki, K.; Huebner, C.D.; Park, Y.-L. The detection of tree of heaven (Ailanthus altissima) using drones and optical sensors: Implications for the management of invasive plants and insects. Drones 2023, 8, 1. [Google Scholar] [CrossRef]
- Haykin, S. Neural Networks: A Comprehensive Foundation; Prentice Hall PTR: Upper Saddle River, NJ, USA, 1998. [Google Scholar]
- Raffel, C.; Shazeer, N.; Roberts, A.; Lee, K.; Narang, S.; Matena, M.; Zhou, Y.; Li, W.; Liu, P.J. Exploring the limits of transfer learning with a unified text-to-text transformer. J. Mach. Learn. Res. 2020, 21, 1–67. [Google Scholar]
- Zhang, H.; Cisse, M.; Dauphin, Y.N.; Lopez-Paz, D. mixup: Beyond Empirical Risk Minimization. In Proceedings of the International Conference on Learning Representations, Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar]
- Yun, S.; Han, D.; Oh, S.J.; Chun, S.; Choe, J.; Yoo, Y. Cutmix: Regularization strategy to train strong classifiers with localizable features. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 6023–6032. [Google Scholar]
- MMPretrain Contributors. OpenMMLab’s Pre-Training Toolbox and Benchmark. 2023. Available online: https://github.com/open-mmlab/mmpretrain (accessed on 28 April 2024).
- Van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Grevstad, F.S.; Andreas, J.E.; Bourchier, R.S.; Shaw, R.; Winston, R.L.; Randall, C.B. Biology and Biological Control of Knotweeds; United States Department of Agriculture, Forest Health Assessment and Applied Sciences Team: Fort Collins, CO, USA, 2018. [Google Scholar]
- Park, Y.L.; Gururajan, S.; Thistle, H.; Chandran, R.; Reardon, R. Aerial release of Rhinoncomimus latipes (Coleoptera: Curculionidae) to control Persicaria perfoliata (Polygonaceae) using an unmanned aerial system. Pest Manag. Sci. 2018, 74, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Shaw, R.H.; Bryner, S.; Tanner, R. The life history and host range of the Japanese knotweed psyllid, Aphalara itadori Shinji: Potentially the first classical biological weed control agent for the European Union. Biol. Control 2009, 49, 105–113. [Google Scholar] [CrossRef]
- Kussul, N.; Lavreniuk, M.; Skakun, S.; Shelestov, A. Deep learning classification of land cover and crop types using remote sensing data. IEEE Geosci. Remote Sens. Lett. 2017, 14, 778–782. [Google Scholar] [CrossRef]
- Agilandeeswari, L.; Prabukumar, M.; Radhesyam, V.; Phaneendra, K.L.B.; Farhan, A. Crop classification for agricultural applications in hyperspectral remote sensing images. Appl. Sci. 2022, 12, 1670. [Google Scholar] [CrossRef]
- Orynbaikyzy, A.; Gessner, U.; Conrad, C. Crop type classification using a combination of optical and radar remote sensing data: A review. Int. J. Remote Sens. 2019, 40, 6553–6595. [Google Scholar] [CrossRef]
- Batchuluun, G.; Nam, S.H.; Park, K.R. Deep learning-based plant classification and crop disease classification by thermal camera. J. King Saud Univ.-Comput. Inf. Sci. 2022, 34, 10474–10486. [Google Scholar] [CrossRef]
- Barnes, M.L.; Yoder, L.; Khodaee, M. Detecting winter cover crops and crop residues in the Midwest US using machine learning classification of thermal and optical imagery. Remote Sens. 2021, 13, 1998. [Google Scholar] [CrossRef]
- Chandel, N.S.; Rajwade, Y.A.; Dubey, K.; Chandel, A.K.; Subeesh, A.; Tiwari, M.K. Water stress identification of winter wheat crop with state-of-the-art AI techniques and high-resolution thermal-RGB imagery. Plants 2022, 11, 3344. [Google Scholar] [CrossRef]
- Gadiraju, K.K.; Ramachandra, B.; Chen, Z.; Vatsavai, R.R. Multimodal deep learning-based crop classification using multispectral and multitemporal satellite imagery. In Proceedings of the 26th International Conference on Knowledge Discovery & Data Mining, Virtual, 6–10 July 2020; Association for Computing Machinery: New York, NY, USA, 2020; pp. 3234–3242. [Google Scholar]
- Siesto, G.; Fernández-Sellers, M.; Lozano-Tello, A. Crop classification of satellite imagery using synthetic multitemporal and multispectral images in convolutional neural networks. Remote Sens. 2021, 13, 3378. [Google Scholar] [CrossRef]
- Khan, S.D.; Basalamah, S.; Lbath, A. Weed–Crop segmentation in drone images with a novel encoder–decoder framework enhanced via attention modules. Remote Sens. 2023, 15, 5615. [Google Scholar] [CrossRef]
- Genze, N.; Wirth, M.; Schreiner, C.; Ajekwe, R.; Grieb, M.; Grimm, D.G. Improved weed segmentation in UAV imagery of sorghum fields with a combined deblurring segmentation model. Plant Methods 2023, 19, 87. [Google Scholar] [CrossRef]
- Valicharla, S.K.; Li, X.; Greenleaf, J.; Turcotte, R.; Hayes, C.; Park, Y.-L. Precision detection and assessment of ash death and decline caused by the emerald ash borer using drones and deep learning. Plants 2023, 12, 798. [Google Scholar] [CrossRef]
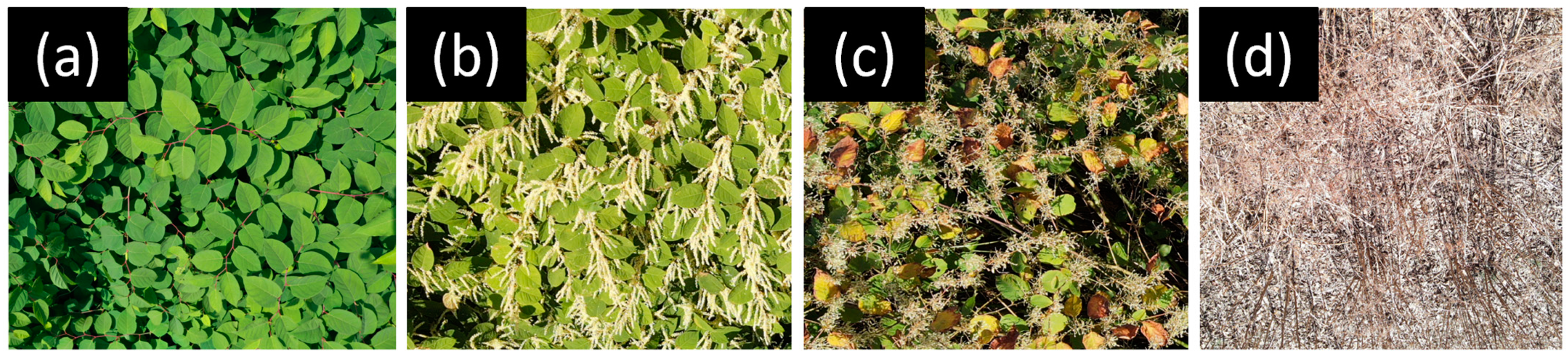
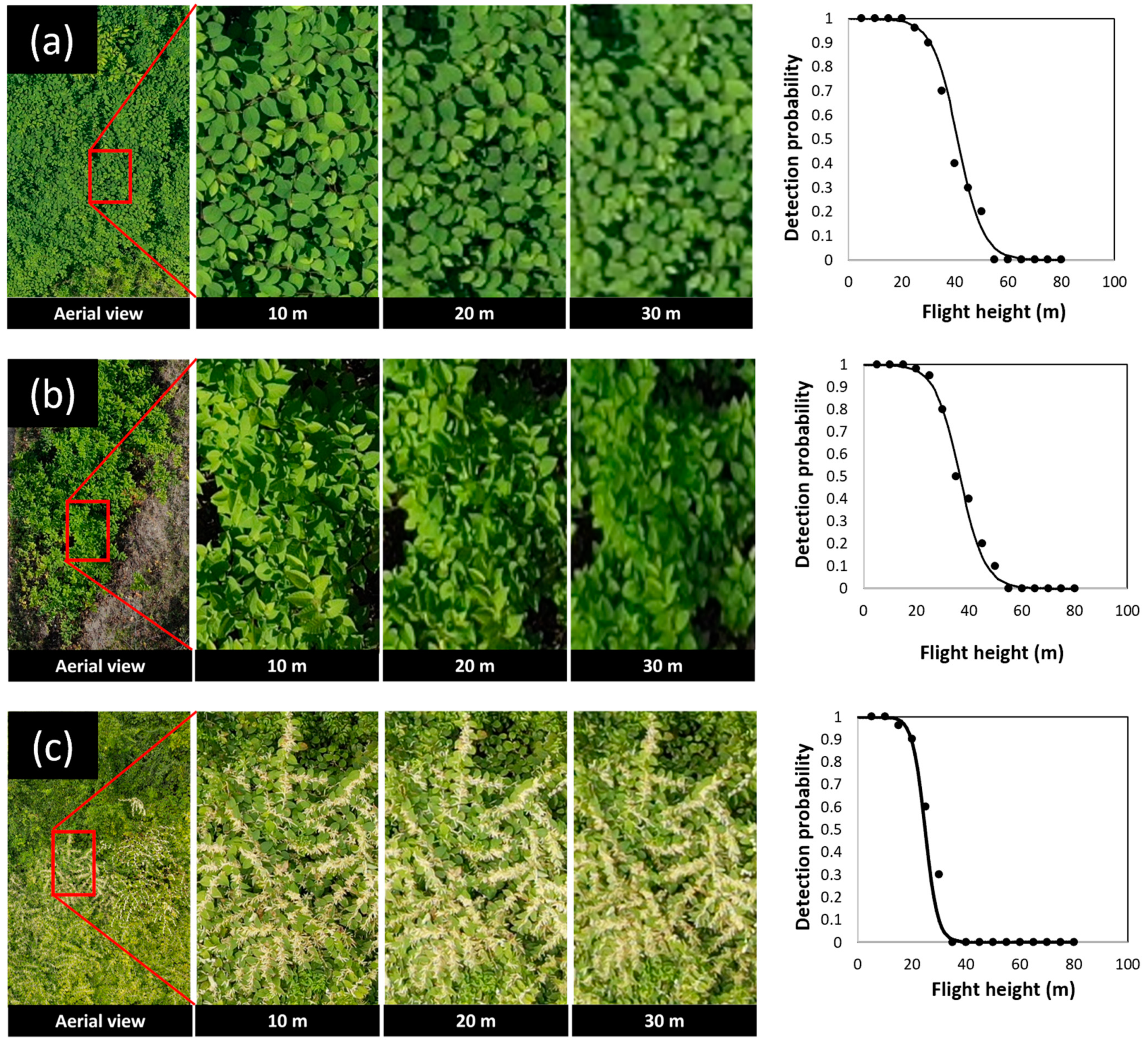
| Category | Training Images | Validation Images | Testing Images | Total Images |
|---|---|---|---|---|
| Giant knotweed at the vegetative stage | 5351 | 1528 | 764 | 7643 |
| Japanese knotweed at the vegetative stage | 7538 | 2154 | 1075 | 10,767 |
| Knotweeds at the flowering stage | 5827 | 1665 | 833 | 8325 |
| Species and Stages | Temperature | t | df | p | ||
|---|---|---|---|---|---|---|
| Knotweed | Surrounding Vegetation | Ambient Air | ||||
| Japanese knotweed | ||||||
| Vegetative stage | 37.06 ± 1.36 °C | 37.21 ± 1.87 °C | 28 °C | 0.2051 | 18 | 0.8398 |
| Flowering stage | 24.87 ± 1.36 °C | 25.65 ± 1.65 °C | 16 °C | 1.1536 | 18 | 0.2638 |
| Giant knotweed | ||||||
| Vegetative stage | 14.34 ± 0.21 °C | 14.56 ± 0.37 °C | 11 °C | 1.6352 | 18 | 0.1194 |
| Method | Backbone | Accuracy | Precision | Recall |
|---|---|---|---|---|
| ResNet50 [23] | 98.9141 | 99.0193 | 98.7977 | |
| CNNs | VGG16 [22] | 97.7907 | 98.0992 | 97.5329 |
| EfficientNet [25] | 99.1013 | 99.1718 | 99.0284 | |
| DeiT [28] | 98.7268 | 99.9377 | 98.5418 | |
| Transformers | ViT-B [27] | 99.3072 | 99.2945 | 99.3251 |
| Swin-T [29] | 99.4758 | 99.4713 | 99.4836 |
| True Label | Predicted Label | ||
|---|---|---|---|
| Giant Knotweed at the Vegetative Stage | Knotweeds at the Flowering Stage | Japanese Knotweed at the Vegetative Stage | |
| Giant knotweed at the vegetative stage | 731 | 33 | 0 |
| Japanese knotweed at the vegetative stage | 0 | 0 | 833 |
| Knotweeds at the flowering stage | 20 | 1055 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valicharla, S.K.; Karimzadeh, R.; Naharki, K.; Li, X.; Park, Y.-L. Detection and Multi-Class Classification of Invasive Knotweeds with Drones and Deep Learning Models. Drones 2024, 8, 293. https://doi.org/10.3390/drones8070293
Valicharla SK, Karimzadeh R, Naharki K, Li X, Park Y-L. Detection and Multi-Class Classification of Invasive Knotweeds with Drones and Deep Learning Models. Drones. 2024; 8(7):293. https://doi.org/10.3390/drones8070293
Chicago/Turabian StyleValicharla, Sruthi Keerthi, Roghaiyeh Karimzadeh, Kushal Naharki, Xin Li, and Yong-Lak Park. 2024. "Detection and Multi-Class Classification of Invasive Knotweeds with Drones and Deep Learning Models" Drones 8, no. 7: 293. https://doi.org/10.3390/drones8070293
APA StyleValicharla, S. K., Karimzadeh, R., Naharki, K., Li, X., & Park, Y.-L. (2024). Detection and Multi-Class Classification of Invasive Knotweeds with Drones and Deep Learning Models. Drones, 8(7), 293. https://doi.org/10.3390/drones8070293







