On the Scale Invariance in State of the Art CNNs Trained on ImageNet †
Abstract
:1. Introduction
2. Related Work
3. Materials and Methods
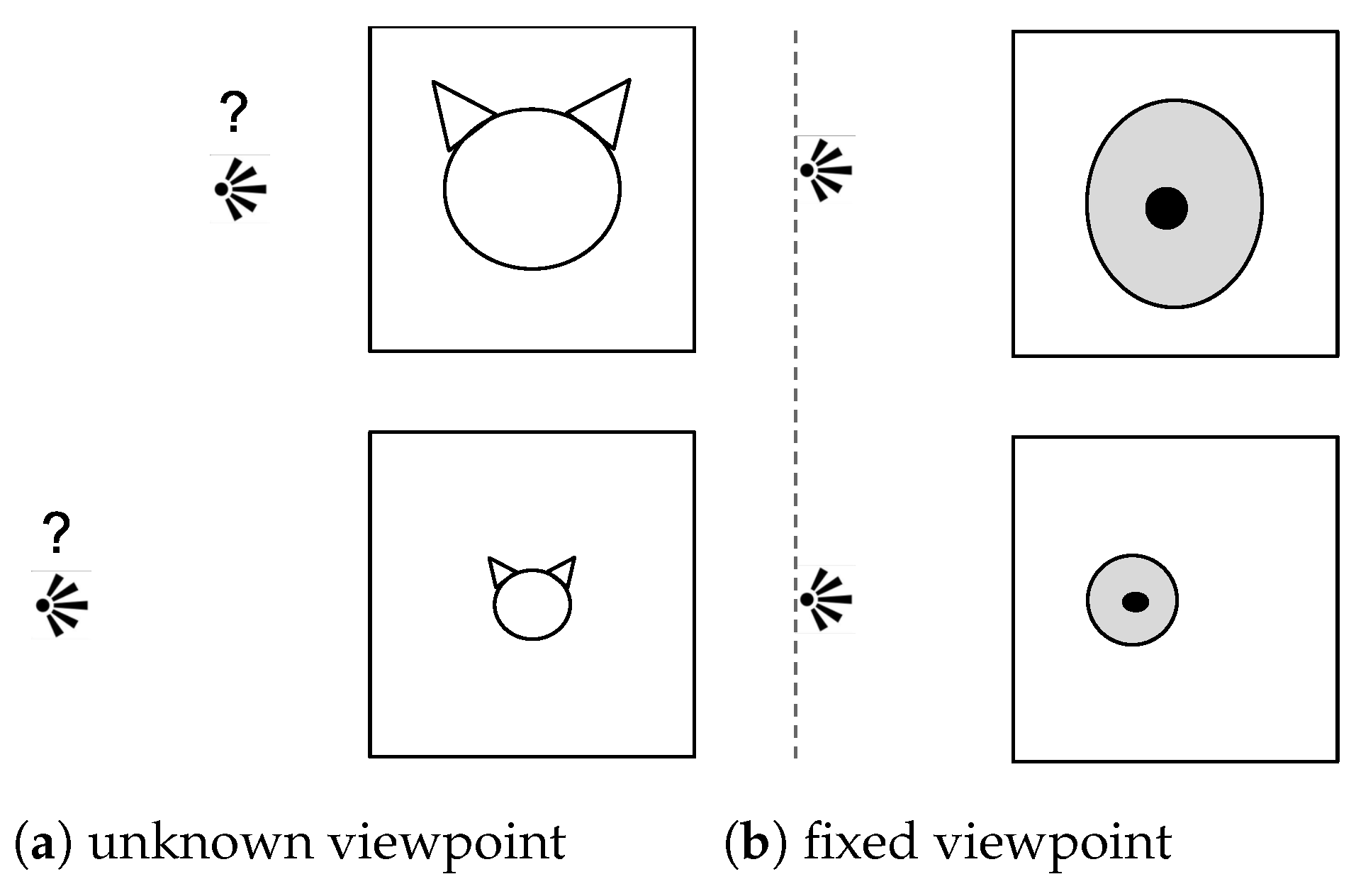
3.1. Notations
3.2. Datasets
3.3. Network Architectures
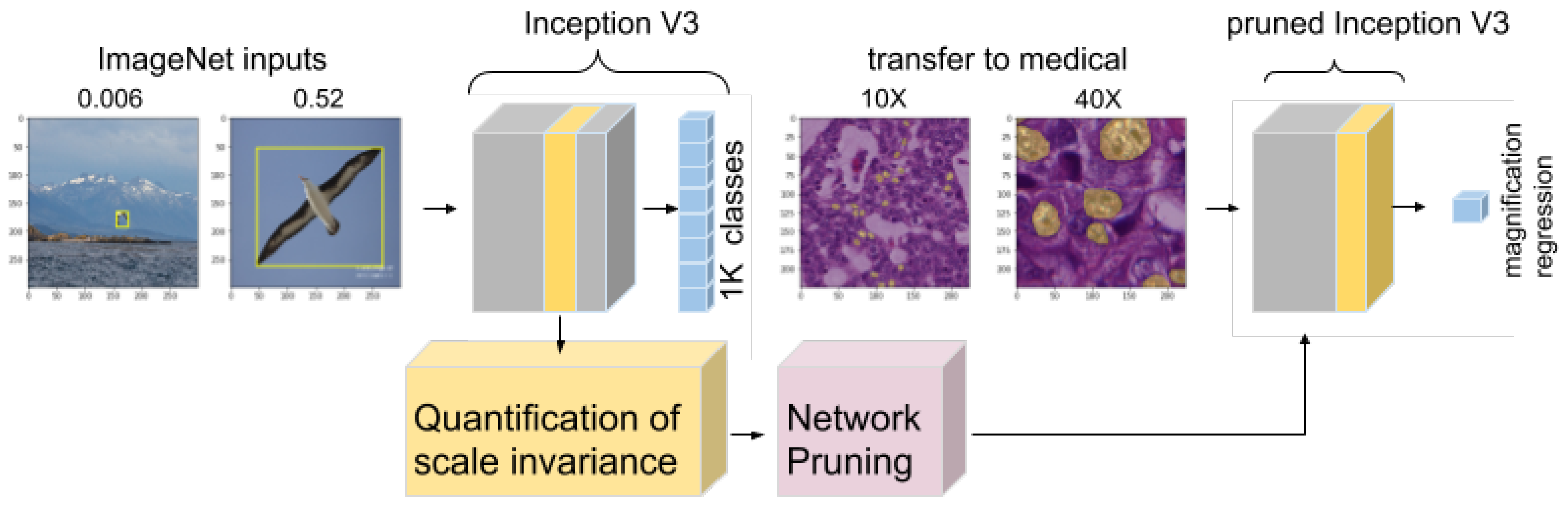
3.4. Quantification of the Scale and Pruning Strategy
3.5. Evaluation
3.6. Experimental Setups
4. Results
4.1. Invariance of the Predictions to Resizing
4.2. Experiments on Noise Inputs
4.3. Layer-wise Quantification of Scale Covariance
4.4. Improvement of Transfer to Histopathology
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CNN | Convolutional Neural Networks |
| RCVs | Regression Concept Vectors |
| CAV | Concept Activation Vectors |
Appendix A
Appendix A.1. Extended Regression Results


Appendix A.2. Detailed Determination Coefficients
| Model | InceptionV3 ImageNet | InceptionV3 Random Weights | InceptionV3 Random Scale Measures |
|---|---|---|---|
| mixed0 | −0.72 | −283 | −5.75 |
| mixed1 | −2.39 | −331 | −20.1 |
| mixed2 | −3.12 | −14.4 | −20.6 |
| mixed3 | 0.59 | −0.47 | −1.81 |
| mixed4 | 0.69 | −0.41 | −1.66 |
| mixed5 | 0.70 | −0.53 | −1.68 |
| mixed6 | 0.79 | −0.54 | −1.62 |
| mixed7 | 0.73 | −0.66 | −1.45 |
| mixed8 | 0.84 | −0.06 | −0.98 |
| mixed9 | 0.77 | 0.06 | −0.73 |
| mixed10 | 0.54 | 0.08 | −1.24 |
| pre-soft. | −0.12 | −0.13 | −3.90 |
| softmax | −3861 | −0.13 | −11,077 ± 26,421 |
Appendix A.3. Visualization of Early Layer Features


References
- Szeliski, R. Computer Vision: Algorithms and Applications; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Kanazawa, A.; Sharma, A.; Jacobs, D.W. Locally Scale-Invariant Convolutional Neural Networks. arXiv 2014, arXiv:1412.5104. [Google Scholar]
- Marcos, D.; Kellenberger, B.; Lobry, S.; Tuia, D. Scale equivariance in CNNs with vector fields. arXiv 2018, arXiv:1807.11783. [Google Scholar]
- Worrall, D.E.; Welling, M. Deep Scale-spaces: Equivariance Over Scale. arXiv 2019, arXiv:1905.11697. [Google Scholar]
- Ghosh, R.; Gupta, A.K. Scale Steerable Filters for Locally Scale-Invariant Convolutional Neural Networks. arXiv 2019, arXiv:1906.03861. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2818–2826. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Lenc, K.; Vedaldi, A. Understanding image representations by measuring their equivariance and equivalence. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 991–999. [Google Scholar]
- Huh, M.; Agrawal, P.; Efros, A.A. What makes ImageNet good for transfer learning? arXiv 2016, arXiv:1608.08614. [Google Scholar]
- Graziani, M.; Andrearczyk, V.; Müller, H. Visualizing and Interpreting Feature Reuse of Pretrained CNNs for Histopathology. Irish Mach. Vis. Image Process. (IMVIP) 2019. Available online: https://iprcs.scss.tcd.ie/IMVIP.2019.html (accessed on 2 April 2021).
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? arXiv 2014, arXiv:1411.1792. [Google Scholar]
- Cohen, T.; Welling, M. Group equivariant convolutional networks. In Proceedings of the 33rd International Conference on Machine Learning (ICML 2016), New York City, NY, USA, 19–24 June 2016; pp. 2990–2999. [Google Scholar]
- Andrearczyk, V.; Fageot, J.; Oreiller, V.; Montet, X.; Depeursinge, A. Local rotation invariance in 3D CNNs. Med. Image Anal. 2020, 65, 101756. [Google Scholar] [CrossRef] [PubMed]
- Bejnordi, B.E.; Zuidhof, G.; Balkenhol, M.; Hermsen, M.; Bult, P.; van Ginneken, B.; Karssemeijer, N.; Litjens, G.; van der Laak, J. Context-aware stacked convolutional neural networks for classification of breast carcinomas in whole-slide histopathology images. J. Med. Imaging 2017, 4, 044504. [Google Scholar] [CrossRef] [PubMed]
- Wan, T.; Cao, J.; Chen, J.; Qin, Z. Automated grading of breast cancer histopathology using cascaded ensemble with combination of multi-level image features. Neurocomputing 2017, 229, 34–44. [Google Scholar] [CrossRef]
- Otálora, S.; Atzori, M.; Andrearczyk, V.; Müller, H. Image Magnification Regression Using DenseNet for Exploiting Histopathology Open Access Content. In Proceedings of the MICCAI 2018—Computational Pathology Workshop (COMPAY), Granada, Spain, 16–20 September 2018. [Google Scholar]
- Hu, Z.; Tang, J.; Wang, Z.; Zhang, K.; Zhang, L.; Sun, Q. Deep learning for image-based cancer detection and diagnosis—A survey. Pattern Recognit. 2018, 83, 134–149. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, H.C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raghu, M.; Zhang, C.; Kleinberg, J.; Bengio, S. Transfusion: Understanding transfer learning with applications to medical imaging. arXiv 2019, arXiv:1902.07208. [Google Scholar]
- Graziani, M.; Lompech, T.; Depeursinge, A.; Andrearczyk, V. Interpretable CNN Pruning for Preserving Scale-Covariant Features in Medical Imaging. In Proceedings of the iMIMIC at MICCAI, Lima, Peru, 4–8 October 2020. [Google Scholar]
- Graziani, M.; Andrearczyk, V.; Müller, H. Regression concept vectors for bidirectional explanations in histopathology. In Understanding and Interpreting Machine Learning in Medical Image Computing Applications; Springer: Cham, Switzerland, 2018; pp. 124–132. [Google Scholar]
- Alain, G.; Bengio, Y. Understanding intermediate layers using linear classifier probes. arXiv 2016, arXiv:1610.01644. [Google Scholar]
- Graziani, M.; Andrearczyk, V.; Marchand-Maillet, S.; Müller, H. Concept attribution: Explaining CNN decisions to physicians. Comput. Biol. Med. 2020, 2020, 103865. [Google Scholar]
- Müller, H.; Andrearczyk, V.; del Toro, O.J.; Dhrangadhariya, A.; Schaer, R.; Atzori, M. Studying Public Medical Images from the Open Access Literature and Social Networks for Model Training and Knowledge Extraction. In Proceedings of the 26th International Conference, MMM 2020, Daejeon, Korea, 5–8 January 2020; pp. 553–564. [Google Scholar]
- Van Noord, N.; Postma, E. Learning scale-variant and scale-invariant features for deep image classification. Pattern Recognit. 2017, 61, 583–592. [Google Scholar] [CrossRef] [Green Version]
- Bruna, J.; Mallat, S. Invariant scattering convolution networks. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 1872–1886. [Google Scholar] [CrossRef] [Green Version]
- Yosinski, J.; Clune, J.; Nguyen, A.; Fuchs, T.; Lipson, H. Understanding neural networks through deep visualization. arXiv 2015, arXiv:1506.06579. [Google Scholar]
- Aubry, M.; Russell, B.C. Understanding deep features with computer-generated imagery. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 2875–2883. [Google Scholar]
- Worrall, D.E.; Garbin, S.J.; Turmukhambetov, D.; Brostow, G.J. Interpretable transformations with encoder-decoder networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 5726–5735. [Google Scholar]
- Azulay, A.; Weiss, Y. Why do deep convolutional networks generalize so poorly to small image transformations? arXiv 2018, arXiv:1805.12177. [Google Scholar]
- Kanbak, C.; Moosavi-Dezfooli, S.M.; Frossard, P. Geometric robustness of deep networks: Analysis and improvement. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4441–4449. [Google Scholar]
- Molchanov, P.; Tyree, S.; Karras, T.; Aila, T.; Kautz, J. Pruning convolutional neural networks for resource efficient inference. arXiv 2016, arXiv:1611.06440. [Google Scholar]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. Ssd: Single shot multibox detector. In Proceedings of the 14th European Conference, Amsterdam, The Netherlands, 11–14 October 2016; pp. 21–37. [Google Scholar]
- Wang, P.; Wang, J.; Li, Y.; Li, L.; Zhang, H. Adaptive pruning of transfer learned deep convolutional neural network for classification of cervical pap smear images. IEEE Access 2020, 8, 50674–50683. [Google Scholar] [CrossRef]
- Fernandes, F.E.; Yen, G.G. Pruning of generative adversarial neural networks for medical imaging diagnostics with evolution strategy. Inf. Sci. 2021, 558, 91–102. [Google Scholar] [CrossRef]
- Lipton, Z.C. The Mythos of Model Interpretability. Queue 2018, 16, 30:31–30:57. [Google Scholar] [CrossRef]
- Kim, B.; Wattenberg, M.; Gilmer, J.; Cai, C.; Wexler, J.; Viegas, F.; Sayres, R. Interpretability Beyond Feature Attribution: Quantitative Testing with Concept Activation Vectors (TCAV). In Proceedings of the International Conference on Machine Learning, Stockholm, Sweden, 10–15 July 2018; pp. 2673–2682. [Google Scholar]
- Graziani, M.; Müller, H.; Andrearczyk, V. Interpreting Intentionally Flawed Models with Linear Probes. In Proceedings of the SDL-CV workshop at the IEEE International International Conference on Computer Vision, Seoul, Korea, 27–28 October 2019. [Google Scholar]
- Graziani, M.; Brown, J.; Andrearczyck, V.; Yildiz, V.; Campbell, J.P.; Erdogmus, D.; Ioannidis, S.; Chiang, M.F.; Kalpathy-Kramer, J.; Muller, H. Improved Interpretability for Computer-Aided Severity Assessment of Retinopathy of Prematurity. In Proceedings of the Medical Imaging 2019: Computer-Aided Diagnosis, San Diego, CA, USA, 16–21 February 2019. [Google Scholar]
- Yeche, H.; Harrison, J.; Berthier, T. UBS: A Dimension-Agnostic Metric for Concept Vector Interpretability Applied to Radiomics. In Interpretability of Machine Intelligence in Medical Image Computing and Multimodal Learning for Clinical Decision Support; Springer: Berlin/Heidelberg, Germany, 2019; pp. 12–20. [Google Scholar]
- Everingham, M.; Van Gool, L.; Williams, C.K.I.; Winn, J.; Zisserman, A. The PASCAL Visual Object Classes Challenge 2008 (VOC2008) Results. Available online: http://host.robots.ox.ac.uk/pascal/VOC/voc2008/index.html (accessed on 2 April 2021).
- Janowczyk, A.; Madabhushi, A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J. Pathol. Inform. 2016, 7, 29. [Google Scholar] [CrossRef]
- Touvron, H.; Vedaldi, A.; Douze, M.; Jégou, H. Fixing the train-test resolution discrepancy. arXiv 2019, arXiv:1906.06423. [Google Scholar]
- Yan, E.; Huan, Y. Do CNNs Encode Data Augmentations? arXiv 2020, arXiv:2003.08773. [Google Scholar]
- Bau, D.; Zhou, B.; Khosla, A.; Oliva, A.; Torralba, A. Network dissection: Quantifying interpretability of deep visual representations. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 6541–6549. [Google Scholar]
- Olah, C.; Mordvintsev, A.; Schubert, L. Feature Visualization. Distill 2017. Available online: https://distill.pub/2017/feature-visualization (accessed on 2 April 2021). [CrossRef]







| Split/# Patches | 5X | 8X | 10X | 15X | 20X | 30X | 40X | Total |
|---|---|---|---|---|---|---|---|---|
| Train | 94 | 2174 | 4141 | 7293 | 9002 | 10,736 | 11,638 | 45,078 |
| Validation | 8 | 588 | 1197 | 2132 | 2604 | 3504 | 3150 | 12,733 |
| Test | 36 | 428 | 900 | 1728 | 2198 | 2802 | 3166 | 11,208 |
| Total | 138 | 3190 | 6238 | 11,153 | 13,804 | 16,592 | 17,904 | 69,019 |
| Model | Layer | MAE (std) | Kappa (std) |
|---|---|---|---|
| pretrained IV3 | mixed10 | 81.85 (11.08) | 0.435 (0.02) |
| from scratch IV3 | mixed10 | 82.30 (17.92) | 0.560 (0.09) |
| pruned IV3 | mixed8 | 54.93 (4.32) | 0.571 (0.05) |
| pretrained ResNet50 | add16 | 70.08 (12.49) | 0.610 (0.03) |
| from scratch ResNet50 | add16 | 95.66 (21.39) | 0.461 (0.09) |
| pruned ResNet50 | add15 | 54.76 (3.10) | 0.623 (0.04) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Graziani, M.; Lompech, T.; Müller, H.; Depeursinge, A.; Andrearczyk, V. On the Scale Invariance in State of the Art CNNs Trained on ImageNet. Mach. Learn. Knowl. Extr. 2021, 3, 374-391. https://doi.org/10.3390/make3020019
Graziani M, Lompech T, Müller H, Depeursinge A, Andrearczyk V. On the Scale Invariance in State of the Art CNNs Trained on ImageNet. Machine Learning and Knowledge Extraction. 2021; 3(2):374-391. https://doi.org/10.3390/make3020019
Chicago/Turabian StyleGraziani, Mara, Thomas Lompech, Henning Müller, Adrien Depeursinge, and Vincent Andrearczyk. 2021. "On the Scale Invariance in State of the Art CNNs Trained on ImageNet" Machine Learning and Knowledge Extraction 3, no. 2: 374-391. https://doi.org/10.3390/make3020019
APA StyleGraziani, M., Lompech, T., Müller, H., Depeursinge, A., & Andrearczyk, V. (2021). On the Scale Invariance in State of the Art CNNs Trained on ImageNet. Machine Learning and Knowledge Extraction, 3(2), 374-391. https://doi.org/10.3390/make3020019







