Climate Change and Soil Health: Explainable Artificial Intelligence Reveals Microbiome Response to Warming
Abstract
1. Introduction
2. Materials and Methods
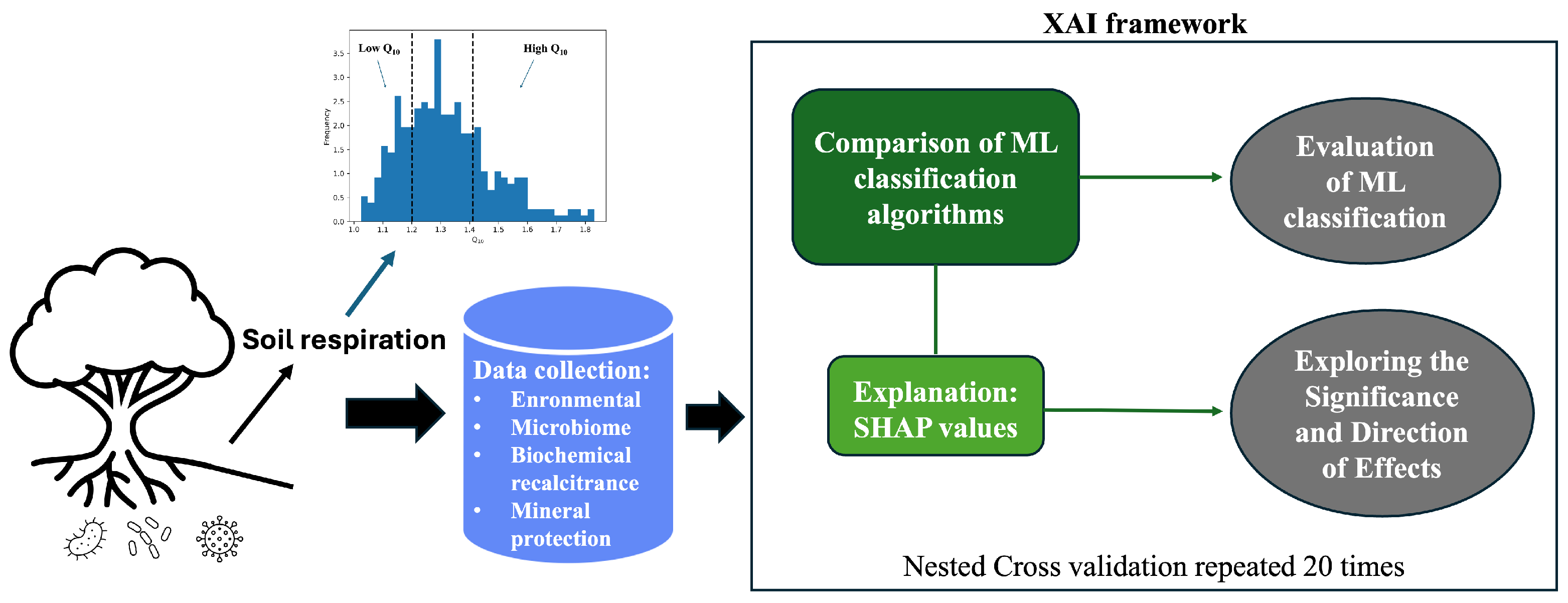
2.1. Data Preparation
2.2. Machine Learning-Based Classification
- C ∈ {1.0, 0.1, 0.01, 0.5},
- kernel ∈ {‘linear’, ‘poly’, ‘rbf’, ‘sigmoid’}.
- n_neighbors ∈ {3, 5, 7, 9, 11},
- weights ∈ {‘uniform’, ‘distance’},
- metric ∈ {‘euclidean’, ‘manhattan’}.
- penalty ∈ {‘l1’,‘l2’,‘elasticnet’},
- C ∈ {1.0, 0.1, 0.01, 0.5},
- solver ∈ {‘lbfgs’, ‘liblinear’, ‘saga’}.
- max_depth ∈ {None, 3, 5, 7, 9},
- criterion ∈ { ‘gini’, ‘entropy’, ‘log_loss’}.
- max_depth ∈ {None, 3, 5, 7, 9},
- n_estimators ∈ {50, 100, 150, 200, 250}.
- Accuracy:The proportion of correctly classified instances among the total instances
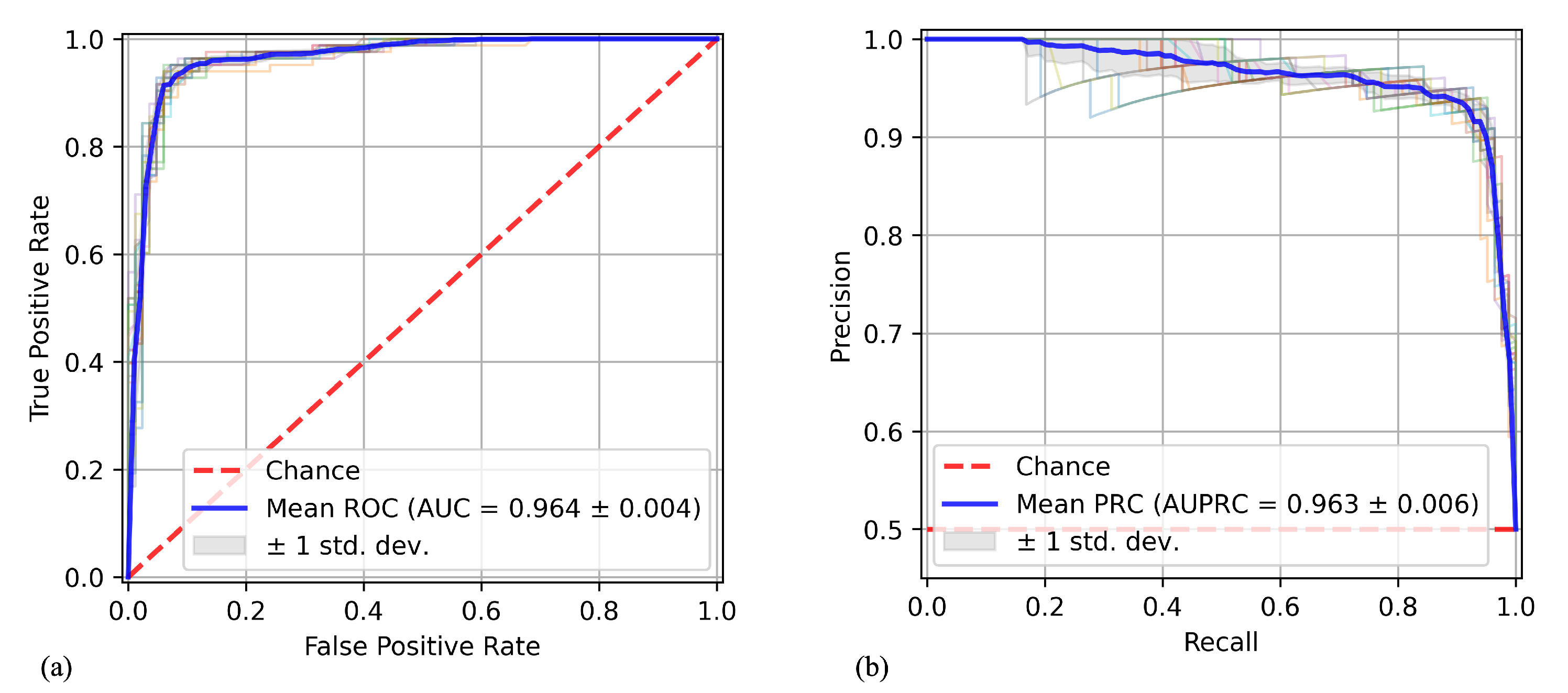
- Area Under the ROC Curve (AUC-ROC):The ROC (Receiver Operating Characteristic) curve and AUC (Area Under the Curve) are assessment tools employed to gauge the effectiveness of a binary classification model. The ROC curve presents a graphical depiction of how sensitivity (true positives) and specificity (true negatives) change across various classification thresholds. Essentially, it illustrates the balance between accurately identifying positive and negative instances by the model. The AUC quantifies the overall performance of the model by measuring the area under the ROC curve: a value closer to 1 signifies superior model performance, while a value around 0.5 suggests random classification. In summary, these metrics are vital for evaluating and contrasting the classification ability of binary models [33].
- Area Under the PRC Curve (AUC-PRC):The Precision-Recall Curve (PRC) and Area Under the Curve (AUC) are tools used to evaluate the performance of a binary classification model. The PRC represents the relationship between precision (true positive rate) and recall (sensitivity) of the model at different decision thresholds. Precision measures the fraction of instances classified as positive that are actually positive, while recall measures the fraction of actual positive instances in the dataset that are correctly identified by the model. The Area Under the PRC Curve provides an aggregated measure of the model’s performance in terms of precision and recall. A larger area indicates better performance, with a maximum area of 1 corresponding to a perfect model [34].
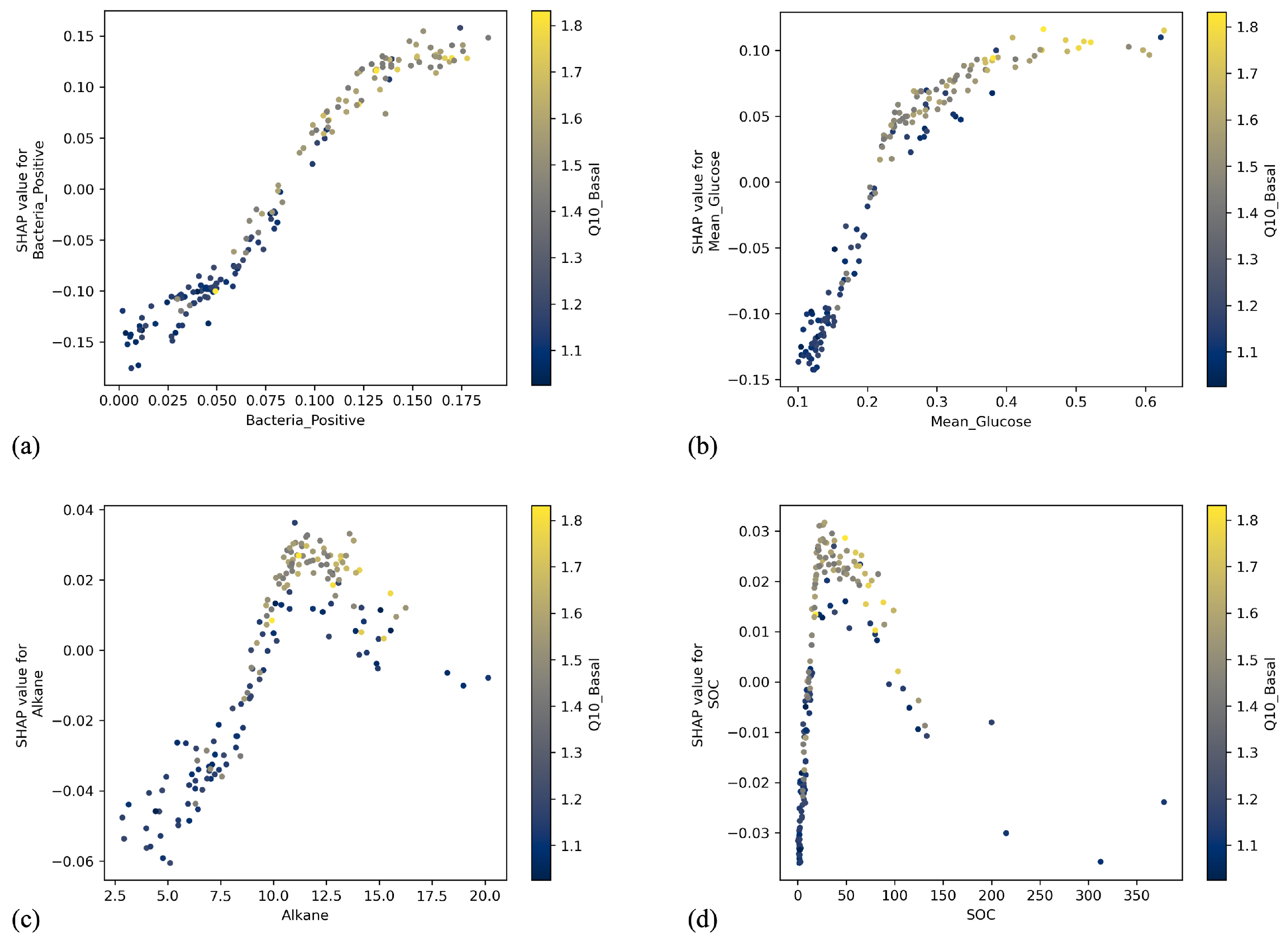
2.3. Explainable Artificial Intelligence (XAI)
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allen, D.E.; Singh, B.P.; Dalal, R.C. Soil health indicators under climate change: A review of current knowledge. In Soil Health and Climate Change; Springer: Berlin/Heidelberg, Germany, 2011; pp. 25–45. [Google Scholar]
- Lal, R. Soil health and climate change: An overview. In Soil Health and Climate Change; Springer: Berlin/Heidelberg, Germany, 2011; pp. 3–24. [Google Scholar]
- Patil, A.; Lamnganbi, M. Impact of climate change on soil health: A review. Int. J. Chem. Stud 2018, 6, 2399–2404. [Google Scholar]
- Haaf, D.; Six, J.; Doetterl, S. Global patterns of geo-ecological controls on the response of soil respiration to warming. Nat. Clim. Change 2021, 11, 623–627. [Google Scholar] [CrossRef]
- Reeve, J.R.; Hoagland, L.A.; Villalba, J.J.; Carr, P.M.; Atucha, A.; Cambardella, C.; Davis, D.R.; Delate, K. Organic farming, soil health, and food quality: Considering possible links. Adv. Agron. 2016, 137, 319–367. [Google Scholar]
- Crowther, T.W.; Van den Hoogen, J.; Wan, J.; Mayes, M.A.; Keiser, A.; Mo, L.; Averill, C.; Maynard, D.S. The global soil community and its influence on biogeochemistry. Science 2019, 365, eaav0550. [Google Scholar] [CrossRef] [PubMed]
- Mahecha, M.D.; Reichstein, M.; Carvalhais, N.; Lasslop, G.; Lange, H.; Seneviratne, S.I.; Vargas, R.; Ammann, C.; Arain, M.A.; Cescatti, A.; et al. Global convergence in the temperature sensitivity of respiration at ecosystem level. Science 2010, 329, 838–840. [Google Scholar] [CrossRef] [PubMed]
- Meyer, N.; Welp, G.; Amelung, W. The temperature sensitivity (Q10) of soil respiration: Controlling factors and spatial prediction at regional scale based on environmental soil classes. Glob. Biogeochem. Cycles 2018, 32, 306–323. [Google Scholar] [CrossRef]
- Rolnick, D.; Donti, P.L.; Kaack, L.H.; Kochanski, K.; Lacoste, A.; Sankaran, K.; Ross, A.S.; Milojevic-Dupont, N.; Jaques, N.; Waldman-Brown, A.; et al. Tackling climate change with machine learning. ACM Comput. Surv. (CSUR) 2022, 55, 1–96. [Google Scholar] [CrossRef]
- Huntingford, C.; Jeffers, E.S.; Bonsall, M.B.; Christensen, H.M.; Lees, T.; Yang, H. Machine learning and artificial intelligence to aid climate change research and preparedness. Environ. Res. Lett. 2019, 14, 124007. [Google Scholar] [CrossRef]
- Wilhelm, R.C.; van Es, H.M.; Buckley, D.H.. Predicting measures of soil health using the microbiome and supervised machine learning. Soil Biol. Biochem. 2022, 164, 108472. [Google Scholar]
- Papoutsoglou, G.; Tarazona, S.; Lopes, M.B.; Klammsteiner, T.; Ibrahimi, E.; Eckenberger, J.; Novielli, P.; Tonda, A.; Simeon, A.; Shigdel, R.; et al. Machine learning approaches in microbiome research: Challenges and best practices. Front. Microbiol. 2023, 14, 1261889. [Google Scholar] [CrossRef]
- Di Gilio, A.; Catino, A.; Lombardi, A.; Palmisani, J.; Facchini, L.; Mongelli, T.; Varesano, N.; Bellotti, R.; Galetta, D.; de Gennaro, G.; et al. Breath analysis for early detection of malignant pleural mesothelioma: Volatile organic compounds (VOCs) determination and possible biochemical pathways. Cancers 2020, 12, 1262. [Google Scholar] [CrossRef] [PubMed]
- Cascio, D.; Taormina, V.; Cipolla, M.; Bruno, S.; Fauci, F.; Raso, G. A multi-process system for HEp-2 cells classification based on SVM. Pattern Recognit. Lett. 2016, 82, 56–63. [Google Scholar] [CrossRef]
- Biecek, P.; Burzykowski, T. Explanatory Model Analysis: Explore, Explain, and Examine Predictive Models; Chapman and Hall/CRC: Boca Raton, FL, USA, 2021. [Google Scholar]
- Ghalebikesabi, S.; Ter-Minassian, L.; DiazOrdaz, K.; Holmes, C.C. On locality of local explanation models. Adv. Neural Inf. Process. Syst. 2021, 34, 18395–18407. [Google Scholar]
- Li, Z. Extracting spatial effects from machine learning model using local interpretation method: An example of SHAP and XGBoost. Comput. Environ. Urban Syst. 2022, 96, 101845. [Google Scholar] [CrossRef]
- Sáez-Sandino, T.; García-Palacios, P.; Maestre, F.T.; Plaza, C.; Guirado, E.; Singh, B.K.; Wang, J.; Cano-Díaz, C.; Eisenhauer, N.; Gallardo, A.; et al. The soil microbiome governs the response of microbial respiration to warming across the globe. Nat. Clim. Change 2023, 13, 1382–1387. [Google Scholar] [CrossRef]
- Nottingham, A.T.; Whitaker, J.; Turner, B.L.; Salinas, N.; Zimmermann, M.; Malhi, Y.; Meir, P. Climate warming and soil carbon in tropical forests: Insights from an elevation gradient in the Peruvian Andes. Bioscience 2015, 65, 906–921. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.P.; Cherry, R.S.; Schlesinger, W.H. The Q10 relationship of microbial respiration in a temperate forest soil. Soil Biol. Biochem. 1996, 28, 1067–1072. [Google Scholar] [CrossRef]
- Steyerberg, E.W.; Steyerberg, E.W. Coding of categorical and continuous predictors. In Clinical Prediction Models: A Practical Approach to Development, Validation, and Updating; Springer: Cham, Switzerland, 2019; pp. 175–190. [Google Scholar]
- Ibrahimi, E.; Lopes, M.B.; Dhamo, X.; Simeon, A.; Shigdel, R.; Hron, K.; Stres, B.; D’Elia, D.; Berland, M.; Marcos-Zambrano, L.J. Overview of data preprocessing for machine learning applications in human microbiome research. Front. Microbiol. 2023, 14, 1250909. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, M.M.; Mahmud, M.P.; Saha, P.K.; Gupta, K.D.; Siddique, Z. Effect of data scaling methods on machine learning algorithms and model performance. Technologies 2021, 9, 52. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Guo, G.; Wang, H.; Bell, D.; Bi, Y.; Greer, K. KNN model-based approach in classification. In On The Move to Meaningful Internet Systems 2003: CoopIS, DOA, and ODBASE. OTM; Lecture Notes in Computer Science; Meersman, R., Tari, Z., Schmidt, D.C., Eds.; Springer: Berlin/Heidelberg, Germany, 2003; Volume 2888, pp. 986–996. [Google Scholar]
- LaValley, M.P. Logistic regression. Circulation 2008, 117, 2395–2399. [Google Scholar] [CrossRef] [PubMed]
- Swain, P.H.; Hauska, H. The decision tree classifier: Design and potential. IEEE Trans. Geosci. Electron. 1977, 15, 142–147. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely randomized trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Aydın, Y.; Işıkdağ, Ü.; Bekdaş, G.; Nigdeli, S.M.; Geem, Z.W. Use of machine learning techniques in soil classification. Sustainability 2023, 15, 2374. [Google Scholar] [CrossRef]
- Ferrer, L. Analysis and comparison of classification metrics. arXiv 2022, arXiv:2209.05355. [Google Scholar]
- Ozenne, B.; Subtil, F.; Maucort-Boulch, D. The precision–recall curve overcame the optimism of the receiver operating characteristic curve in rare diseases. J. Clin. Epidemiol. 2015, 68, 855–859. [Google Scholar] [CrossRef]
- Wen, P.; Xu, Q.; Yang, Z.; He, Y.; Huang, Q. Algorithm-Dependent Generalization of AUPRC Optimization: Theory and Algorithm. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 5062–5079. [Google Scholar] [CrossRef]
- Novielli, P.; Romano, D.; Magarelli, M.; Bitonto, P.D.; Diacono, D.; Chiatante, A.; Lopalco, G.; Sabella, D.; Venerito, V.; Filannino, P.; et al. Explainable Artificial Intelligence for Microbiome Data Analysis in Colorectal Cancer Biomarker Identification. Front. Microbiol. 2024, 15, 1348974. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.I. A unified approach to interpreting model predictions. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar] [CrossRef]
- Ali, R.S.; Poll, C.; Kandeler, E. Dynamics of soil respiration and microbial communities: Interactive controls of temperature and substrate quality. Soil Biol. Biochem. 2018, 127, 60–70. [Google Scholar] [CrossRef]
- Jansson, J.K.; Hofmockel, K.S. Soil microbiomes and climate change. Nat. Rev. Microbiol. 2020, 18, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wu, H.; Wang, Z.; Semenov, M.V.; Ye, J.; Yin, L.; Wang, X.; Kravchenko, I.; Semenov, V.; Kuzyakov, Y.; et al. Linkages between the temperature sensitivity of soil respiration and microbial life strategy are dependent on sampling season. Soil Biol. Biochem. 2022, 172, 108758. [Google Scholar] [CrossRef]
- Tong, D.; Li, Z.; Xiao, H.; Nie, X.; Liu, C.; Zhou, M. How do soil microbes exert impact on soil respiration and its temperature sensitivity? Environ. Microbiol. 2021, 23, 3048–3058. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, W.; Drury, C.; Tan, C.; Fox, C.; Yang, X. Use of indicators and pore volume-function characteristics to quantify soil physical quality. Geoderma 2009, 152, 252–263. [Google Scholar] [CrossRef]
- Popolizio, S.; Stellacci, A.M.; Giglio, L.; Barca, E.; Spagnuolo, M.; Castellini, M. Seasonal and soil use dependent variability of physical and hydraulic properties: An assessment under minimum tillage and no-tillage in a long-term experiment in southern Italy. Agronomy 2022, 12, 3142. [Google Scholar] [CrossRef]
- Reynolds, W.; Bowman, B.; Drury, C.; Tan, C.; Lu, X. Indicators of good soil physical quality: Density and storage parameters. Geoderma 2002, 110, 131–146. [Google Scholar] [CrossRef]

| Variable | Description | Units |
|---|---|---|
| Longitude_c | Longitude | Decimal degree |
| Forest | Presence of forest | unitless |
| MAP_wc2 | Mean annual precipitation | mm |
| MAT_wc2 | Mean annual temperature | °C |
| Plant_richness | Number of species | unitless |
| Plant_cover_v3 | Proportion of the plant that extends into the soil | unitless |
| Soil_pH | pH of soil | unitless |
| Soil_salinity | Electrical conductivity | dS m−1 |
| Fine_texture | Percentage of clay and silt in the soil | g 100 g−1 dry soil |
| Soil_P | Soil total phosphorus | mg P kg −1 dry soil |
| Soil_CN | Ratio between total organic carbon and total N | unitless |
| SOC | Soil organic carbon | g C kg−1 soil |
| MAOC/POC_Ratio | Ratio between proportion of mineral-associated organic C and proportion of particulate organic C | unitless |
| Aromatic | Percentage of aromatic | unitless |
| Alkanes | Percentage of alkane | unitless |
| Polysaccharide | Percentage of polysaccharide | unitless |
| Amide | Percentage of amide | unitless |
| Mean_Glucose | Glucose-induced soil respiration | g CO2-C g−1 soil h−1 |
| Richness_bacteria | Richness of bacteria | Number of zOTUs |
| Richness_fungi | Richness of fungi | Number of zOTUs |
| Richness_protist | Richness of protist | Number of zOTUs |
| Bacteria_Negative | Standardized proportion of bacteria taxa negatively associated with Q10 | unitless |
| Fungi_Negative | Standardized proportion of fungi taxa negatively associated with Q10 | unitless |
| Protists_Negative | Standardized proportion of protist taxa negatively associated with Q10 | unitless |
| Bacteria_Positive | Standardized proportion of bacteria taxa positively associated with Q10 | unitless |
| Fungi_Positive | Standardized proportion of fungi taxa positively associated with Q10 | unitless |
| Protists_Positive | Standardized proportion of protist taxa positively associated with Q10 | unitless |
| ML Classifier | Accuracy | AUROC | AUPRC |
|---|---|---|---|
| Extra Trees | 0.923 ± 0.009 | 0.964 ± 0.004 | 0.963 ± 0.006 |
| XGBoost | 0.899 ± 0.008 | 0.932 ± 0.006 | 0.899 ± 0.012 |
| Random Forest | 0.920 ± 0.007 | 0.951 ± 0.005 | 0.939 ± 0.013 |
| Decision Tree | 0.838 ± 0.022 | 0.862 ± 0.027 | 0.861 ± 0.031 |
| SVM | 0.901 ± 0.013 | 0.946 ± 0.010 | 0.930 ± 0.029 |
| KNN | 0.883 ± 0.016 | 0.947 ± 0.007 | 0.951 ± 0.006 |
| Logistic Regression | 0.901 ± 0.010 | 0.941 ± 0.010 | 0.923 ± 0.029 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Novielli, P.; Magarelli, M.; Romano, D.; de Trizio, L.; Di Bitonto, P.; Monaco, A.; Amoroso, N.; Stellacci, A.M.; Zoani, C.; Bellotti, R.; et al. Climate Change and Soil Health: Explainable Artificial Intelligence Reveals Microbiome Response to Warming. Mach. Learn. Knowl. Extr. 2024, 6, 1564-1578. https://doi.org/10.3390/make6030075
Novielli P, Magarelli M, Romano D, de Trizio L, Di Bitonto P, Monaco A, Amoroso N, Stellacci AM, Zoani C, Bellotti R, et al. Climate Change and Soil Health: Explainable Artificial Intelligence Reveals Microbiome Response to Warming. Machine Learning and Knowledge Extraction. 2024; 6(3):1564-1578. https://doi.org/10.3390/make6030075
Chicago/Turabian StyleNovielli, Pierfrancesco, Michele Magarelli, Donato Romano, Lorenzo de Trizio, Pierpaolo Di Bitonto, Alfonso Monaco, Nicola Amoroso, Anna Maria Stellacci, Claudia Zoani, Roberto Bellotti, and et al. 2024. "Climate Change and Soil Health: Explainable Artificial Intelligence Reveals Microbiome Response to Warming" Machine Learning and Knowledge Extraction 6, no. 3: 1564-1578. https://doi.org/10.3390/make6030075
APA StyleNovielli, P., Magarelli, M., Romano, D., de Trizio, L., Di Bitonto, P., Monaco, A., Amoroso, N., Stellacci, A. M., Zoani, C., Bellotti, R., & Tangaro, S. (2024). Climate Change and Soil Health: Explainable Artificial Intelligence Reveals Microbiome Response to Warming. Machine Learning and Knowledge Extraction, 6(3), 1564-1578. https://doi.org/10.3390/make6030075











