Whole-Transcriptome Sequencing: A Powerful Tool for Vascular Tissue Engineering and Endothelial Mechanobiology
Abstract
:1. Background
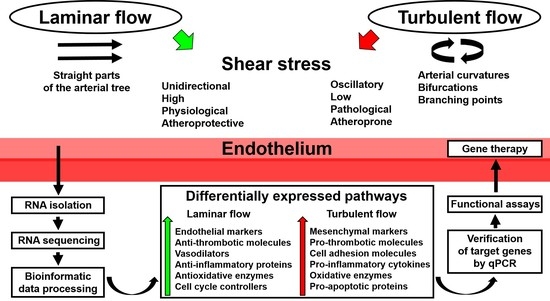
2. Application of Whole-Transcriptome Sequencing to Endothelial Mechanobiology Studies
3. Application of Whole-Transcriptome Sequencing to Vascular Tissue Engineering Studies
4. Conclusions and Future Directions
Acknowledgments
Conflicts of Interest
References
- GBD 2016 Causes of Death Collaborators. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1151–1210. [Google Scholar] [CrossRef] [PubMed]
- GBD 2016 DALYs and HALE Collaborators. Global, regional, and national disability-adjusted life-years (DALYs) for 333 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1260–1344. [Google Scholar] [CrossRef] [PubMed]
- Friedman, A.A.; Letai, A.; Fisher, D.E.; Flaherty, K.T. Precision medicine for cancer with next-generation functional diagnostics. Nat. Rev. Cancer 2015, 15, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Pauli, C.; Hopkins, B.D.; Prandi, D.; Shaw, R.; Fedrizzi, T.; Sboner, A.; Sailer, V.; Augello, M.; Puca, L.; Rosati, R.; et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017, 7, 462–477. [Google Scholar] [CrossRef] [PubMed]
- Dimitrakopoulos, L.; Prassas, I.; Diamandis, E.P.; Charames, G.S. Onco-proteogenomics: Multi-omics level data integration for accurate phenotype prediction. Crit. Rev. Clin. Lab. Sci. 2017, 54, 414–432. [Google Scholar] [CrossRef] [PubMed]
- Laurila, P.P.; Surakka, I.; Sarin, A.P.; Yetukuri, L.; Hyötyläinen, T.; Söderlund, S.; Naukkarinen, J.; Tang, J.; Kettunen, J.; Mirel, D.B.; et al. Genomic, transcriptomic, and lipidomic profiling highlights the role of inflammation in individuals with low high-density lipoprotein cholesterol. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Würtz, P.; Raiko, J.R.; Magnussen, C.G.; Soininen, P.; Kangas, A.J.; Tynkkynen, T.; Thomson, R.; Laatikainen, R.; Savolainen, M.J.; Laurikka, J.; et al. High-throughput quantification of circulating metabolites improves prediction of subclinical atherosclerosis. Eur. Heart J. 2012, 33, 2307–2316. [Google Scholar] [CrossRef] [PubMed]
- Döring, Y.; Noels, H.; Weber, C. The use of high-throughput technologies to investigate vascular inflammation and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Qiao, D.; Lange, C.; Beaty, T.H.; Crapo, J.D.; Barnes, K.C.; Bamshad, M.; Hersh, C.P.; Morrow, J.; Pinto-Plata, V.M.; Marchetti, N.; et al. Exome sequencing analysis in severe, early-onset chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2016, 193, 1353–1363. [Google Scholar] [CrossRef] [PubMed]
- Khurana, R.; Ranches, G.; Schafferer, S.; Lukasser, M.; Rudnicki, M.; Mayer, G.; Hüttenhofer, A. Identification of urinary exosomal noncoding RNAs as novel biomarkers in chronic kidney disease. RNA 2017, 23, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Kalim, S.; Rhee, E.P. An overview of renal metabolomics. Kidney Int. 2017, 91, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Vivante, A.; Hildebrandt, F. Exploring the genetic basis of early-onset chronic kidney disease. Nat. Rev. Nephrol. 2016, 12, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Ju, W.; Greene, C.S.; Eichinger, F.; Nair, V.; Hodgin, J.B.; Bitzer, M.; Lee, Y.S.; Zhu, Q.; Kehata, M.; Li, M.; et al. Defining cell-type specificity at the transcriptional level in human disease. Genome Res. 2013, 23, 1862–1873. [Google Scholar] [CrossRef] [PubMed]
- Guasch-Ferré, M.; Hruby, A.; Toledo, E.; Clish, C.B.; Martínez-González, M.A.; Salas-Salvadó, J.; Hu, F.B. Metabolomics in prediabetes and diabetes: A systematic review and meta-analysis. Diabetes Care 2016, 39, 833–846. [Google Scholar] [CrossRef] [PubMed]
- De Bruin, C.; Dauber, A. Insights from exome sequencing for endocrine disorders. Nat. Rev. Endocrinol. 2015, 11, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Wahl, S.; Drong, A.; Lehne, B.; Loh, M.; Scott, W.R.; Kunze, S.; Tsai, P.C.; Ried, J.S.; Zhang, W.; Yang, Y.; et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature 2017, 541, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Johar, A.S.; Anaya, J.M.; Andrews, D.; Patel, H.R.; Field, M.; Goodnow, C.; Arcos-Burgos, M. Candidate gene discovery in autoimmunity by using extreme phenotypes, next generation sequencing and whole exome capture. Autoimmun. Rev. 2015, 14, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Norman, P.J.; Hollenbach, J.A.; Nemat-Gorgani, N.; Marin, W.M.; Norberg, S.J.; Ashouri, E.; Jayaraman, J.; Wroblewski, E.E.; Trowsdale, J.; Rajalingam, R.; et al. Defining KIR and HLA Class I genotypes at highest resolution via high-throughput sequencing. Am. J. Hum. Genet. 2016, 99, 375–391. [Google Scholar] [CrossRef] [PubMed]
- Parikshak, N.N.; Gandal, M.J.; Geschwind, D.H. Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat. Rev. Genet. 2015, 16, 441–458. [Google Scholar] [CrossRef] [PubMed]
- Chen-Plotkin, A.S. Unbiased approaches to biomarker discovery in neurodegenerative diseases. Neuron 2014, 84, 594–607. [Google Scholar] [CrossRef] [PubMed]
- Jansen, I.E.; Ye, H.; Heetveld, S.; Lechler, M.C.; Michels, H.; Seinstra, R.I.; Lubbe, S.J.; Drouet, V.; Lesage, S.; Majounie, E.; et al. Discovery and functional prioritization of Parkinson’s disease candidate genes from large-scale whole exome sequencing. Genome Biol. 2017, 18, 22. [Google Scholar] [CrossRef] [PubMed]
- Berg, E.L. Systems biology in drug discovery and development. Drug Discov. Today 2014, 19, 113–125. [Google Scholar] [CrossRef] [PubMed]
- De Chassey, B.; Meyniel-Schicklin, L.; Vonderscher, J.; André, P.; Lotteau, V. Virus-host interactomics: New insights and opportunities for antiviral drug discovery. Genome Med. 2014, 6, 115. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, L.P.; Long, O.S.; Cobanoglu, M.C.; Benson, J.A.; Luke, C.J.; Miedel, M.T.; Hale, P.; Perlmutter, D.H.; Bahar, I.; Silverman, G.A.; et al. A genome-wide RNAi screen identifies potential drug targets in a C. elegans model of α1-antitrypsin deficiency. Hum. Mol. Genet. 2014, 23, 5123–5132. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.S.; Maron, B.A.; Loscalzo, J. Systems medicine: Evolution of systems biology from bench to bedside. Wiley Interdiscip. Rev. Syst. Biol. Med. 2015, 7, 141–161. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.H.; Shimoni, Y.; Yang, W.S.; Subramaniam, P.; Iyer, A.; Nicoletti, P.; Rodríguez Martínez, M.; López, G.; Mattioli, M.; Realubit, R.; et al. Elucidating compound mechanism of action by network perturbation analysis. Cell 2015, 162, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Backus, K.M.; Correia, B.E.; Lum, K.M.; Forli, S.; Horning, B.D.; González-Páez, G.E.; Chatterjee, S.; Lanning, B.R.; Teijaro, J.R.; Olson, A.J.; et al. Proteome-wide covalent ligand discovery in native biological systems. Nature 2016, 534, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Lapek, J.D., Jr.; Greninger, P.; Morris, R.; Amzallag, A.; Pruteanu-Malinici, I.; Benes, C.H.; Haas, W. Detection of dysregulated protein-association networks by high-throughput proteomics predicts cancer vulnerabilities. Nat. Biotechnol. 2017, 35, 983–989. [Google Scholar] [CrossRef] [PubMed]
- Bush, E.C.; Ray, F.; Alvarez, M.J.; Realubit, R.; Li, H.; Karan, C.; Califano, A.; Sims, P.A. PLATE-Seq for genome-wide regulatory network analysis of high-throughput screens. Nat. Commun. 2017, 8, 105. [Google Scholar] [CrossRef] [PubMed]
- Zampieri, M.; Sekar, K.; Zamboni, N.; Sauer, U. Frontiers of high-throughput metabolomics. Curr. Opin. Chem. Biol. 2017, 36, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, V.D.; Bruno, A.; Tomasello, G.; Damiano, G.; Lo Monte, A.I. Bioengineered vascular scaffolds: The state of the art. Int. J. Artif. Organs 2014, 37, 503–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocco, K.A.; Maxfield, M.W.; Best, C.A.; Dean, E.W.; Breuer, C.K. In vivo applications of electrospun tissue-engineered vascular grafts: A review. Tissue Eng. Part B Rev. 2014, 20, 628–640. [Google Scholar] [CrossRef] [PubMed]
- Chong, D.S.; Lindsey, B.; Dalby, M.J.; Gadegaard, N.; Seifalian, A.M.; Hamilton, G. Luminal surface engineering, ‘micro and nanopatterning’: Potential for self endothelialising vascular grafts? Eur. J. Vasc. Endovasc. Surg. 2014, 47, 566–576. [Google Scholar] [CrossRef] [PubMed]
- Niu, G.; Sapoznik, E.; Soker, S. Bioengineered blood vessels. Expert Opin. Biol. Ther. 2014, 14, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Feng, Y.; Guo, J.; Wang, H.; Li, Q.; Yang, J.; Hao, X.; Lv, J.; Ma, N.; Li, W. Surface modification and endothelialization of biomaterials as potential scaffolds for vascular tissue engineering applications. Chem. Soc. Rev. 2015, 44, 5680–5742. [Google Scholar] [CrossRef] [PubMed]
- Tara, S.; Rocco, K.A.; Hibino, N.; Sugiura, T.; Kurobe, H.; Breuer, C.K.; Shinoka, T. Vessel bioengineering. Circ. J. 2014, 78, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Sarwal, M.M. Transplant genetics and genomics. Nat. Rev. Genet. 2017, 18, 309–326. [Google Scholar] [CrossRef] [PubMed]
- Rehm, H.L. Evolving health care through personal genomics. Nat. Rev. Genet. 2017, 18, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.R.; Johnson, T.; Warren, L.; Hughes, A.R.; Chissoe, S.L.; Xu, C.F.; Waterworth, D.M. The genetics of drug efficacy: Opportunities and challenges. Nat. Rev. Genet. 2016, 17, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Willis, J.C.; Lord, G.M. Immune biomarkers: The promises and pitfalls of personalized medicine. Nat. Rev. Immunol. 2015, 15, 323–329. [Google Scholar] [CrossRef] [PubMed]
- DeVita, V.T., Jr.; Eggermont, A.M.; Hellman, S.; Kerr, D.J. Clinical cancer research: The past, present and the future. Nat. Rev. Clin. Oncol. 2014, 11, 663–669. [Google Scholar] [CrossRef] [PubMed]
- André, N.; Carré, M.; Pasquier, E. Metronomics: Towards personalized chemotherapy? Nat. Rev. Clin. Oncol. 2014, 11, 413–431. [Google Scholar] [CrossRef] [PubMed]
- Schilsky, R.L. Implementing personalized cancer care. Nat. Rev. Clin. Oncol. 2014, 11, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Völzke, H.; Schmidt, C.O.; Baumeister, S.E.; Ittermann, T.; Fung, G.; Krafczyk-Korth, J.; Hoffmann, W.; Schwab, M.; Meyer zu Schwabedissen, H.E.; Dörr, M.; et al. Personalized cardiovascular medicine: Concepts and methodological considerations. Nat. Rev. Cardiol. 2013, 10, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Shantsila, E.; Watson, T.; Lip, G.Y. Endothelial progenitor cells in cardiovascular disorders. J. Am. Coll. Cardiol. 2007, 49, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Liu, S.; Zhang, K.; Chen, J.; Huang, N. Endothelialization of implanted cardiovascular biomaterial surfaces: The development from in vitro to in vivo. J. Biomed. Mater. Res. A 2014, 102, 3754–3772. [Google Scholar] [CrossRef] [PubMed]
- Melchiorri, A.J.; Bracaglia, L.G.; Kimerer, L.K.; Hibino, N.; Fisher, J.P. In Vitro endothelialization of biodegradable vascular grafts via endothelial progenitor cell seeding and maturation in a tubular perfusion system bioreactor. Tissue Eng. Part C Methods 2016, 22, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, A.K.; Kumar, U.; Hardikar, A.A.; Poddar, P.; Nair, P.D.; Hardikar, A.A. Human blood vessel-derived endothelial progenitors for endothelialization of small diameter vascular prosthesis. PLoS ONE 2009, 4, e7718. [Google Scholar] [CrossRef] [PubMed]
- Ingram, D.A.; Mead, L.E.; Tanaka, H.; Meade, V.; Fenoglio, A.; Mortell, K.; Pollok, K.; Ferkowicz, M.J.; Gilley, D.; Yoder, M.C. Identification of a novel hierarchy of endothelial progenitor cells using human peripheral and umbilical cord blood. Blood 2004, 104, 2752–2760. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.M.; Zalos, G.; Halcox, J.P.; Schenke, W.H.; Waclawiw, M.A.; Quyyumi, A.A.; Finkel, T. Circulating endothelial progenitor cells, vascular function, and cardiovascular risk. N. Engl. J. Med. 2003, 348, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Takahashi, T.; Asahara, T.; Ohura, N.; Sokabe, T.; Kamiya, A.; Ando, J. Proliferation, differentiation, and tube formation by endothelial progenitor cells in response to shear stress. J. Appl. Physiol. 2003, 95, 2081–2088. [Google Scholar] [CrossRef] [PubMed]
- Yoder, M.C. Human endothelial progenitor cells. Cold. Spring. Harb. Perspect. Med. 2012, 2, a006692. [Google Scholar] [CrossRef] [PubMed]
- Asahara, T.; Murohara, T.; Sullivan, A.; Silver, M.; van der Zee, R.; Li, T.; Witzenbichler, B.; Schatteman, G.; Isner, J.M. Isolation of putative progenitor endothelial cells for angiogenesis. Science 1997, 275, 964–967. [Google Scholar] [CrossRef] [PubMed]
- Boyer, M.; Townsend, L.E.; Vogel, L.M.; Falk, J.; Reitz-Vick, D.; Trevor, K.T.; Villalba, M.; Bendick, P.J.; Glover, J.L. Isolation of endothelial cells and their progenitor cells from human peripheral blood. J. Vasc. Surg. 2000, 31, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Peters, E.B. Endothelial progenitor cells for the vascularization of engineered tissues. Tissue Eng. Part. B Rev. 2018, 24, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Shirota, T.; He, H.; Yasui, H.; Matsuda, T. Human endothelial progenitor cell-seeded hybrid graft: Proliferative and antithrombogenic potentials in vitro and fabrication processing. Tissue Eng. 2003, 9, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Riha, G.M.; Yan, S.; Li, M.; Chai, H.; Yang, H.; Yao, Q.; Chen, C. Shear stress induces endothelial differentiation from a murine embryonic mesenchymal progenitor cell line. Arterioscler. Thromb. Vasc. Biol. 2005, 25, 1817–1823. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.A.; Wallace, C.S.; Angelos, M.; Truskey, G.A. Characterization of umbilical cord blood-derived late outgrowth endothelial progenitor cells exposed to laminar shear stress. Tissue Eng. Part A 2009, 15, 3575–3587. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.B.; Khan, S.; Lapidos, K.A.; Ameer, G.A. Toward engineering a human neoendothelium with circulating progenitor cells. Stem Cells 2010, 28, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Angelos, M.G.; Brown, M.A.; Satterwhite, L.L.; Levering, V.W.; Shaked, N.T.; Truskey, G.A. Dynamic adhesion of umbilical cord blood endothelial progenitor cells under laminar shear stress. Biophys. J. 2010, 99, 3545–3554. [Google Scholar] [CrossRef] [PubMed]
- Achneck, H.E.; Jamiolkowski, R.M.; Jantzen, A.E.; Haseltine, J.M.; Lane, W.O.; Huang, J.K.; Galinat, L.J.; Serpe, M.J.; Lin, F.H.; Li, M.; et al. The biocompatibility of titanium cardiovascular devices seeded with autologous blood-derived endothelial progenitor cells: EPC-seeded antithrombotic Ti implants. Biomaterials 2011, 32, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Sekiguchi, H.; Ii, M.; Jujo, K.; Yokoyama, A.; Hagiwara, N.; Asahara, T. Improved culture-based isolation of differentiating endothelial progenitor cells from mouse bone marrow mononuclear cells. PLoS ONE 2011, 6, e28639. [Google Scholar] [CrossRef] [PubMed]
- Rössig, L.; Urbich, C.; Brühl, T.; Dernbach, E.; Heeschen, C.; Chavakis, E.; Sasaki, K.; Aicher, D.; Diehl, F.; Seeger, F.; et al. Histone deacetylase activity is essential for the expression of HoxA9 and for endothelial commitment of progenitor cells. J. Exp. Med. 2005, 201, 1825–1835. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Sokabe, T.; Watabe, T.; Miyazono, K.; Yamashita, J.K.; Obi, S.; Ohura, N.; Matsushita, A.; Kamiya, A.; Ando, J. Fluid shear stress induces differentiation of Flk-1-positive embryonic stem cells into vascular endothelial cells in vitro. Am. J. Physiol. Heart Circ. Physiol. 2005, 288, H1915–H1924. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Xiao, Q.; Margariti, A.; Zhang, Z.; Zampetaki, A.; Patel, S.; Capogrossi, M.C.; Hu, Y.; Xu, Q. HDAC3 is crucial in shear- and VEGF-induced stem cell differentiation toward endothelial cells. J. Cell. Biol. 2006, 174, 1059–1069. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Bai, L.; Yan, Z.Q.; Wang, Y.H.; Jiang, Z.L. Shear stress and vascular smooth muscle cells promote endothelial differentiation of endothelial progenitor cells via activation of Akt. Clin. Biomech. (Bristol, Avon) 2008, 23 (Suppl. 1), S118–S124. [Google Scholar] [CrossRef] [PubMed]
- Masumura, T.; Yamamoto, K.; Shimizu, N.; Obi, S.; Ando, J. Shear stress increases expression of the arterial endothelial marker ephrinB2 in murine ES cells via the VEGF-Notch signaling pathways. Arterioscler. Thromb. Vasc. Biol. 2009, 29, 2125–2131. [Google Scholar] [CrossRef] [PubMed]
- Boon, R.A.; Urbich, C.; Fischer, A.; Fontijn, R.D.; Seeger, F.H.; Koyanagi, M.; Horrevoets, A.J.; Dimmeler, S. Kruppel-like factor 2 improves neovascularization capacity of aged proangiogenic cells. Eur. Heart J. 2011, 32, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Zhang, X.; Guan, X.; Li, H.; Li, X.; Lu, H.; Cheng, M. Shear stress augments the endothelial cell differentiation marker expression in late EPCs by upregulating integrins. Biochem. Biophys. Res. Commun. 2012, 425, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.B.; Yan, Z.Q.; Yao, Q.P.; Shen, B.R.; Wang, J.Y.; Gao, L.Z.; Li, Y.Q.; Yuan, H.T.; Qi, Y.X.; Jiang, Z.L. Association of SIRT1 expression with shear stress induced endothelial progenitor cell differentiation. J. Cell. Biochem. 2012, 113, 3663–3671. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Yamamoto, K.; Ando, J.; Matsumoto, K.; Matsuda, T. Arterial shear stress augments the differentiation of endothelial progenitor cells adhered to VEGF-bound surfaces. Biochem. Biophys. Res. Commun. 2012, 423, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Obi, S.; Masuda, H.; Shizuno, T.; Sato, A.; Yamamoto, K.; Ando, J.; Abe, Y.; Asahara, T. Fluid shear stress induces differentiation of circulating phenotype endothelial progenitor cells. Am. J. Physiol. Cell. Physiol. 2012, 303, C595–C606. [Google Scholar] [CrossRef] [PubMed]
- Egorova, A.D.; DeRuiter, M.C.; de Boer, H.C.; van de Pas, S.; Gittenberger-de Groot, A.C.; van Zonneveld, A.J.; Poelmann, R.E.; Hierck, B.P. Endothelial colony-forming cells show a mature transcriptional response to shear stress. In Vitro Cell. Dev. Biol. Anim. 2012, 48, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.B.; Qu, M.J.; Wu, L.L.; Shen, Y.; Yan, Z.Q.; Zhang, P.; Qi, Y.X.; Jiang, Z.L. MicroRNA-34a targets Forkhead box j2 to modulate differentiation of endothelial progenitor cells in response to shear stress. J. Mol. Cell. Cardiol. 2014, 74, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.P.; Xiao, S.; Tang, Y.B.; Tan, Z.; Tang, H.; Ren, Z.; Zeng, H.; Yang, Z. Shear stress-mediated upregulation of GTP cyclohydrolase/tetrahydrobiopterin pathway ameliorates hypertension-related decline in reendothelialization capacity of endothelial progenitor cells. J. Hypertens. 2017, 35, 784–797. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Tao, J.; Wang, J.M.; Tu, C.; Xu, M.G.; Wang, Y.; Pan, S.R. Shear stress contributes to t-PA mRNA expression in human endothelial progenitor cells and nonthrombogenic potential of small diameter artificial vessels. Biochem. Biophys. Res. Commun. 2006, 342, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Yang, Z.; Wang, J.M.; Tu, C.; Pan, S.R. Effects of fluid shear stress on eNOS mRNA expression and NO production in human endothelial progenitor cells. Cardiology 2006, 106, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Sreerekha, P.R.; Krishnan, L.K. Cultivation of endothelial progenitor cells on fibrin matrix and layering on dacron/polytetrafluoroethylene vascular grafts. Artif. Organs 2006, 30, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Yang, Z.; Wang, J.M.; Wang, L.C.; Luo, C.F.; Tang, A.L.; Dong, Y.G.; Ma, H. Shear stress increases Cu/Zn SOD activity and mRNA expression in human endothelial progenitor cells. J. Hum. Hypertens. 2007, 21, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, J.M.; Wang, L.C.; Chen, L.; Tu, C.; Luo, C.F.; Tang, A.L.; Wang, S.M.; Tao, J. In vitro shear stress modulates antithrombogenic potentials of human endothelial progenitor cells. J. Thromb. Thrombolysis 2007, 23, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Lund, T.; Hermansen, S.E.; Andreasen, T.V.; Olsen, J.O.; Østerud, B.; Myrmel, T.; Ytrehus, K. Shear stress regulates inflammatory and thrombogenic gene transcripts in cultured human endothelial progenitor cells. Thromb. Haemost. 2010, 104, 582–591. [Google Scholar] [CrossRef] [PubMed]
- Gimbrone, M.A., Jr.; Garcia-Cardena, G. Endothelial Cell Dysfunction and the Pathobiology of Atherosclerosis. Circ. Res. 2016, 118, 620–636. [Google Scholar] [CrossRef] [PubMed]
- Jensen, H.A.; Mehta, J.L. Endothelial cell dysfunction as a novel therapeutic target in atherosclerosis. Expert Rev. Cardiovasc. Ther. 2016, 14, 1021–1033. [Google Scholar] [CrossRef] [PubMed]
- Cahill, P.A.; Redmond, E.M. Vascular endothelium - Gatekeeper of vessel health. Atherosclerosis 2016, 248, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Yurdagul, A., Jr.; Finney, A.C.; Woolard, M.D.; Orr, A.W. The arterial microenvironment: The where and why of atherosclerosis. Biochem. J. 2016, 473, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.A.; Shantsila, E.; Varma, C.; Lip, G.Y. Current understanding of atherogenesis. Am. J. Med. 2017, 130, 268–282. [Google Scholar] [CrossRef] [PubMed]
- Metaxa, E.; Meng, H.; Kaluvala, S.R.; Szymanski, M.P.; Paluch, R.A.; Kolega, J. Nitric oxide-dependent stimulation of endothelial cell proliferation by sustained high flow. Am. J. Physiol. Heart Circ. Physiol. 2008, 295, H736–H742. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Baker, B.M.; Chen, C.S.; Schwartz, M.A. Endothelial cell sensing of flow direction. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 2130–2136. [Google Scholar] [CrossRef] [PubMed]
- Baeyens, N.; Mulligan-Kehoe, M.J.; Corti, F.; Simon, D.D.; Ross, T.D.; Rhodes, J.M.; Wang, T.Z.; Mejean, C.O.; Simons, M.; Humphrey, J.; et al. Syndecan 4 is required for endothelial alignment in flow and atheroprotective signaling. Proc. Natl. Acad. Sci. USA 2014, 111, 17308–17313. [Google Scholar] [CrossRef] [PubMed]
- Dancu, M.B.; Berardi, D.E.; Vanden Heuvel, J.P.; Tarbell, J.M. Asynchronous shear stress and circumferential strain reduces endothelial NO synthase and cyclooxygenase-2 but induces endothelin-1 gene expression in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 2088–2094. [Google Scholar] [CrossRef] [PubMed]
- Amaya, R.; Pierides, A.; Tarbell, J.M. The Interaction between fluid wall shear stress and solid circumferential strain affects endothelial gene expression. PLoS ONE 2015, 10, e0129952. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Xia, W.H.; Zhang, Y.Y.; Xu, S.Y.; Liu, X.; Zhang, X.Y.; Yu, B.B.; Qiu, Y.X.; Tao, J. Shear stress-induced activation of Tie2-dependent signaling pathway enhances reendothelialization capacity of early endothelial progenitor cells. J. Mol. Cell. Cardiol. 2012, 52, 1155–1163. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Luo, J.Y.; Li, B.; Tian, X.Y.; Chen, L.; Huang, Y.; Liu, J.; Deng, D.; Lau, C.W.; Wan, S.; et al. Integrin-YAP/TAZ-JNK cascade mediates atheroprotective effect of unidirectional shear flow. Nature 2016, 540, 579–582. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Yeh, Y.T.; Nguyen, P.; Limqueco, E.; Lopez, J.; Thorossian, S.; Guan, K.L.; Li, Y.J.; Chien, S. Flow-dependent YAP/TAZ activities regulate endothelial phenotypes and atherosclerosis. Proc. Natl. Acad. Sci. USA 2016, 113, 11525–11530. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Koroleva, M.; Yin, M.; Jin, Z.G. Atheroprotective laminar flow inhibits Hippo pathway effector YAP in endothelial cells. Transl. Res. 2016, 176, 18–28.e2. [Google Scholar] [CrossRef] [PubMed]
- Nigro, P.; Abe, J.; Berk, B.C. Flow shear stress and atherosclerosis: A matter of site specificity. Antioxid. Redox Signal. 2011, 15, 1405–1414. [Google Scholar] [CrossRef] [PubMed]
- Hosoya, T.; Maruyama, A.; Kang, M.I.; Kawatani, Y.; Shibata, T.; Uchida, K.; Warabi, E.; Noguchi, N.; Itoh, K.; Yamamoto, M. Differential responses of the Nrf2-Keap1 system to laminar and oscillatory shear stresses in endothelial cells. J. Biol. Chem. 2005, 280, 27244–27250. [Google Scholar] [CrossRef] [PubMed]
- Saxena, A.; Carninci, P. Whole-transcriptome analysis: What are we still missing? Wiley Interdiscip. Rev. Syst. Biol. Med. 2011, 3, 527–543. [Google Scholar] [CrossRef] [PubMed]
- Costa, V.; Aprile, M.; Esposito, R.; Ciccodicola, A. RNA-Seq and human complex diseases: Recent accomplishments and future perspectives. Eur. J. Hum. Genet. 2013, 21, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Zhou, X.; Li, R.; Michal, J.J.; Zhang, S.; Dodson, M.V.; Zhang, Z.; Harland, R.M. Whole-transcriptome analysis with sequencing: Methods, challenges and potential solutions. Cell. Mol. Life Sci. 2015, 72, 3425–3439. [Google Scholar] [CrossRef] [PubMed]
- Fledderus, J.O.; van Thienen, J.V.; Boon, R.A.; Dekker, R.J.; Rohlena, J.; Volger, O.L.; Bijnens, A.P.; Daemen, M.J.; Kuiper, J.; van Berkel, T.J.; et al. Prolonged shear stress and KLF2 suppress constitutive proinflammatory transcription through inhibition of ATF2. Blood 2007, 109, 4249–4257. [Google Scholar] [CrossRef] [PubMed]
- Aoki, T.; Yamamoto, K.; Fukuda, M.; Shimogonya, Y.; Fukuda, S.; Narumiya, S. Sustained expression of MCP-1 by low wall shear stress loading concomitant with turbulent flow on endothelial cells of intracranial aneurysm. Acta Neuropathol. Commun. 2016, 4, 48. [Google Scholar] [CrossRef] [PubMed]
- Fledderus, J.O.; Boon, R.A.; Volger, O.L.; Hurttila, H.; Ylä-Herttuala, S.; Pannekoek, H.; Levonen, A.L.; Horrevoets, A.J. KLF2 primes the antioxidant transcription factor Nrf2 for activation in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 1339–1346. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Youn, S.W.; Cho, H.J.; Kwon, Y.W.; Lee, S.W.; Kim, S.J.; Park, Y.B.; Oh, B.H.; Kim, H.S. FOXO1 impairs whereas statin protects endothelial function in diabetes through reciprocal regulation of Kruppel-like factor 2. Cardiovasc. Res. 2013, 97, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Gracia-Sancho, J.; Villarreal, G., Jr.; Zhang, Y.; García-Cardeña, G. Activation of SIRT1 by resveratrol induces KLF2 expression conferring an endothelial vasoprotective phenotype. Cardiovasc. Res. 2010, 85, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Xu, S.; Liu, P.; Koroleva, M.; Zhang, S.; Si, S.; Jin, Z.G. Suberanilohydroxamic Acid as a Pharmacological Kruppel-Like Factor 2 Activator That Represses Vascular Inflammation and Atherosclerosis. J. Am. Heart Assoc. 2017, 6, e007134. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Liu, P.; Xu, S.; Koroleva, M.; Zhang, S.; Si, S.; Jin, Z.G. Tannic acid as a plant-derived polyphenol exerts vasoprotection via enhancing KLF2 expression in endothelial cells. Sci. Rep. 2017, 7, 6686. [Google Scholar] [CrossRef] [PubMed]
- Campolo, J.; Vozzi, F.; Penco, S.; Cozzi, L.; Caruso, R.; Domenici, C.; Ahluwalia, A.; Rial, M.; Marraccini, P.; Parodi, O. Vascular injury post stent implantation: Different gene expression modulation in human umbilical vein endothelial cells (HUVECs) model. PLoS ONE 2014, 9, e90213. [Google Scholar] [CrossRef] [PubMed]
- Doddaballapur, A.; Michalik, K.M.; Manavski, Y.; Lucas, T.; Houtkooper, R.H.; You, X.; Chen, W.; Zeiher, A.M.; Potente, M.; Dimmeler, S.; et al. Laminar shear stress inhibits endothelial cell metabolism via KLF2-mediated repression of PFKFB3. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Riising, E.M.; Comet, I.; Leblanc, B.; Wu, X.; Johansen, J.V.; Helin, K. Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol. Cell 2014, 55, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Maleszewska, M.; Vanchin, B.; Harmsen, M.C.; Krenning, G. The decrease in histone methyltransferase EZH2 in response to fluid shear stress alters endothelial gene expression and promotes quiescence. Angiogenesis 2016, 19, 9–24. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Martin, M.; Zhang, J.; Huang, H.Y.; Bai, L.; Zhang, J.; Kang, J.; He, M.; Li, J.; Maurya, M.R.; Gupta, S.; et al. Krüppel-Like Factor 4 Regulation of Cholesterol-25-Hydroxylase and Liver X Receptor Mitigates Atherosclerosis Susceptibility. Circulation 2017, 136, 1315–1330. [Google Scholar] [CrossRef] [PubMed]
- Hauser, S.; Jung, F.; Pietzsch, J. human endothelial cell models in biomaterial research. Trends Biotechnol. 2017, 35, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Qiao, C.; Meng, F.; Jang, I.; Jo, H.; Chen, Y.E.; Zhang, J. Deep transcriptomic profiling reveals the similarity between endothelial cells cultured under static and oscillatory shear stress conditions. Physiol. Genomics 2016, 48, 660–666. [Google Scholar] [CrossRef] [PubMed]
- McCormick, M.E.; Manduchi, E.; Witschey, W.R.T.; Gorman, R.C.; Gorman, J.H., 3rd; Jiang, Y.Z.; Stoeckert, C.J., Jr.; Barker, A.J.; Yoon, S.; Markl, M.; et al. Spatial phenotyping of the endocardial endothelium as a function of intracardiac hemodynamic shear stress. J. Biomech. 2017, 50, 11–19. [Google Scholar] [CrossRef] [PubMed]
- McCormick, M.E.; Manduchi, E.; Witschey, W.R.; Gorman, R.C.; Gorman, J.H., 3rd; Jiang, Y.Z.; Stoeckert, C.J., Jr.; Barker, A.J.; Markl, M.; Davies, P.F. Integrated Regional Cardiac Hemodynamic Imaging and RNA Sequencing Reveal Corresponding Heterogeneity of Ventricular Wall Shear Stress and Endocardial Transcriptome. J. Am. Heart Assoc. 2016, 5, e003170. [Google Scholar] [CrossRef] [PubMed]
- Reitsma, S.; Slaaf, D.W.; Vink, H.; van Zandvoort, M.A.; oude Egbrink, M.G. The endothelial glycocalyx: Composition, functions, and visualization. Pflugers. Arch. 2007, 454, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Ajami, N.E.; Gupta, S.; Maurya, M.R.; Nguyen, P.; Li, J.Y.; Shyy, J.Y.; Chen, Z.; Chien, S.; Subramaniam, S. Systems biology analysis of longitudinal functional response of endothelial cells to shear stress. Proc. Natl. Acad. Sci. USA 2017, 114, 10990–10995. [Google Scholar] [CrossRef] [PubMed]
- Öörni, K.; Rajamäki, K.; Nguyen, S.D.; Lähdesmäki, K.; Plihtari, R.; Lee-Rueckert, M.; Kovanen, P.T. Acidification of the intimal fluid: The perfect storm for atherogenesis. J. Lipid Res. 2015, 56, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.S.; Wang, K.C.; Quon, S.; Nguyen, P.; Chang, T.Y.; Chen, Z.; Li, Y.S.; Subramaniam, S.; Shyy, J.; Chien, S. LINC00341 exerts an anti-inflammatory effect on endothelial cells by repressing VCAM1. Physiol. Genomics 2017, 49, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Davies, P.F.; Remuzzi, A.; Gordon, E.J.; Dewey, C.F., Jr.; Gimbrone, M.A., Jr. Turbulent fluid shear stress induces vascular endothelial cell turnover in vitro. Proc. Natl. Acad. Sci. USA 1986, 83, 2114–2117. [Google Scholar] [CrossRef] [PubMed]
- White, C.R.; Haidekker, M.; Bao, X.; Frangos, J.A. Temporal gradients in shear, but not spatial gradients, stimulate endothelial cell proliferation. Circulation 2001, 103, 2508–2513. [Google Scholar] [CrossRef] [PubMed]
- Lerner-Marmarosh, N.; Yoshizumi, M.; Che, W.; Surapisitchat, J.; Kawakatsu, H.; Akaike, M.; Ding, B.; Huang, Q.; Yan, C.; Berk, B.C.; et al. Inhibition of tumor necrosis factor-α-induced SHP-2 phosphatase activity by shear stress: A mechanism to reduce endothelial inflammation. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 1775–1781. [Google Scholar] [CrossRef] [PubMed]
- Von Offenberg Sweeney, N.; Cummins, P.M.; Birney, Y.A.; Cullen, J.P.; Redmond, E.M.; Cahill, P.A. Cyclic strain-mediated regulation of endothelial matrix metalloproteinase-2 expression and activity. Cardiovasc. Res. 2004, 63, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Morrow, D.; Cullen, J.P.; Cahill, P.A.; Redmond, E.M. Cyclic strain regulates the Notch/CBF-1 signaling pathway in endothelial cells: Role in angiogenic activity. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, N.; Yamamoto, K.; Obi, S.; Kumagaya, S.; Masumura, T.; Shimano, Y.; Naruse, K.; Yamashita, J.K.; Igarashi, T.; Ando, J. Cyclic strain induces mouse embryonic stem cell differentiation into vascular smooth muscle cells by activating PDGF receptor β. J. Appl. Physiol. (1985) 2008, 104, 766–772. [Google Scholar] [CrossRef] [PubMed]
- Ando, J.; Yamamoto, K. Effects of shear stress and stretch on endothelial function. Antioxid. Redox Signal. 2011, 15, 1389–1403. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.B.; Zhang, J.; Xin, S.Y.; Liu, C.; Wang, C.Y.; Zhao, D.; Zhang, Z.R. Mechanosensitive properties in the endothelium and their roles in the regulation of endothelial function. J. Cardiovasc. Pharmacol. 2013, 61, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Zhang, K.; Wang, Y.; Cao, J.; Zhang, F.; Zhou, Q.; Dong, R. Identification of a microRNA signature in endothelial cells with mechanical stretch stimulation. Mol. Med. Rep. 2015, 12, 3525–3530. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; Zhang, K.; Wang, Y.L.; Zhang, F.; Cao, J.; Zheng, J.B.; Zhang, H.J. MiR-551b-5p Contributes to pathogenesis of Vein Graft failure via upregulating early growth response-1 expression. Chin. Med. J. (Engl.) 2017, 130, 1578–1585. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Guan, X.; Li, H.; Cui, X.; Zhang, X.; Li, X.; Jing, X.; Wu, H.; Avsar, E. Shear stress regulates late EPC differentiation via mechanosensitive molecule-mediated cytoskeletal rearrangement. PLoS ONE 2013, 8, e67675. [Google Scholar] [CrossRef] [PubMed]
- Kingshott, P.; Andersson, G.; McArthur, S.L.; Griesser, H.J. Surface modification and chemical surface analysis of biomaterials. Curr. Opin. Chem. Biol. 2011, 15, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Bendrea, A.D.; Cianga, L.; Cianga, I. Review paper: Progress in the field of conducting polymers for tissue engineering applications. J. Biomater. Appl. 2011, 26, 3–84. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.F.; Lai, E.S.; Ribeiro, A.J.; Pan, S.; Pruitt, B.L.; Fuller, G.G.; Cooke, J.P. Spatial patterning of endothelium modulates cell morphology, adhesiveness and transcriptional signature. Biomaterials 2013, 34, 2928–2937. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Wernig, M.; Meissner, A.; Foreman, R.; Brambrink, T.; Ku, M.; Hochedlinger, K.; Bernstein, B.E.; Jaenisch, R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 2007, 448, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Park, I.H.; Zhao, R.; West, J.A.; Yabuuchi, A.; Huo, H.; Ince, T.A.; Lerou, P.H.; Lensch, M.W.; Daley, G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 2008, 451, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Narazaki, G.; Uosaki, H.; Teranishi, M.; Okita, K.; Kim, B.; Matsuoka, S.; Yamanaka, S.; Yamashita, J.K. Directed and systematic differentiation of cardiovascular cells from mouse induced pluripotent stem cells. Circulation 2008, 118, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Mauritz, C.; Schwanke, K.; Reppel, M.; Neef, S.; Katsirntaki, K.; Maier, L.S.; Nguemo, F.; Menke, S.; Haustein, M.; Hescheler, J.; et al. Generation of functional murine cardiac myocytes from induced pluripotent stem cells. Circulation 2008, 118, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, K.H.; Joshi, P.A.; Lai, E.S.; Gujar, P.; Joubert, L.M.; Chen, B.; Huang, N.F. Bilayered vascular graft derived from human induced pluripotent stem cells with biomimetic structure and function. Regen. Med. 2015, 10, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, S.; One, J.; Siewert, J.; Teodosescu, S.; Zhao, L.; Dimitrievska, S.; Qian, H.; Huang, A.H.; Niklason, L. Tissue-engineered vascular grafts created from human induced pluripotent stem cells. Stem Cells Transl. Med. 2014, 3, 1535–1543. [Google Scholar] [CrossRef] [PubMed]
- Gui, L.; Dash, B.C.; Luo, J.; Qin, L.; Zhao, L.; Yamamoto, K.; Hashimoto, T.; Wu, H.; Dardik, A.; Tellides, G.; et al. Implantable tissue-engineered blood vessels from human induced pluripotent stem cells. Biomaterials 2016, 102, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Sivarapatna, A.; Ghaedi, M.; Le, A.V.; Mendez, J.J.; Qyang, Y.; Niklason, L.E. Arterial specification of endothelial cells derived from human induced pluripotent stem cells in a biomimetic flow bioreactor. Biomaterials 2015, 53, 621–633. [Google Scholar] [CrossRef] [PubMed]
- Sivarapatna, A.; Ghaedi, M.; Xiao, Y.; Han, E.; Aryal, B.; Zhou, J.; Fernandez-Hernando, C.; Qyang, Y.; Hirschi, K.K.; Niklason, L.E. Engineered Microvasculature in PDMS Networks Using Endothelial Cells Derived from HumanInduced Pluripotent Stem Cells. Cell. Transplant. 2017, 26, 1365–1379. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chu, L.F.; Hou, Z.; Schwartz, M.P.; Hacker, T.; Vickerman, V.; Swanson, S.; Leng, N.; Nguyen, B.K.; Elwell, A.; et al. Functional characterization of human pluripotent stem cell-derived arterial endothelial cells. Proc. Natl. Acad. Sci. USA 2017, 114, E6072–E6078. [Google Scholar] [CrossRef] [PubMed]
- Belair, D.G.; Whisler, J.A.; Valdez, J.; Velazquez, J.; Molenda, J.A.; Vickerman, V.; Lewis, R.; Daigh, C.; Hansen, T.D.; Mann, D.A.; et al. Human vascular tissue models formed from human induced pluripotent stem cell derived endothelial cells. Stem Cell. Rev. 2015, 11, 511–525. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, K.H.; Hong, G.; Lee, J.C.; Patel, J.; Edwards, B.; Zaitseva, T.S.; Paukshto, M.V.; Dai, H.; Cooke, J.P.; Woo, Y.J.; et al. Aligned-braided nanofibrillar scaffold with endothelial cells enhances arteriogenesis. ACS Nano 2015, 9, 6900–6908. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, Y.K.; Yin, R.T.; Shang, M.R.; Shirure, V.S.; Moya, M.L.; George, S.C. Human Induced Pluripotent Stem Cell-Derived Endothelial Cells for Three-Dimensional Microphysiological Systems. Tissue Eng. Part. C Methods 2017, 23, 474–484. [Google Scholar] [CrossRef] [PubMed]
- Ellis, B.W.; Acun, A.; Can, U.I.; Zorlutuna, P. Human iPSC-derived myocardium-on-chip with capillary-like flow for personalized medicine. Biomicrofluidics 2017, 11, 024105. [Google Scholar] [CrossRef] [PubMed]
- Youssef, A.A.; Ross, E.G.; Bolli, R.; Pepine, C.J.; Leeper, N.J.; Yang, P.C. The promise and challenge of induced pluripotent stem cells for cardiovascular applications. JACC Basic Transl. Sci. 2016, 1, 510–523. [Google Scholar] [CrossRef] [PubMed]
- Xu, S. Transcriptome profiling in systems vascular medicine. Front. Pharmacol. 2017, 8, 563. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kutikhin, A.G.; Sinitsky, M.Y.; Yuzhalin, A.E.; Velikanova, E.A. Whole-Transcriptome Sequencing: A Powerful Tool for Vascular Tissue Engineering and Endothelial Mechanobiology. High-Throughput 2018, 7, 5. https://doi.org/10.3390/ht7010005
Kutikhin AG, Sinitsky MY, Yuzhalin AE, Velikanova EA. Whole-Transcriptome Sequencing: A Powerful Tool for Vascular Tissue Engineering and Endothelial Mechanobiology. High-Throughput. 2018; 7(1):5. https://doi.org/10.3390/ht7010005
Chicago/Turabian StyleKutikhin, Anton G., Maxim Yu. Sinitsky, Arseniy E. Yuzhalin, and Elena A. Velikanova. 2018. "Whole-Transcriptome Sequencing: A Powerful Tool for Vascular Tissue Engineering and Endothelial Mechanobiology" High-Throughput 7, no. 1: 5. https://doi.org/10.3390/ht7010005
APA StyleKutikhin, A. G., Sinitsky, M. Y., Yuzhalin, A. E., & Velikanova, E. A. (2018). Whole-Transcriptome Sequencing: A Powerful Tool for Vascular Tissue Engineering and Endothelial Mechanobiology. High-Throughput, 7(1), 5. https://doi.org/10.3390/ht7010005