Conformational Design and Characterisation of a Truncated Diamine Oxidase from Arthrobacter globiformis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Homology Modelling of Diamine Oxidase
2.2. Analysing the Diamine Oxidase Domains
2.3. Molecular Dynamics (MD) Simulation and Ligand Docking Analysis
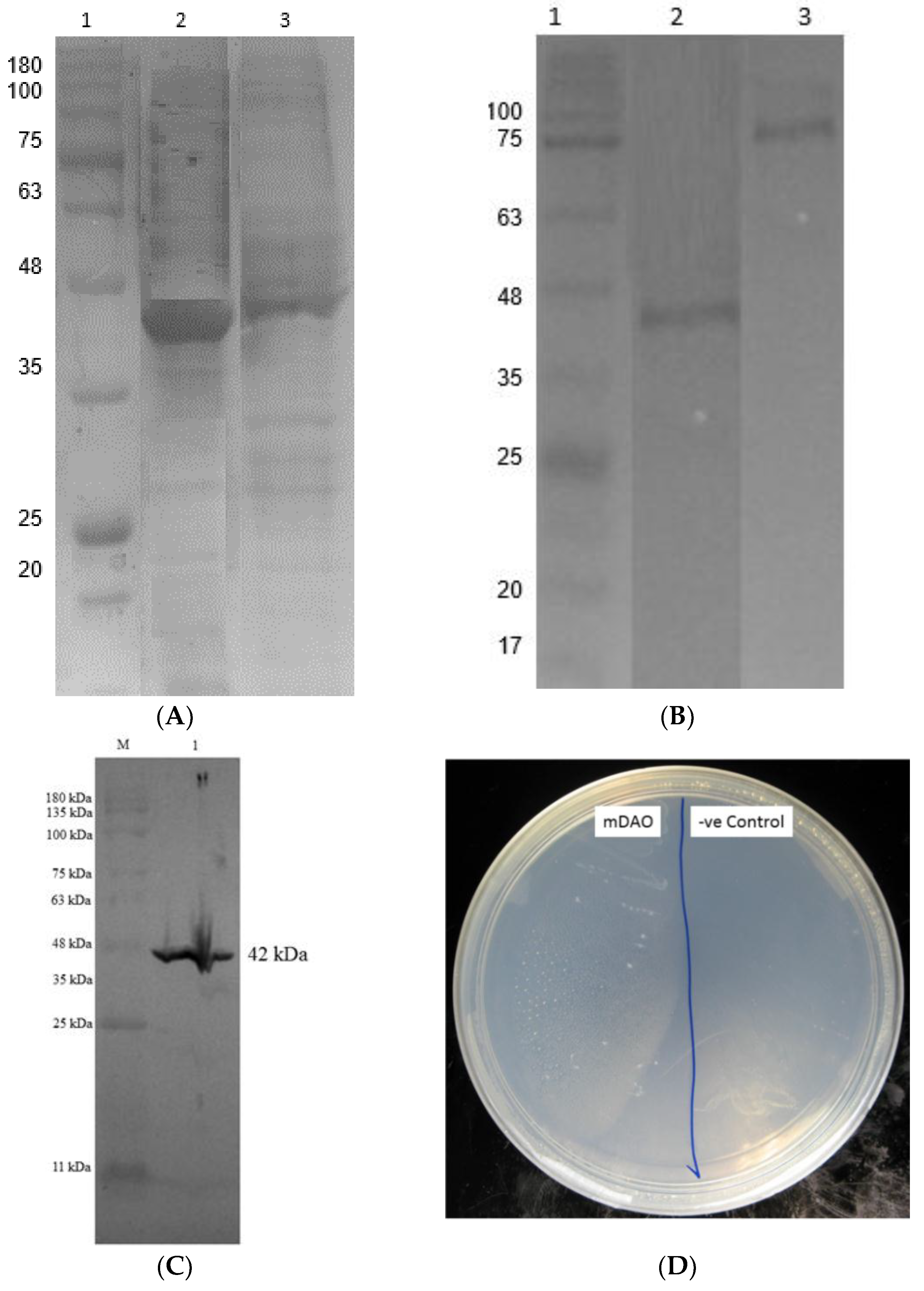
2.4. Cloning and Expression of Mini DAO
2.5. Purification of Mini DAO
2.6. Measurement of Mini DAO Activity and Stability
3. Results and Discussion
3.1. Molecular Analysis of Mini Diamine Oxidase from A. globiformis
3.2. Homology Modelling of Mini Diamine Oxidase
3.3. Molecular Dynamic Simulation and Protein Docking Analysis
3.4. Expressing of Recombinant Mini DAO in E. coli Cells
3.5. Purification of Fusion Mini DAO
3.6. Characterisation of Mini DAO
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DAO | Diamine Oxidase |
| DNA | Deoxyribonucleic acid |
| YASARA | Yet Another Scientific Artificial Reality Application |
| PDB | Protein Data Bank |
| 3D | Three-dimensional |
| MD | Molecular dynamics |
| NVT | (constant number of particles, volume, and temperature |
| SDF | Structure Data Format |
| OD | Optical Density |
| IPTG | isopropyl-β-d-1-thiogalactopyranoside |
| PAGE | polyacrylamide gel electrophoresis |
| SDS-PAGE | sodium dodecyl sulphate polyacrylamide gel electrophoresis |
| ODA | Dianisidine |
| NMR | Nuclear magnetic resonance |
| AGAO | Arhrobacter globiformis |
| hDAO | Human diamine oxidase |
| CDD | Conserved domain database |
| RMSD | Root-Mean-Square deviation |
References
- Souers, A.J.; Ellman, J.A. β-turn mimetic library synthesis: Scaffolds and application. Tetrahedron 2001, 57, 7431–7448. [Google Scholar] [CrossRef]
- Li, R.; Dowd, V.; Stewart, D.J.; Burton, S.J.; Lowe, C.R. Design, synthesis and application of a protein a mimetic. Nat. Biotechnol. 1998, 16, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Mason, J.M. Design and development of peptide mimetics as antagonists for therapeutic intervention. Future Med. Chem. 2010, 2, 1813–1822. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.B.; Raleigh, D.P.; Lombardi, A.; De Grado, W.F. De novo design of helical bundles as models for understanding protein folding and function. Acc. Chem. Res. 2000, 33, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Braisted, A.C.; Wells, J.A. Minimizing a binding domain from protein. Proc. Natl. Acad. Sci. USA 1996, 93, 5688–5692. [Google Scholar] [CrossRef] [PubMed]
- McGrath, A.P.; Hilmer, K.M.; Collyer, C.A.; Sherphard, E.M.; Elmore, B.O.; Brown, D.E.; Dooley, D.M.; Guss, J.M. Structure and inhibition of human diame oxidase. Biochemistry 2009, 48, 9810–9822. [Google Scholar] [CrossRef] [PubMed]
- Tanizawa, K.; Matsuzaki, R.; Shimizu, E.; Yorifuji, T.; Fukui, T. Cloning and sequencing of phenylethylamine oxidase from arthrobacter globiformis and implication of tyr-382 as the precursor to its covalently bound quinone cofactor. Biochem. Biophys. Res. Commun. 1994, 199, 1096–1102. [Google Scholar] [CrossRef] [PubMed]
- Best, C.H. The disappearance of histamine from autolysin lung tissue. J. Physiol. 1929, 67, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Stesina, L.N.; Akopyan, Z.I.; Gorkin, V.Z. Modification of catalytic properties of amine oxidases. FEBS Lett. 1971, 16, 349–351. [Google Scholar] [CrossRef] [Green Version]
- McGrath, A.P.; Hilmer, K.M.; Collyer, C.A.; Dooley, D.M.; Guss, J.M. A new crystal form of human diamine oxidase. Acta Crytallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 137–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casset, F.; Roux, F.; Mouchet, P.; Bes, C.; Chardes, T.; Granier, C.; Mani, J.C.; Pugnière, M.; Laune, D.; Pau, B.; et al. A peptide mimetic of an anti-cd4 monoclonal antibody by rational design. Biochem. Biophys. Res. Commun. 2003, 307, 198–205. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Buchanan, R.E.; Gibbons, N.R. Bergey’s Manual of Determinative Bacteriology, 8th ed.; Williams & Wilkins: Baltimore, MA, USA, 1974. [Google Scholar]
- Butt, A.M.; Batool, M.; Tong, Y. Homology modeling, comparative genomics and functional annotation of mycoplasma genitalium hypothetical protein mg_237. Bioinformation 2011, 7, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Urh, M.; Simpson, D.; Zhao, K. Affinity chromatography: General methods. Methods Enzymol. 2009, 463, 417–438. [Google Scholar] [CrossRef] [PubMed]
- Aarsen, P.N.; Kemp, A. Rapid spectrophotometric micro method for determination of histaminase activity. Nature 1964, 204, 1195. [Google Scholar] [CrossRef] [PubMed]
- Yeung, N.; Lin, Y.; Gao, Y.; Zhao, X.; Russell, B.S.; Lei, L.; Miner, K.D.; Robinson, H.; Lu, Y. Rational design of a structural and functional nitric oxide reductase. Nature 2009, 462, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Nabuurs, S.B.; Vriend, G. Homology modeling. Methods Biochem. Anal. 2003, 44, 509–523. [Google Scholar] [PubMed]
- Singh, R.; Chaturvedi, N.; Singh, V.K. In-silico study of herbal compounds (baicalin, curcumin and dronabinol) as novel mao inhibitors for parkinson’s disease treatment. Int. J. Life Sci. Pharma Res. 2012, 2, 81–98. [Google Scholar]
- Kumar, S. Comparative modelling and molecular docking of orphan human Cyp4v2 protein with fatty acid substrates: Insights into substrate specificity. Bioinformation 2011, 7, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Di Paolo, M.L.; Lunelli, M.; Fuxreiter, M.; Rigo, A.; Simon, I.; Scarpa, M. Active site residue involvement in monoamine or diamine oxidation catalysed by pea seedling amine oxidase. FEBS J. 2011, 278, 1232–1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollingsworth, S.A.; Karplus, P.A. A fresh look at the ramachandran plot and the occurrence of standard structures in proteins. Biomol. Concepts 2010, 1, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, B.; Prasad, S. Homology modeling and functional annotation of bubaline pregnancy associated glycoprotein 2. J. Anim. Sci. Biotechnol. 2012, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.Q.; O’Hern, C.S.; Regan, L. Revisiting the ramachandran plot from a new angle. Protein Sci. 2011, 20, 1166–1171. [Google Scholar] [CrossRef] [PubMed]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Smialowski, P.; Martin-Galiano, A.J.; Mikolajka, A.; Girschick, T.; Holak, T.A.; Frishman, D. Protein solubility: Sequence based prediction and experimental verification. Bioinformatics 2007, 23, 2536–2542. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, D.L.; Harrison, R.G. Predicting the solubility of recombinant protein in Escherichia coli. N. Y.: Bio/Technol. 1991, 9, 443–448. [Google Scholar] [CrossRef]
- Wang, X.; Snoeyink, J. Multiple structure alignment by optimal rmsd implies that the average structure is a consensus. Comput. Syst. Bioinform. Conf. 2006, 79–87. [Google Scholar]
- Sharma, J.; Ramanathan, K.; Sethumadhavan, R. Identification of potential inhibitors against acetylcholinesterase associated with alzheimer’s diseases: A molecular docking approach. J. Comput. Method Mol. Des. 2011, 1, 44–51. [Google Scholar]
- Nguyen, E.D.; Norn, C.; Frimurer, T.M.; Meiler, J. Assessment and challenges of ligand docking into comparative models of g-protein coupled receptors. PLoS ONE 2013, 8, E67302. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.H.; Spies, M.A.; Culpepper, M.B.; Murakawa, T.; Hirota, S.; Okajima, T.; Tanizawa, K.; Mure, M. Trapping of a dopaquinone intermediate in the tpq cofactor biogenesis in a copper-containing amine oxidase from Arthrobacter globiformis. J. Am. Chem. Soc. 2007, 129, 11524–11534. [Google Scholar] [CrossRef] [PubMed]
- Keerti Gupta, A.; Kumar, V.; Dubey, A.; Verma, A.K. Kinetic characterisation and effect of immobilized thermostable β-glucosidase in alginate gel beads on sugarcane juice. Biochemistry 2014, 2014, 1–8. [Google Scholar] [CrossRef]
- Elovaara, H.; Kidron, H.; Parkash, V.; Nymalm, Y.; Bligt, E.; Ollikka, P.; Smith, D.J.; Pihlavisto, M.; Salmi, M.; Jalkanen, S.; et al. Identification of two imidazole binding sites and key residues for substrate specificity in human primary amine oxidase aoc3. Biochemistry 2011, 50, 5507–5520. [Google Scholar] [CrossRef] [PubMed]
- Alexeeva, M.; Enright, A.; Dawson, M.J.; Mahmoudian, M.; Turner, N.J. Deracemization of alpha-methylbenzylamine using an enzyme obtained by in vitro evolution. Angew. Chem. Int. Ed. 2002, 41, 3177–3180. [Google Scholar] [CrossRef]
- Sekiguchi, Y.; Makita, H.; Yamamura, A.; Matsumoto, K. A thermostable histamine oxidase from Arthrobacter crytallopoietes KAIT-007. J. Biosci. Bioeng. 2004, 97, 104–110. [Google Scholar] [CrossRef]
- Choi, Y.H.; Matsuzaki, R.; Fukui, T.; Shimizu, E.; Yorifuji, T.; Sato, H.; Ozaki, Y.; Tanizawa, K. Copper/topa quinine-containing histamine oxidase from Arthobacter globiformis molecular cloning and sequencing, overproduction of precursor enzyme, and generation of topo-quinone cofactor. J. Biol. Chem. 1995, 270, 4712–4720. [Google Scholar] [CrossRef] [PubMed]
- Yamada, H.; Adachi, O.; Ogata, K. Amine oxidases of microorganisms: Purification and crystallization of amine oxidase of Aspergillus niger. Agric. Biol. Chem. 1965, 29, 649–654. [Google Scholar] [CrossRef]
- Figuera-Losada, M.; LoGrasso, P.V. Enzyme kinetics and interaction studies for human jnk1β1 and substrates activating transcription factor 2 (atf2) and c-jun n-terminal kinase (c-Jun). J. Biol. Chem. 2012, 287, 13291–13302. [Google Scholar] [CrossRef] [PubMed]











| Protein | Number of Residues in Favoured Region (98.0% Expected) | Number of Residues in Allowed Region (~2.0% Expected) | Number of Residues in Outlier Region |
|---|---|---|---|
| HDAO | 97.7 | 2.3 | 0 |
| AGAO | 96.4 | 3.5 | 0 |
| D4 | 96.7 | 3.3 | 0 |
| Mini DAO | 98.0 | 2.0 | 1 |
| Substrates | Natives | D04 | D03 | D02 | Mini DAO |
|---|---|---|---|---|---|
| Histamine | 8.92 | 7.45 | 4.11 | 3.66 | 8.13 |
| Putresine | 7.04 | 6.42 | 4.82 | 4.04 | 6.92 |
| Cadaverine | 6.04 | 5.56 | 6.39 | 3.92 | 5.99 |
| Spermidine | 5.92 | 5.46 | 4.52 | 3.64 | 5.99 |
| Spermine | 5.16 | 4.60 | 4.36 | 3.50 | 5.13 |
| Purification Step | Total Protein (mg) | Total Activity (U) | Specific Activity (U/mg) | Fold | Yield (%) |
|---|---|---|---|---|---|
| Crude | 19.613 | 438.929 | 22.380 | 1 | 100 |
| Ni2+-Sepharose | 11.872 | 498.265 | 41.970 | 1.875 | 88 |
| Properties | Native DAO | Mini DAO |
|---|---|---|
| Molecular weight (SDS-PAGE) | ~74 kDa | ~24 kDa (32% smaller) |
| Temperature | 35 °C | 40 °C |
| pH | 7 | 7 |
| Substrate specificity | Histamine (C5) | Histamine (C5), Spermidine (C7) |
| Half-life | 50 min at 37 °C, no activity at 50 °C | 80 min at 50 °C |
| Km Value | 0.274 mM | 1.3 mM |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Razali, N.N.; Hashim, N.H.; Leow, A.T.C.; Salleh, A.B. Conformational Design and Characterisation of a Truncated Diamine Oxidase from Arthrobacter globiformis. High-Throughput 2018, 7, 21. https://doi.org/10.3390/ht7030021
Razali NN, Hashim NH, Leow ATC, Salleh AB. Conformational Design and Characterisation of a Truncated Diamine Oxidase from Arthrobacter globiformis. High-Throughput. 2018; 7(3):21. https://doi.org/10.3390/ht7030021
Chicago/Turabian StyleRazali, Nur Nadia, Nur Hafizah Hashim, Adam Thean Chor Leow, and Abu Bakar Salleh. 2018. "Conformational Design and Characterisation of a Truncated Diamine Oxidase from Arthrobacter globiformis" High-Throughput 7, no. 3: 21. https://doi.org/10.3390/ht7030021
APA StyleRazali, N. N., Hashim, N. H., Leow, A. T. C., & Salleh, A. B. (2018). Conformational Design and Characterisation of a Truncated Diamine Oxidase from Arthrobacter globiformis. High-Throughput, 7(3), 21. https://doi.org/10.3390/ht7030021





