Soil Microbial Responses to Aflatoxin Exposure: Consequences for Biomass, Activity and Catabolic Functionality
Abstract
:1. Introduction
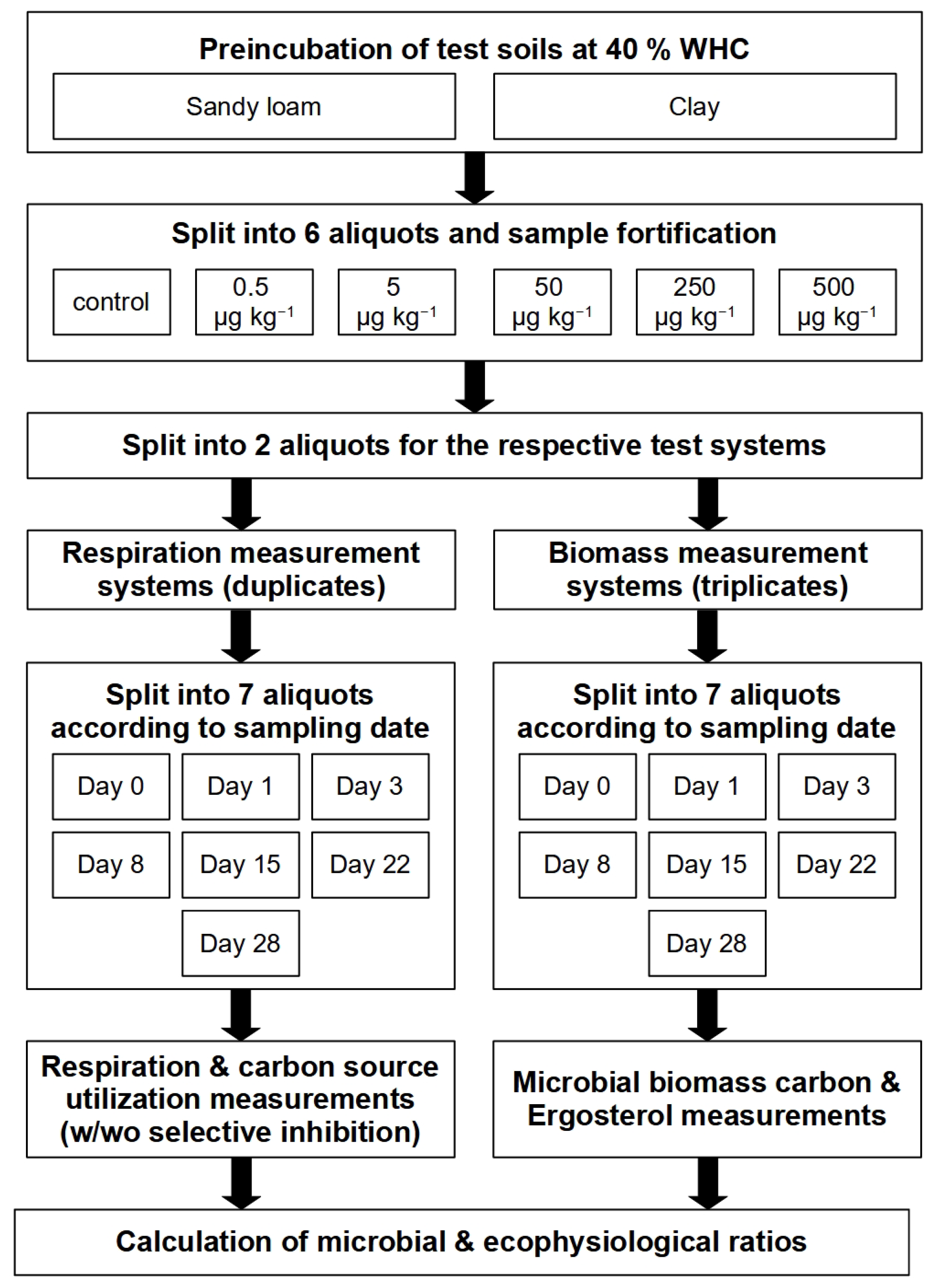
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Description of Test Soils
2.3. Aflatoxin B1 Concentrations, Soil Incubation and Sampling
2.4. Soil Microbial and Fungal Biomass
2.5. Determination of Microbial and Fungal Respiration and Catabolic Profiles
2.6. Soil Microbial Indices and Ecophysiological Ratios
2.7. Data Analyses
3. Results and Discussion
3.1. Biomass Responses to Aflatoxin Exposure
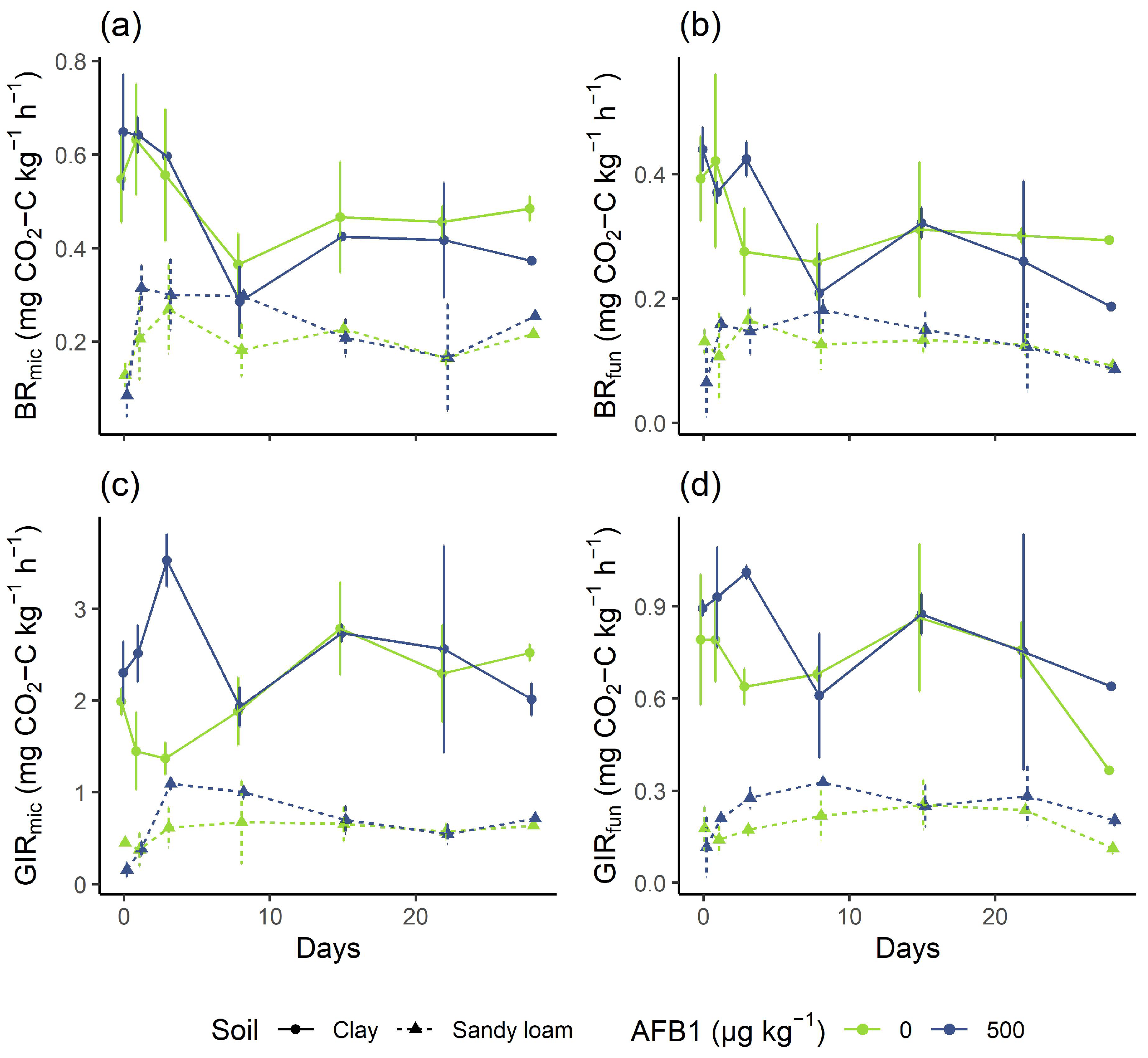
3.2. Response of Microbial Activity to Aflatoxin Exposure
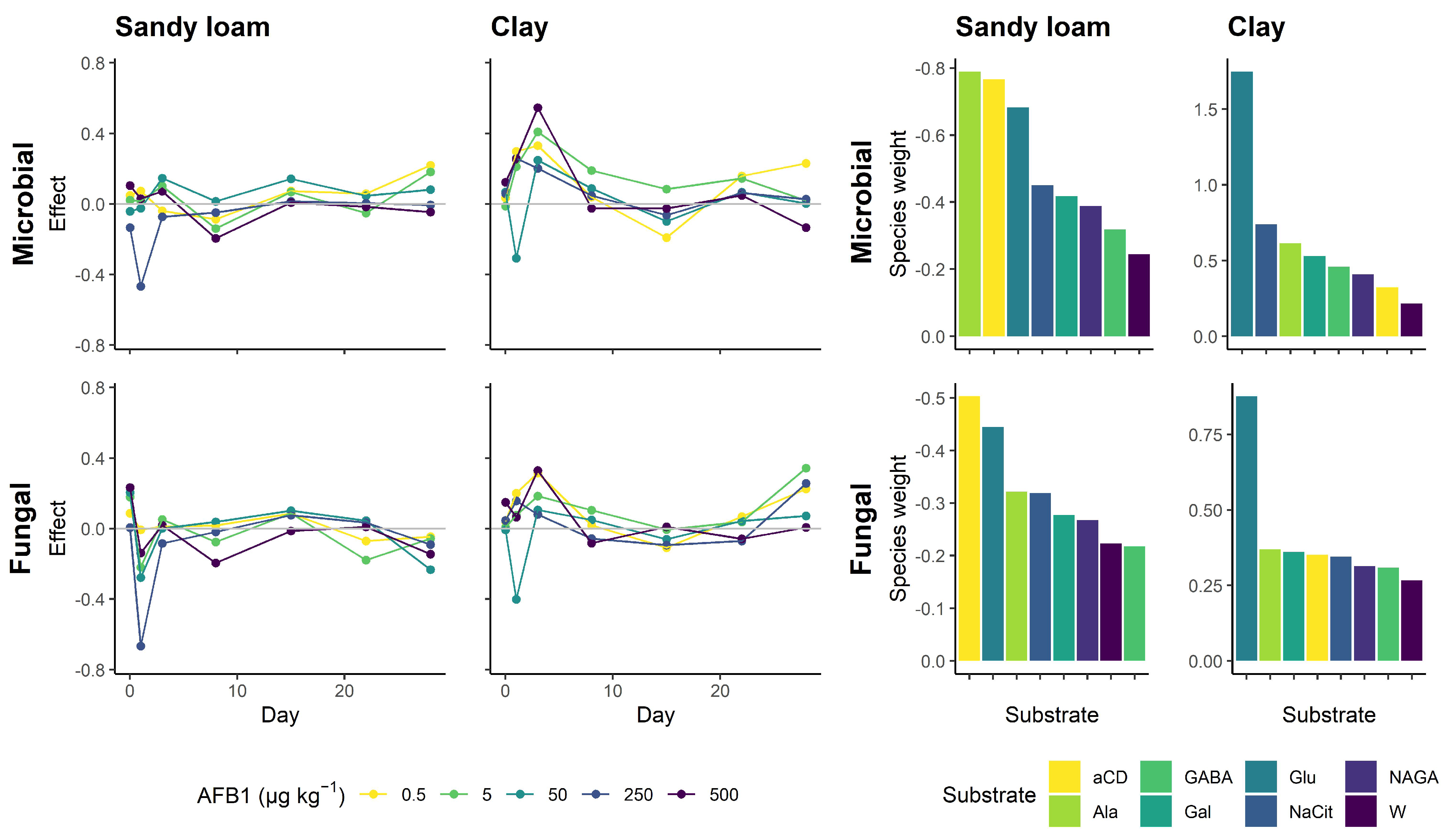
3.3. Carbon Source Utilization Patterns
3.4. Soil Microbial and Ecophysiological Ratios
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Calibration Figures


Appendix B. Microbial Responses to Aflatoxin B1




Appendix C. Principal Component Analysis
| Day | AFB1 (µg kg) | Sandy Loam | Clay | ||
|---|---|---|---|---|---|
| Microbial | Fungal | Microbial | Fungal | ||
| 0 | 0.5 | 0.048 | 0.086 | 0.034 | 0.04 |
| 5 | 0.022 | 0.179 | −0.013 | 0.007 | |
| 50 | −0.042 | 0.204 | 0.067 | −0.007 | |
| 250 | −0.135 | 0.006 | 0.059 | 0.046 | |
| 500 | 0.104 | 0.233 | 0.124 | 0.15 | |
| 1 | 0.5 | 0.073 | −0.007 | 0.298 | 0.201 |
| 5 | 0.016 | −0.219 | 0.211 | 0.075 | |
| 50 | −0.025 | −0.278 | −0.308 | −0.4 | |
| 250 | −0.468 | −0.667 | 0.258 | 0.157 | |
| 500 | 0.029 | −0.138 | 0.254 | 0.065 | |
| 3 | 0.5 | −0.039 | 0.012 | 0.331 | 0.316 |
| 5 | 0.101 | 0.051 | 0.409 | 0.185 | |
| 50 | 0.146 | 0.002 | 0.248 | 0.106 | |
| 250 | −0.073 | −0.083 | 0.201 | 0.081 | |
| 500 | 0.072 | 0.019 | 0.545 | 0.329 | |
| 8 | 0.5 | −0.087 | 0.018 | 0.039 | 0.019 |
| 5 | −0.139 | −0.076 | 0.19 | 0.104 | |
| 50 | 0.014 | 0.038 | 0.088 | 0.049 | |
| 250 | −0.048 | −0.019 | 0.047 | −0.057 | |
| 500 | −0.194 | −0.195 | −0.024 | −0.082 | |
| 15 | 0.5 | 0.073 | 0.085 | −0.191 | −0.108 |
| 5 | 0.068 | 0.087 | 0.084 | −0.006 | |
| 50 | 0.143 | 0.102 | −0.099 | −0.061 | |
| 250 | 0.014 | 0.076 | −0.063 | −0.094 | |
| 500 | 0.008 | −0.013 | −0.026 | 0.009 | |
| 22 | 0.5 | 0.058 | −0.071 | 0.158 | 0.068 |
| 5 | −0.052 | −0.179 | 0.145 | 0.038 | |
| 50 | 0.046 | 0.046 | 0.067 | 0.043 | |
| 250 | 0.004 | 0.033 | 0.062 | −0.071 | |
| 500 | −0.017 | 0.01 | 0.049 | −0.059 | |
| 28 | 0.5 | 0.22 | −0.046 | 0.23 | 0.224 |
| 5 | 0.181 | −0.057 | 0.019 | 0.343 | |
| 50 | 0.082 | −0.232 | 0.003 | 0.072 | |
| 250 | −0.005 | −0.091 | 0.027 | 0.256 | |
| 500 | −0.046 | −0.145 | −0.134 | 0.007 | |
| Substrate | Sandy Loam | Clay | ||
|---|---|---|---|---|
| Microbial | Fungal | Microbial | Fungal | |
| aCD | −0.77 | −0.5 | 0.32 | 0.35 |
| Ala | −0.79 | −0.32 | 0.62 | 0.37 |
| GABA | −0.32 | −0.22 | 0.46 | 0.31 |
| Gal | −0.42 | −0.28 | 0.53 | 0.36 |
| Glu | −0.68 | −0.44 | 1.75 | 0.88 |
| NaCit | −0.45 | −0.32 | 0.74 | 0.34 |
| NAGA | −0.39 | −0.27 | 0.41 | 0.31 |
| W | −0.24 | −0.22 | 0.22 | 0.27 |
Appendix D. Test Statistics of the Multiple Regression Models
| Response | Soil | Predictor | B | SE | t | p |
|---|---|---|---|---|---|---|
| Cmic | clay | Intercept | 151 | 7.48 | 20.173 | <0.001 |
| Time | 1.2 | 0.5 | 2.41 | 0.017 | ||
| AFB1 | 0.147 | 0.0327 | 4.516 | <0.001 | ||
| Time:AFB1 | −0.00642 | 0.00218 | −2.942 | 0.004 | ||
| sandy loam | Intercept | 76.7 | 3.74 | 20.513 | <0.001 | |
| Time | 0.703 | 0.231 | 3.038 | 0.003 | ||
| AFB1 | −0.0119 | 0.0163 | −0.731 | 0.466 | ||
| Time:AFB1 | −0.00082 | 0.00101 | −0.813 | 0.418 | ||
| ERG | clay | Intercept | 3.93 | 0.0303 | 129.616 | <0.001 |
| Time | −0.00894 | 0.00203 | −4.411 | <0.001 | ||
| AFB1 | −0.00035 | 0.000132 | −2.644 | 0.009 | ||
| Time:AFB1 | 1.051 | 0.296 | ||||
| sandy loam | Intercept | 0.788 | 0.0154 | 51.069 | <0.001 | |
| Time | −0.00732 | 0.00103 | −7.097 | <0.001 | ||
| AFB1 | −0.274 | 0.784 | ||||
| Time:AFB1 | −1.205 | 0.231 | ||||
| BRmic | clay | Intercept | 0.552 | 0.0265 | 20.866 | <0.001 |
| Time | −0.00301 | 0.00177 | −1.704 | 0.092 | ||
| AFB1 | 0.000115 | 0.104 | 0.918 | |||
| Time:AFB1 | −1.395 | 0.167 | ||||
| sandy loam | Intercept | 0.235 | 0.0189 | 12.428 | <0.001 | |
| Time | −0.00151 | 0.00128 | −1.18 | 0.241 | ||
| AFB1 | 1.178 | 0.242 | ||||
| Time:AFB1 | −2.66e−06 | 5.54e−06 | −0.48 | 0.632 | ||
| BRfun | clay | Intercept | 0.37 | 0.0161 | 22.894 | <0.001 |
| Time | −0.00294 | 0.00108 | −2.722 | 0.008 | ||
| AFB1 | 0.323 | 0.748 | ||||
| Time:AFB1 | −1.735 | 0.087 | ||||
| sandy loam | Intercept | 0.141 | 0.0106 | 13.315 | <0.001 | |
| Time | −0.00065 | 0.000714 | −0.91 | 0.366 | ||
| AFB1 | 0.654 | 0.515 | ||||
| Time:AFB1 | −0.757 | 0.451 | ||||
| GIRmic | clay | Intercept | 2.02 | 0.109 | 18.577 | <0.001 |
| Time | 0.0292 | 0.00726 | 4.026 | <0.001 | ||
| AFB1 | 0.00126 | 0.000474 | 2.666 | 0.009 | ||
| Time:AFB1 | −2.682 | 0.009 | ||||
| sandy loam | Intercept | 0.591 | 0.0533 | 11.09 | <0.001 | |
| Time | −0.00327 | 0.0036 | −0.909 | 0.366 | ||
| AFB1 | 0.000214 | 0.000233 | 0.922 | 0.359 | ||
| Time:AFB1 | 0.444 | 0.658 | ||||
| GIRfun | clay | Intercept | 0.765 | 0.0349 | 21.905 | <0.001 |
| Time | −0.000799 | 0.00234 | −0.342 | 0.733 | ||
| AFB1 | 0.000271 | 0.000152 | 1.776 | 0.08 | ||
| Time:AFB1 | −1.214 | 0.228 | ||||
| sandy loam | Intercept | 0.215 | 0.0166 | 12.897 | <0.001 | |
| Time | −0.00152 | 0.00112 | −1.357 | 0.179 | ||
| AFB1 | 1.199 | 0.234 | ||||
| Time:AFB1 | 0.468 | 0.641 | ||||
| ERG:Cmic | clay | Intercept | 38.4 | 1.98 | 19.414 | <0.001 |
| Time | 0.42 | 0.132 | 3.182 | 0.002 | ||
| AFB1 | 0.0428 | 0.00862 | 4.964 | <0.001 | ||
| Time:AFB1 | −0.00178 | 0.000576 | −3.097 | 0.002 | ||
| sandy loam | Intercept | 0.0176 | 0.0011 | 15.998 | <0.001 | |
| Time | 0.817 | 0.416 | ||||
| AFB1 | 0.342 | 0.733 | ||||
| Time:AFB1 | 1.684 | 0.095 | ||||
| BRfun:BRmic | clay | Intercept | 0.671 | 0.0166 | 40.54 | <0.001 |
| Time | −0.00138 | 0.00111 | −1.251 | 0.215 | ||
| AFB1 | 0.773 | 0.442 | ||||
| Time:AFB1 | −1.335 | 0.186 | ||||
| sandy loam | Intercept | 0.623 | 0.0329 | 18.919 | <0.001 | |
| Time | 0.00032 | 0.00223 | 0.144 | 0.886 | ||
| AFB1 | −0.000109 | 0.000144 | −0.755 | 0.452 | ||
| Time:AFB1 | −0.163 | 0.871 | ||||
| GIRfun:GIRmic | clay | Intercept | 0.397 | 0.0159 | 24.961 | <0.001 |
| Time | −0.00513 | 0.00106 | −4.825 | <0.001 | ||
| AFB1 | −0.000111 | 6.94e−05 | −1.595 | 0.115 | ||
| Time:AFB1 | 1.685 | 0.096 | ||||
| sandy loam | Intercept | 0.432 | 0.0362 | 11.932 | <0.001 | |
| Time | −0.00255 | 0.00244 | −1.041 | 0.301 | ||
| AFB1 | 3.5e−05 | 0.000158 | 0.222 | 0.825 | ||
| Time:AFB1 | −0.184 | 0.855 | ||||
| BRmic:GIRmic | clay | Intercept | 0.3 | 0.019 | 15.814 | <0.001 |
| Time | −0.00524 | 0.00127 | −4.125 | <0.001 | ||
| AFB1 | −0.000183 | −2.203 | 0.03 | |||
| Time:AFB1 | 1.112 | 0.27 | ||||
| sandy loam | Intercept | 0.483 | 0.0466 | 10.361 | <0.001 | |
| Time | −0.00174 | 0.00315 | −0.554 | 0.581 | ||
| AFB1 | 0.000203 | −0.068 | 0.946 | |||
| Time:AFB1 | −0.896 | 0.373 | ||||
| BRfun:GIRfun | clay | Intercept | 0.48 | 0.0223 | 21.585 | <0.001 |
| Time | −0.00181 | 0.00149 | −1.214 | 0.228 | ||
| AFB1 | −0.000115 | −1.187 | 0.239 | |||
| Time:AFB1 | −1.212 | 0.229 | ||||
| sandy loam | Intercept | 0.687 | 0.039 | 17.609 | <0.001 | |
| Time | 0.0027 | 0.00264 | 1.023 | 0.31 | ||
| AFB1 | −0.000132 | 0.00017 | −0.777 | 0.439 | ||
| Time:AFB1 | −1.88 | 0.064 | ||||
| qCO2,mic | clay | Intercept | −5.55 | 0.125 | −44.546 | <0.001 |
| Time | −0.0164 | 0.00833 | −1.972 | 0.056 | ||
| AFB1 | −0.000966 | 0.000544 | −1.777 | 0.084 | ||
| Time:AFB1 | 0.559 | 0.58 | ||||
| sandy loam | Intercept | 0.00358 | 0.00033 | 10.85 | <0.001 | |
| Time | −2.987 | 0.005 | ||||
| AFB1 | 2.089 | 0.045 | ||||
| Time:AFB1 | −0.957 | 0.346 | ||||
| qCO2,fun | clay | Intercept | 0.0941 | 0.00426 | 22.092 | <0.001 |
| Time | −0.000556 | 0.000285 | −1.955 | 0.058 | ||
| AFB1 | 0.781 | 0.44 | ||||
| Time:AFB1 | −1.984 | 0.055 | ||||
| sandy loam | Intercept | 0.109 | 0.00814 | 13.389 | <0.001 | |
| Time | −0.00122 | 0.000544 | −2.244 | 0.031 | ||
| AFB1 | 0.429 | 0.67 | ||||
| Time:AFB1 | −0.923 | 0.362 |
Appendix E. Test Statistics of the Monte Carlo Permutation Test
| Fraction | Soil | Day | DF | F | p |
|---|---|---|---|---|---|
| Microbial | sandy loam | 0 | 1 | 0.26 | 0.838 |
| 1 | 1 | 1.15 | 0.382 | ||
| 3 | 1 | 2.36 | 0.105 | ||
| 8 | 1 | 1.48 | 0.209 | ||
| 15 | 1 | 0.13 | 0.838 | ||
| 22 | 1 | 0.24 | 0.846 | ||
| 28 | 1 | 2.7 | 0.117 | ||
| clay | 0 | 1 | 1.51 | 0.21 | |
| 1 | 1 | 0.32 | 0.689 | ||
| 3 | 1 | 5.36 | 0.03 | ||
| 8 | 1 | 0.89 | 0.397 | ||
| 15 | 1 | 0.03 | 0.997 | ||
| 22 | 1 | 0.07 | 0.932 | ||
| 28 | 1 | 3.71 | 0.064 | ||
| Fungal | sandy loam | 0 | 1 | 1.05 | 0.317 |
| 1 | 1 | 2.4 | 0.109 | ||
| 3 | 1 | 2.03 | 0.119 | ||
| 8 | 1 | 1.08 | 0.324 | ||
| 15 | 1 | 0.04 | 0.958 | ||
| 22 | 1 | 1.13 | 0.314 | ||
| 28 | 1 | 5.43 | 0.013 | ||
| clay | 0 | 1 | 0.65 | 0.523 | |
| 1 | 1 | 0.11 | 0.911 | ||
| 3 | 1 | 1.08 | 0.296 | ||
| 8 | 1 | 1.06 | 0.344 | ||
| 15 | 1 | 0.12 | 0.935 | ||
| 22 | 1 | 0.56 | 0.479 | ||
| 28 | 1 | 2.43 | 0.118 |
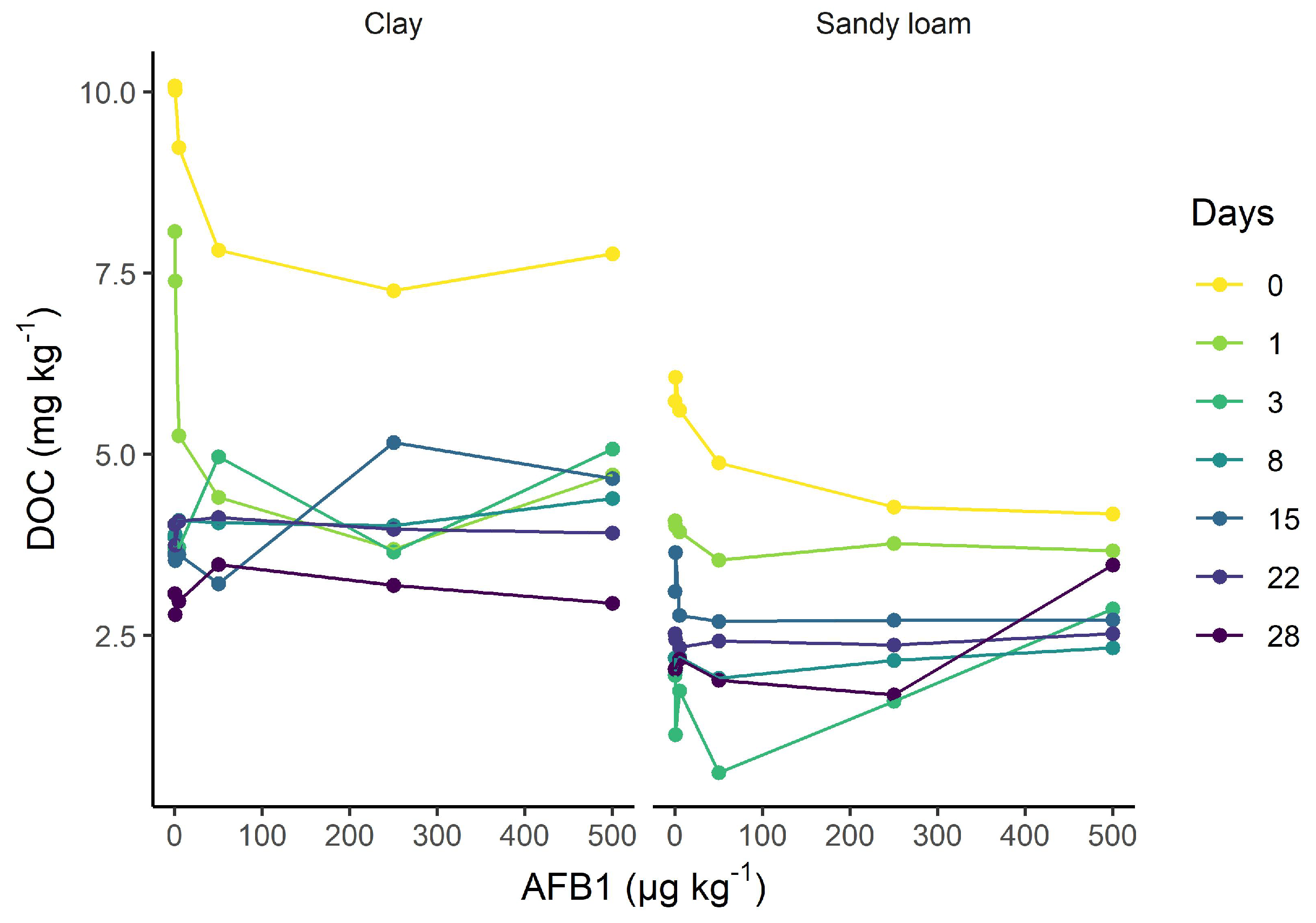
Appendix F. Dissolved Organic Matter in Nonfumigated Samples
References
- Accinelli, C.; Abbas, H.; Zablotowicz, R.; Wilkinson, J. Aspergillus flavus aflatoxin occurrence and expression of aflatoxin biosynthesis genes in soil. Can. J. Microbiol. 2008, 54, 371–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horn, B.W. Ecology and population biology of aflatoxigenic fungi in soil. J. Toxicol. Toxin Rev. 2003, 22, 351–379. [Google Scholar] [CrossRef]
- Jaime-Garcia, R.; Cotty, P.J. Aspergillus flavus in soils and corncobs in south Texas: Implications for management of aflatoxins in corn-cotton rotations. Plant Dis. 2004, 88, 1366–1371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orum, T.V.; Bigelow, D.M.; Nelson, M.R.; Howell, D.R.; Cotty, P.J. Spatial and temporal patterns of Aspergillus flavus strain composition and propagule density in Yuma County, Arizona, soils. Plant Dis. 1997, 81, 911–916. [Google Scholar] [CrossRef] [Green Version]
- Abbas, H.; Wilkinson, J.; Zablotowicz, R.; Accinelli, C.; Abel, C.; Bruns, H.; Weaver, M. Ecology of Aspergillus flavus, regulation of aflatoxin production, and management strategies to reduce aflatoxin contamination of corn. Toxin Rev. 2009, 28, 142–153. [Google Scholar] [CrossRef]
- Elmholt, S. Mycotoxins in the soil environment. In Secondary Metabolites in Soil Ecology; Springer: Berlin/Heidelberg, Germany, 2008; pp. 167–203. [Google Scholar]
- Stoloff, L.; Dantzman, J.; Armbrecht, B. Aflatoxin excretion in wethers. Food Cosmet. Toxicol. 1971, 9, 839–846. [Google Scholar] [CrossRef]
- Fernández, A.; Belío, R.; Ramos, J.J.; Sanz, M.C.; Sáez, T. Aflatoxins and their metabolites in the tissues, faeces and urine from lambs feeding on an aflatoxin-contaminated diet. J. Sci. Food Agric. 1997, 74, 161–168. [Google Scholar] [CrossRef]
- Lüthy, J.; Zweifel, U.; Schlatter, C. Metabolism and tissue distribution of [14C] aflatoxin B1 in pigs. Food Cosmet. Toxicol. 1980, 18, 253–256. [Google Scholar] [CrossRef]
- Stubblefield, R.; Pier, A.; Richard, J.; Shotwell, O. Fate of aflatoxins in tissues, fluids, and excrements from cows dosed orally with aflatoxin B1. Am. J. Vet. Res. 1983, 44, 1750–1752. [Google Scholar]
- Bardon, C.; Piola, F.; Bellvert, F.; Haichar, F.e.Z.; Comte, G.; Meiffren, G.; Pommier, T.; Puijalon, S.; Tsafack, N.; Poly, F. Evidence for biological denitrification inhibition (BDI) by plant secondary metabolites. New Phytol. 2014, 204, 620–630. [Google Scholar] [CrossRef]
- Zhang, Z.; Qiao, M.; Li, D.; Zhao, C.; Li, Y.; Yin, H.; Liu, Q. Effects of two root-secreted phenolic compounds from a subalpine coniferous species on soil enzyme activity and microbial biomass. Chem. Ecol. 2015, 31, 636–649. [Google Scholar] [CrossRef]
- Izhaki, I. Emodin—A secondary metabolite with multiple ecological functions in higher plants. New Phytol. 2002, 155, 205–217. [Google Scholar] [CrossRef] [Green Version]
- Fouché, T.; Claassens, S.; Maboeta, M. Aflatoxins in the soil ecosystem: An overview of its occurrence, fate, effects and future perspectives. Mycotoxin Res. 2020, 36, 303–309. [Google Scholar] [CrossRef]
- Wicklow, D.; Shotwell, O. Intrafungal distribution of aflatoxins among conidia and sclerotia of Aspergillus flavus and Aspergillus parasiticus. Can. J. Microbiol. 1983, 29, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Weckbach, L.; Marth, E. Aflatoxin production by Aspergillus parasiticus in a competitive environment. Mycopathologia 1977, 62, 39–45. [Google Scholar] [CrossRef]
- Cuero, R.; Smith, J.; Lacey, J. Stimulation by Hyphopichia burtonii and Bacillus amyloliquefaciens of aflatoxin production by Aspergillus flavus in irradiated maize and rice grains. Appl. Environ. Microbiol. 1987, 53, 1142–1146. [Google Scholar] [CrossRef] [Green Version]
- Wicklow, D.; Hesseltine, C.; Shotwell, O.; Adams, G. Interference competition and aflatoxin levels in corn. Phytopathology 1980, 78, 68–74. [Google Scholar] [CrossRef] [Green Version]
- Burmeister, H.; Hesseltine, C. Survey of the sensitivity of microorganisms to aflatoxin. Appl. Microbiol. 1966, 14, 403–404. [Google Scholar] [CrossRef]
- Arai, T.; Ito, T.; Koyama, Y. Antimicrobial activity of aflatoxins. J. Bacteriol. 1967, 93, 59–64. [Google Scholar] [CrossRef] [Green Version]
- Angle, J.; Wagner, G. Aflatoxin B1 effects on soil microorganisms. Soil Biol. Biochem. 1981, 13, 381–384. [Google Scholar] [CrossRef]
- Drott, M.T.; Debenport, T.; Higgins, S.A.; Buckley, D.H.; Milgroom, M.G. Fitness cost of aflatoxin production in Aspergillus flavus when competing with soil microbes could maintain balancing selection. MBio 2019, 10, e02782-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pham, V.H.; Kim, J. Cultivation of unculturable soil bacteria. Trends Biotechnol. 2012, 30, 475–484. [Google Scholar] [CrossRef] [PubMed]
- Joergensen, R.G.; Emmerling, C. Methods for evaluating human impact on soil microorganisms based on their activity, biomass, and diversity in agricultural soils. J. Plant Nutr. Soil Sci. 2006, 169, 295–309. [Google Scholar] [CrossRef]
- Blagodatskaya, E.; Kuzyakov, Y. Active microorganisms in soil: Critical review of estimation criteria and approaches. Soil Biol. Biochem. 2013, 67, 192–211. [Google Scholar] [CrossRef]
- Anderson, J.P.; Domsch, K.H. A physiological method for the quantitative measurement of microbial biomass in soils. Soil Biol. Biochem. 1978, 10, 215–221. [Google Scholar] [CrossRef]
- Sassi, M.B.; Dollinger, J.; Renault, P.; Tlili, A.; Bérard, A. The FungiResp method: An application of the MicroResp™ method to assess fungi in microbial communities as soil biological indicators. Ecol. Indic. 2012, 23, 482–490. [Google Scholar] [CrossRef]
- Campbell, C.D.; Chapman, S.J.; Cameron, C.M.; Davidson, M.S.; Potts, J.M. A rapid microtiter plate method to measure carbon dioxide evolved from carbon substrate amendments so as to determine the physiological profiles of soil microbial communities by using whole soil. Appl. Environ. Microbiol. 2003, 69, 3593–3599. [Google Scholar] [CrossRef] [Green Version]
- Creamer, R.; Stone, D.; Berry, P.; Kuiper, I. Measuring respiration profiles of soil microbial communities across Europe using MicroResp™ method. Appl. Soil Ecol. 2016, 97, 36–43. [Google Scholar] [CrossRef]
- Creamer, R.E.; Bellamy, P.; Black, H.I.; Cameron, C.M.; Campbell, C.D.; Chamberlain, P.; Harris, J.; Parekh, N.; Pawlett, M.; Poskitt, J.; et al. An inter-laboratory comparison of multi-enzyme and multiple substrate-induced respiration assays to assess method consistency in soil monitoring. Biol. Fertil. Soils 2009, 45, 623–633. [Google Scholar] [CrossRef]
- Anderson, J.; Domsch, K. Measurement of bacterial and fungal contributions to respiration of selected agricultural and forest soils. Can. J. Microbiol. 1975, 21, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Bailey, V.L.; Smith, J.L.; Bolton, H. Novel antibiotics as inhibitors for the selective respiratory inhibition method of measuring fungal: Bacterial ratios in soil. Biol. Fertil. Soils 2003, 38, 154–160. [Google Scholar] [CrossRef]
- Kang, F.; Ge, Y.; Hu, X.; Goikavi, C.; Waigi, M.G.; Gao, Y.; Ling, W. Understanding the sorption mechanisms of aflatoxin B1 to kaolinite, illite, and smectite clays via a comparative computational study. J. Hazard. Mater. 2016, 320, 80–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mertz, D.; Edward, T.; Lee, D.; Zuber, M. Absorption of aflatoxin by lettuce seedlings grown in soil adulterated with aflatoxin B1. J. Agric. Food Chem. 1981, 29, 1168–1170. [Google Scholar] [CrossRef]
- Angle, J.; Wagner, G. Decomposition of aflatoxin in soil. Soil Sci. Soc. Am. J. 1980, 44, 1237–1240. [Google Scholar] [CrossRef]
- Jaynes, W.; Zartman, R.; Hudnall, W. Aflatoxin B1 adsorption by clays from water and corn meal. Appl. Clay Sci. 2007, 36, 197–205. [Google Scholar] [CrossRef]
- Angle, J. Aflatoxin decomposition in various soils. J. Environ. Sci. Health Part B 1986, 21, 277–288. [Google Scholar] [CrossRef]
- Goldberg, B.; Angle, J. Aflatoxin Movement in Soil; Technical report; Wiley Online Library: Hoboken, NJ, USA, 1985. [Google Scholar]
- Albert, J.; Muñoz, K. Kinetics of microbial and photochemical degradation of aflatoxin B1 in a sandy loam and clay soil. Sci. Rep. 2022, 12, 16849. [Google Scholar] [CrossRef]
- OECD. 217. Soil Microorganisms: Carbon Transformation Test. In OECD Guidelines for the Testing of Chemicals, Section 2; OECD: Paris, France, 2000; Volume 2. [Google Scholar]
- Starr, J.M.; Rushing, B.R.; Selim, M.I. Solvent-dependent transformation of aflatoxin B1 in soil. Mycotoxin Res. 2017, 33, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Northcott, G.L.; Jones, K.C. Spiking hydrophobic organic compounds into soil and sediment: A review and critique of adopted procedures. Environ. Toxicol. Chem. Int. J. 2000, 19, 2418–2430. [Google Scholar] [CrossRef]
- Vance, E.D.; Brookes, P.C.; Jenkinson, D.S. An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 1987, 19, 703–707. [Google Scholar] [CrossRef]
- Joergensen, R.G. The fumigation-extraction method to estimate soil microbial biomass: Calibration of the kEC value. Soil Biol. Biochem. 1996, 28, 25–31. [Google Scholar] [CrossRef]
- Gong, P.; Guan, X.; Witter, E. A rapid method to extract ergosterol from soil by physical disruption. Appl. Soil Ecol. 2001, 17, 285–289. [Google Scholar] [CrossRef]
- Campbell, C.; Grayston, S.; Hirst, D. Use of rhizosphere carbon sources in sole carbon source tests to discriminate soil microbial communities. J. Microbiol. Methods 1997, 30, 33–41. [Google Scholar] [CrossRef]
- Banning, N.; Lalor, B.; Cookson, W.; Grigg, A.; Murphy, D. Analysis of soil microbial community level physiological profiles in native and post-mining rehabilitation forest: Which substrates discriminate? Appl. Soil Ecol. 2012, 56, 27–34. [Google Scholar] [CrossRef]
- Degens, B.; Harris, J. Development of a physiological approach to measuring the catabolic diversity of soil microbial communities. Soil Biol. Biochem. 1997, 29, 1309–1320. [Google Scholar] [CrossRef]
- Anderson, T.H.; Domsch, K.H. Soil microbial biomass: The eco-physiological approach. Soil Biol. Biochem. 2010, 42, 2039–2043. [Google Scholar] [CrossRef]
- Anderson, T.H. Microbial eco-physiological indicators to asses soil quality. Agric. Ecosyst. Environ. 2003, 98, 285–293. [Google Scholar] [CrossRef]
- Insam, H.; Öhlinger, R. Ecophysiological parameters. In Methods in Soil Biology; Springer: Berlin/Heidelberg, Germany, 1996; pp. 306–309. [Google Scholar]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’hara, R.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H.; et al. Package ‘vegan’. Community Ecol. Packag. Version 2013, 2, 1–295. [Google Scholar]
- Steinmetz, Z.; Albert, J.; Kenngott, K. Envalysis: Miscellaneous Functions for Environmental Analyses. R Package Version 0.5.4. 2022. Available online: https://cran.r-project.org/package=envalysis (accessed on 20 January 2023).
- Zuur, A.F.; Ieno, E.N.; Elphick, C.S. A protocol for data exploration to avoid common statistical problems. Methods Ecol. Evol. 2010, 1, 3–14. [Google Scholar] [CrossRef]
- Montgomery, D.C.; Peck, E.A.; Vining, G.G. Introduction to Linear Regression Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2012. [Google Scholar]
- Wakelin, S.; Lombi, E.; Donner, E.; MacDonald, L.; Black, A.; O’Callaghan, M. Application of MicroResp™ for soil ecotoxicology. Environ. Pollut. 2013, 179, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Leys, C.; Ley, C.; Klein, O.; Bernard, P.; Licata, L. Detecting outliers: Do not use standard deviation around the mean, use absolute deviation around the median. J. Exp. Soc. Psychol. 2013, 49, 764–766. [Google Scholar] [CrossRef] [Green Version]
- Box, G.E.; Cox, D.R. An analysis of transformations. J. R. Stat. Soc. Ser. (Methodol.) 1964, 26, 211–243. [Google Scholar] [CrossRef]
- Van den Brink, P.J.; Ter Braak, C.J. Multivariate analysis of stress in experimental ecosystems by principal response curves and similarity analysis. Aquat. Ecol. 1998, 32, 163–178. [Google Scholar] [CrossRef]
- Van den Brink, P.J.; Braak, C.J.T. Principal response curves: Analysis of time-dependent multivariate responses of biological community to stress. Environ. Toxicol. Chem. Int. J. 1999, 18, 138–148. [Google Scholar] [CrossRef]
- Frampton, G.K.; Van den Brink, P.J.; Gould, P.J. Effects of spring precipitation on a temperate arable collembolan community analysed using Principal Response Curves. Appl. Soil Ecol. 2000, 14, 231–248. [Google Scholar] [CrossRef]
- Szöcs, E.; Van den Brink, P.J.; Lagadic, L.; Caquet, T.; Roucaute, M.; Auber, A.; Bayona, Y.; Liess, M.; Ebke, P.; Ippolito, A.; et al. Analysing chemical-induced changes in macroinvertebrate communities in aquatic mesocosm experiments: A comparison of methods. Ecotoxicology 2015, 24, 760–769. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Wang, S.; Wang, C.; Mo, J.; Zhang, W. Is microbial biomass measurement by the chloroform fumigation extraction method biased by experimental addition of N and P? iForest-Biogeosci. For. 2021, 14, 408. [Google Scholar] [CrossRef]
- Schenzel, J.; Goss, K.U.; Schwarzenbach, R.P.; Bucheli, T.D.; Droge, S.T. Experimentally determined soil organic matter–water sorption coefficients for different classes of natural toxins and comparison with estimated numbers. Environ. Sci. Technol. 2012, 46, 6118–6126. [Google Scholar] [CrossRef]
- Ahmed, A.A.; Thiele-Bruhn, S.; Aziz, S.G.; Hilal, R.H.; Elroby, S.A.; Al-Youbi, A.O.; Leinweber, P.; Kühn, O. Interaction of polar and nonpolar organic pollutants with soil organic matter: Sorption experiments and molecular dynamics simulation. Sci. Total Environ. 2015, 508, 276–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, D.; Hyun, S.; Pignatello, J.J.; Lee, L.S. Evidence for π-π electron donor-acceptor interactions between π-donor aromatic compounds and π-acceptor sites in soil organic matter through pH effects on sorption. Environ. Sci. Technol. 2004, 38, 4361–4368. [Google Scholar] [CrossRef]
- Manrique, L.; Jones, C.; Dyke, P. Predicting cation-exchange capacity from soil physical and chemical properties. Soil Sci. Soc. Am. J. 1991, 55, 787–794. [Google Scholar] [CrossRef]
- Manzoni, S.; Taylor, P.; Richter, A.; Porporato, A.; Ågren, G.I. Environmental and stoichiometric controls on microbial carbon-use efficiency in soils. New Phytol. 2012, 196, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Öquist, M.G.; Erhagen, B.; Haei, M.; Sparrman, T.; Ilstedt, U.; Schleucher, J.; Nilsson, M.B. The effect of temperature and substrate quality on the carbon use efficiency of saprotrophic decomposition. Plant Soil 2017, 414, 113–125. [Google Scholar] [CrossRef] [Green Version]
- Jones, D.; Edwards, A. Influence of sorption on the biological utilization of two simple carbon substrates. Soil Biol. Biochem. 1998, 30, 1895–1902. [Google Scholar] [CrossRef]
- Jiang, D.; Huang, Q.; Cai, P.; Rong, X.; Chen, W. Adsorption of Pseudomonas putida on clay minerals and iron oxide. Colloids Surf. B Biointerfaces 2007, 54, 217–221. [Google Scholar] [CrossRef]
- Tietjen, T.; Wetzel, R.G. Extracellular enzyme-clay mineral complexes: Enzyme adsorption, alteration of enzyme activity, and protection from photodegradation. Aquat. Ecol. 2003, 37, 331–339. [Google Scholar] [CrossRef]
- Kaiser, K.; Zech, W. Competitive sorption of dissolved organic matter fractions to soils and related mineral phases. Soil Sci. Soc. Am. J. 1997, 61, 64–69. [Google Scholar] [CrossRef]
- Beck, M.; Robarge, W.; Buol, S. Phosphorus retention and release of anions and organic carbon by two Andisols. Eur. J. Soil Sci. 1999, 50, 157–164. [Google Scholar] [CrossRef]
- Hadacek, F.; Bachmann, G.; Engelmeier, D.; Chobot, V. Hormesis and a chemical raison d’ětre for secondary plant metabolites. Dose Response 2011, 9, 79–116. [Google Scholar] [CrossRef]
- Hashmi, M.Z.; Shen, H.; Zhu, S.; Yu, C.; Shen, C. Growth, bioluminescence and shoal behavior hormetic responses to inorganic and/or organic chemicals: A review. Environ. Int. 2014, 64, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Finotti, E.; Parroni, A.; Zaccaria, M.; Domin, M.; Momeni, B.; Fanelli, C.; Reverberi, M. Aflatoxins are natural scavengers of reactive oxygen species. Sci. Rep. 2021, 11, 16024. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, J.A.; Vilgalys, R.; Jackson, R.B. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 2005, 71, 4117–4120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frostegård, Å.; Tunlid, A.; Bååth, E. Microbial biomass measured as total lipid phosphate in soils of different organic content. J. Microbiol. Methods 1991, 14, 151–163. [Google Scholar] [CrossRef]
- Frostegård, Å.; Tunlid, A.; Bååth, E. Use and misuse of PLFA measurements in soils. Soil Biol. Biochem. 2011, 43, 1621–1625. [Google Scholar] [CrossRef]



| Property | RefeSol 01-A | LUFA 6S |
|---|---|---|
| Soil type | sandy loam | clay |
| Sand (%) | 70.5 | 23.3 |
| Silt (%) | 26.1 | 35.5 |
| Clay (%) | 3.4 | 41.2 |
| Corg (%) | 0.9 | 1.7 |
| Ntot (%) | 0.08 | 0.18 |
| WHC (%) | 29.3 | 42.4 |
| pH (0.01 M CaCl2) | 5.4 | 7.3 |
| Cmic (mg kg−1) | 95 ± 15 | 267 ± 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albert, J.; More, C.; Korz, S.; Muñoz, K. Soil Microbial Responses to Aflatoxin Exposure: Consequences for Biomass, Activity and Catabolic Functionality. Soil Syst. 2023, 7, 23. https://doi.org/10.3390/soilsystems7010023
Albert J, More C, Korz S, Muñoz K. Soil Microbial Responses to Aflatoxin Exposure: Consequences for Biomass, Activity and Catabolic Functionality. Soil Systems. 2023; 7(1):23. https://doi.org/10.3390/soilsystems7010023
Chicago/Turabian StyleAlbert, Julius, Camilla More, Sven Korz, and Katherine Muñoz. 2023. "Soil Microbial Responses to Aflatoxin Exposure: Consequences for Biomass, Activity and Catabolic Functionality" Soil Systems 7, no. 1: 23. https://doi.org/10.3390/soilsystems7010023
APA StyleAlbert, J., More, C., Korz, S., & Muñoz, K. (2023). Soil Microbial Responses to Aflatoxin Exposure: Consequences for Biomass, Activity and Catabolic Functionality. Soil Systems, 7(1), 23. https://doi.org/10.3390/soilsystems7010023






