Decay of Root Debris after Harvesting American Ginseng (Panax quinquefolius) and Changes in Soil Chemistry and Microbiology
Abstract
:1. Introduction
2. Materials and Methods
2.1. Soil and Root Collection
2.2. Root Evaluation
2.3. Soil DNA Extraction and Sequencing
2.4. Microbial Diversity and Abundance
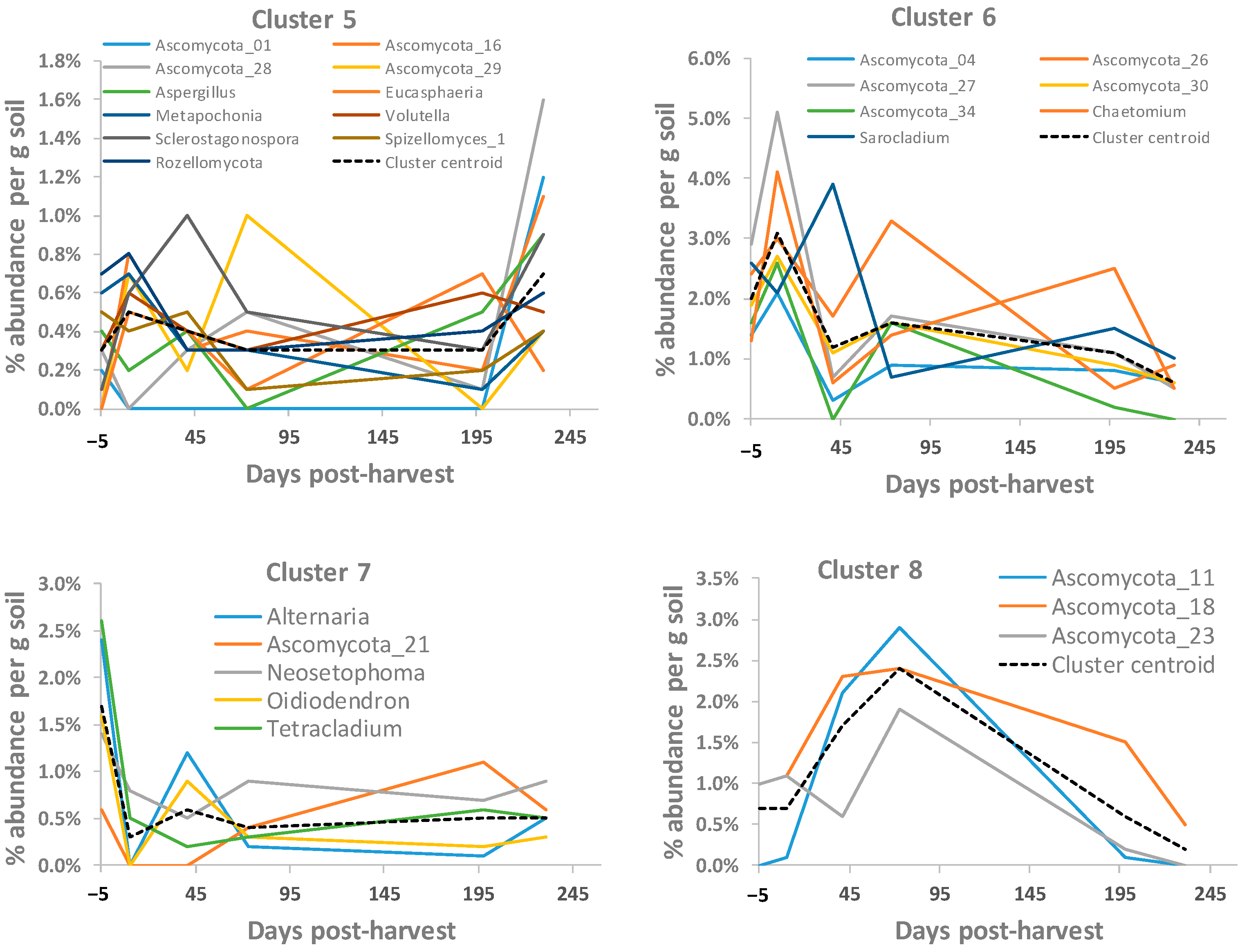
2.5. Cluster Analysis
2.6. Soil Ginsenosides
2.7. Soil Characterization
2.8. Data Analysis
3. Results
3.1. Soil Characterization
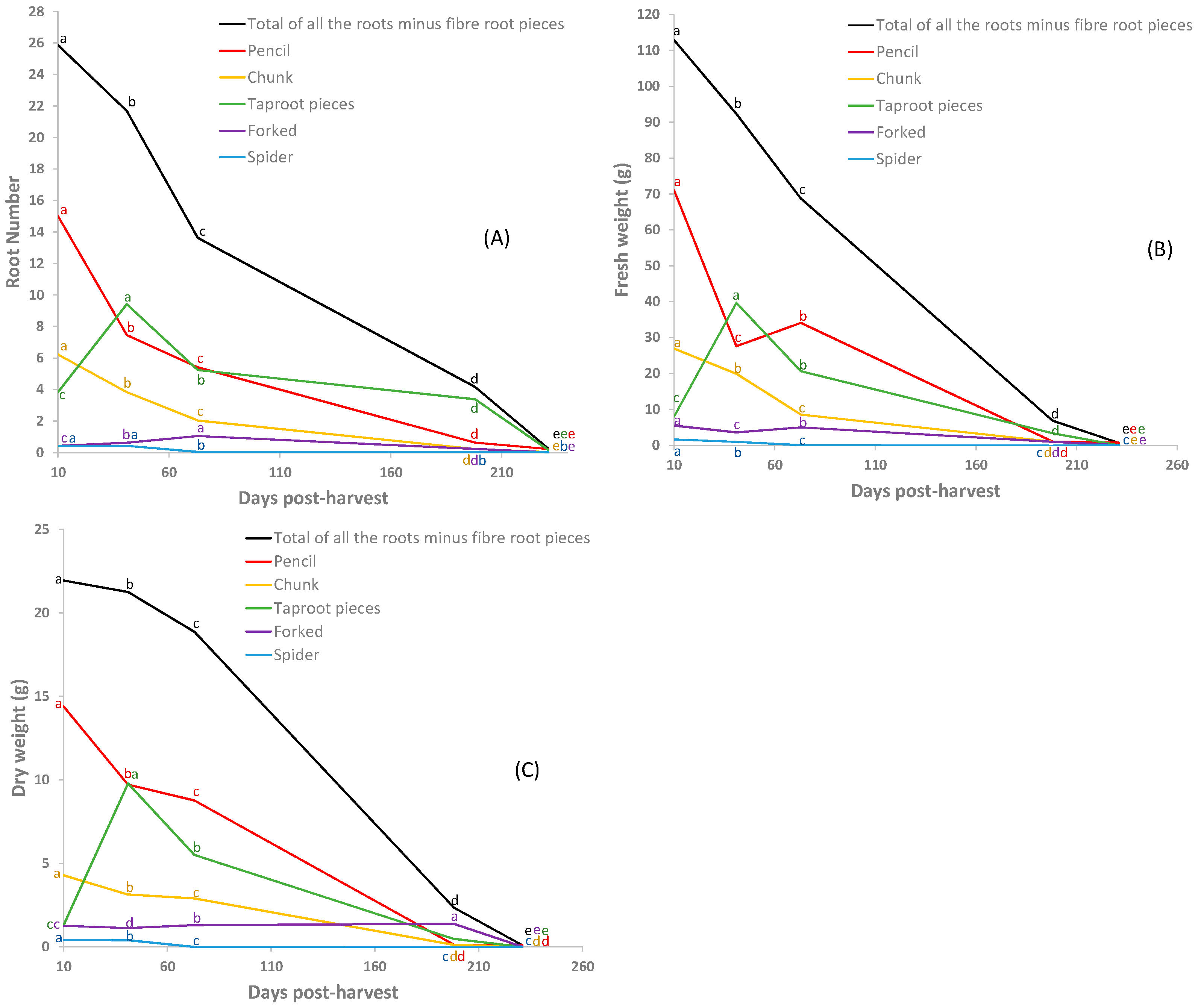
3.2. Root Number and Biomass
3.3. Root Lesions
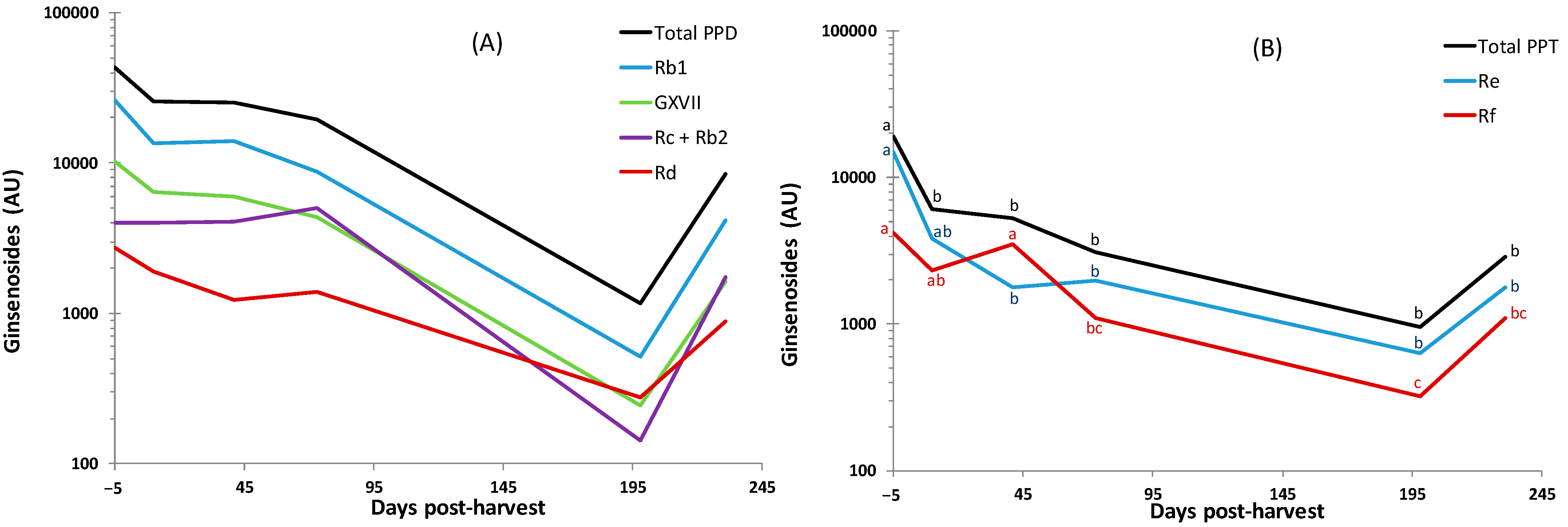
3.4. Soil Ginsenosides
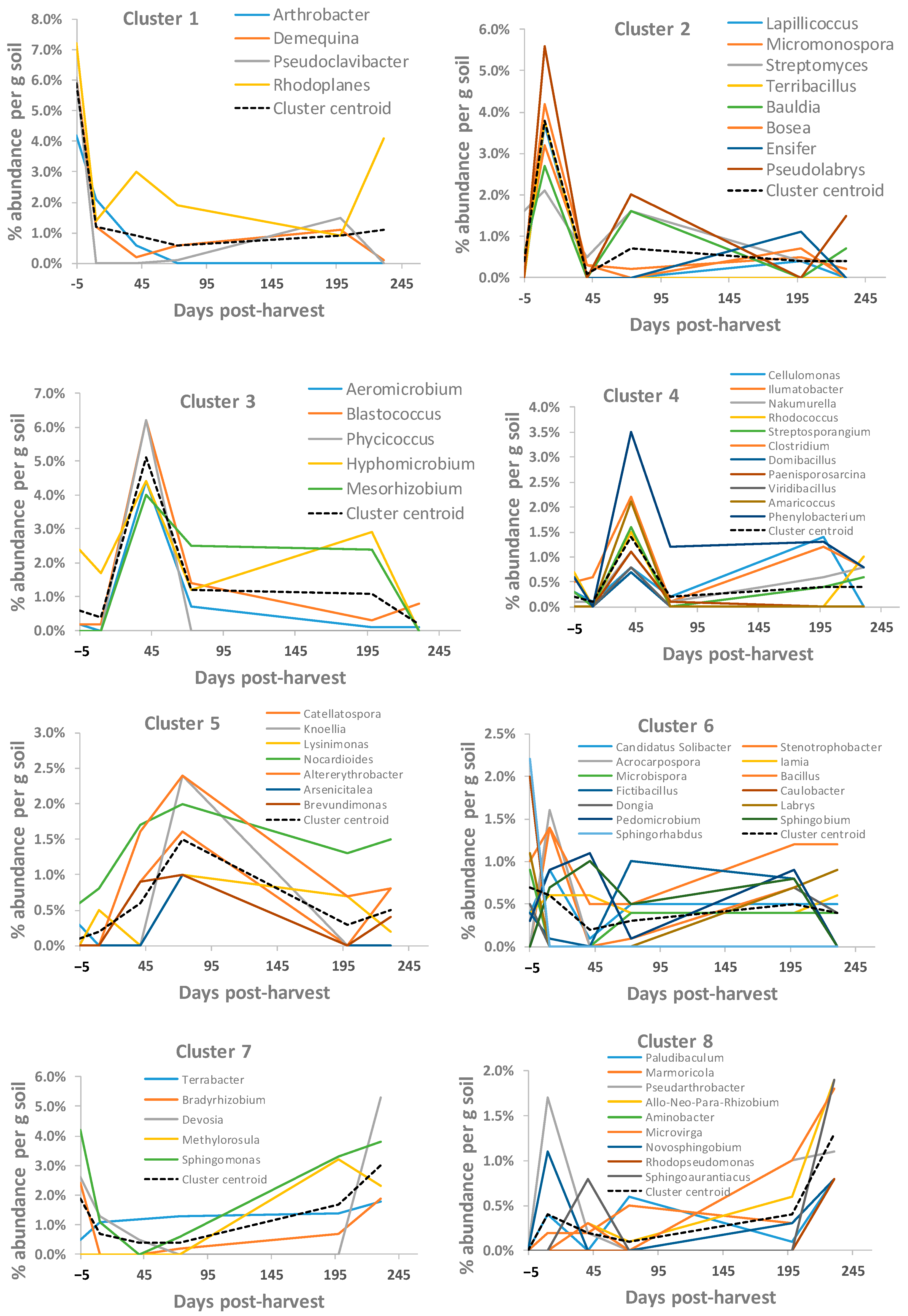
3.5. Soil Bacterial OTUs
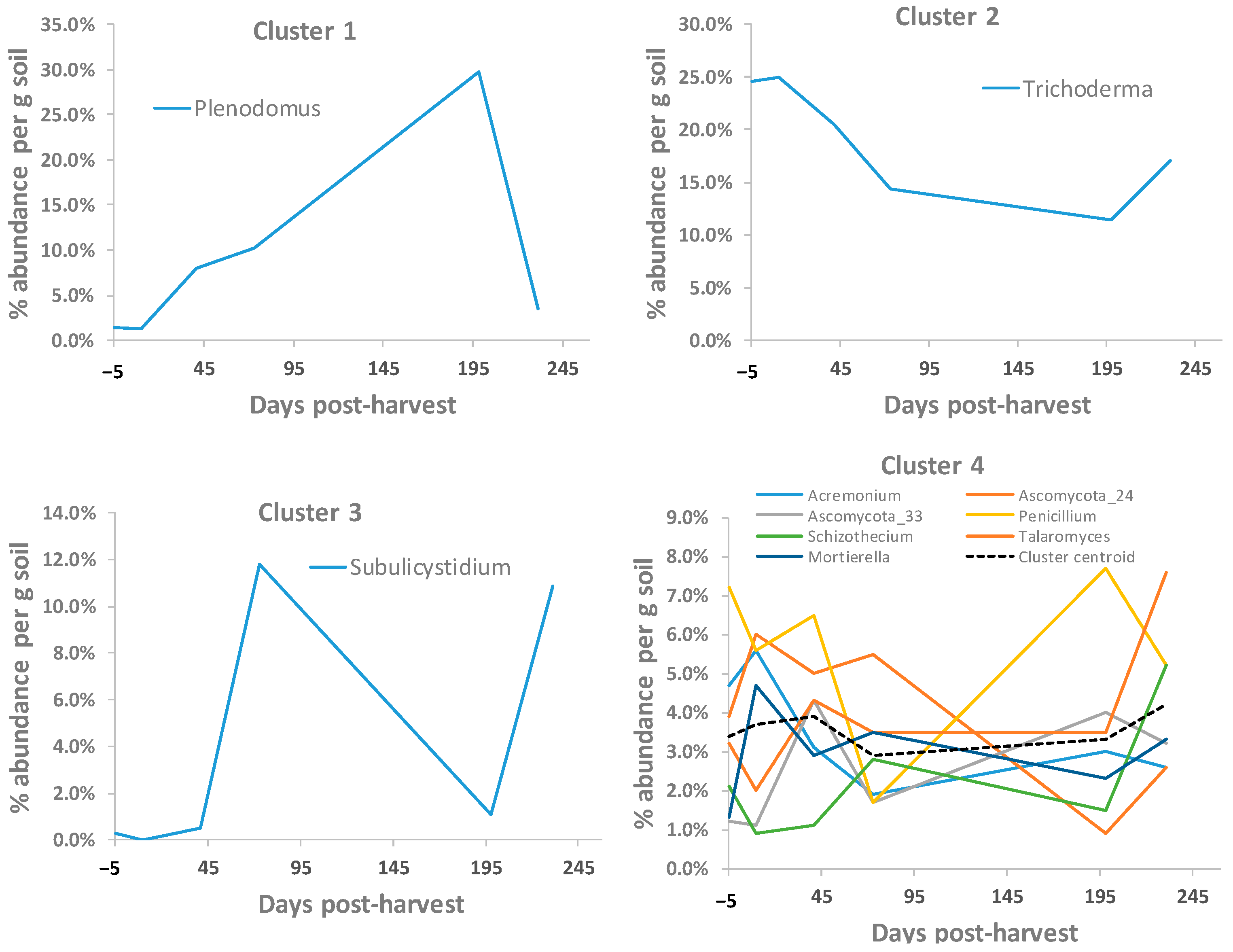
3.6. Soil Fungal OTUs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Batish, D.R.; Singh, H.P.; Kaur, S. Crop allelopathy and its role in ecological agriculture. J. Crop Prod. 2001, 4, 121–161. [Google Scholar] [CrossRef]
- Cipollini, D.; Rigsby, C.M.; Barto, E.K. Microbes as targets and mediators of allelopathy in plants. J. Chem. Ecol. 2012, 38, 714–727. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.I.; Ruegger, P.M.; McKenry, M.V.; Becker, J.O.; Borneman, J. Correlations between root-associated microorganisms and peach replant disease symptoms in a California soil. PLoS ONE 2012, 7, e46420. [Google Scholar] [CrossRef]
- Zhou, X.; Wu, F. P-coumaric acid influenced cucumber rhizosphere soil microbial communities and the growth of Fusarium oxysporum f. sp. cucumerinum Owen. PLoS ONE 2012, 7, e48288. [Google Scholar]
- Patrick, Z.A.; Toussoun, T.A.; Koch, L.W. Effect of crop-residue decomposition products on plant roots. Annu. Rev. Phytopathol. 1964, 2, 267–292. [Google Scholar] [CrossRef]
- Almeida, Á.M.R.; Saraiva, O.F.; Farias, J.R.B.; Gaudêncio, C.A.; Torres, E. Survival of pathogens on soybean debris under no-tillage and conventional tillage systems. Pesqui. Agropecu. Bras. 2001, 36, 1231–1238. [Google Scholar] [CrossRef]
- Krupinsky, J.M.; Bailey, K.L.; McMullen, M.P.; Gossen, B.D.; Turkington, T.K. Managing plant disease risk in diversified cropping systems. Agron. J. 2002, 94, 198–209. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, X.; Xu, Y.; Mei, X.; Jiang, B.; Liao, J.; He, X. Autotoxic ginsenosides in the rhizosphere contribute to the replant failure of Panax notoginseng. PLoS ONE 2015, 10, e0118555. [Google Scholar] [CrossRef]
- Nicol, R.W.; Yousef, L.; Traquair, J.A.; Bernards, M.A. Ginsenosides stimulate the growth of soilborne pathogens of American ginseng. Phytochemistry 2003, 64, 257–264. [Google Scholar] [CrossRef]
- Bernards, M.A.; Yousef, L.F.; Nicol, R.W. The allelopathic potential of ginsenosides. In Allelochemicals: Biological Control of Plant Pathogens and Diseases; Mukerji, K.G., Ed.; Springer: Dordrecht, The Netherlands, 2006; pp. 157–175. [Google Scholar]
- Li, Y.; Dai, S.; Wang, B.; Jiang, Y.; Ma, Y.; Pan, L.; Wu, K.; Huang, X.; Zhang, J.; Cai, Z.; et al. Autotoxic ginsenoside disrupts soil fungal microbiomes by stimulating potentially pathogenic microbes. AEM 2020, 86, e00130-20. [Google Scholar] [CrossRef]
- Bi, X.B.; Yang, J.X.; Gao, W.W. Autotoxicity of phenolic compounds from the soil of American ginseng (Panax quinquefolium L.). Allelopath. J. 2010, 25, 115–122. [Google Scholar]
- Reeleder, R.D.; Roy, R.; Capell, B. Seed and root rots of ginseng (Panax quinquefolius L.) caused by Cylindrocarpon destructans and Fusarium spp. J. Ginseng Res. 2002, 26, 151–158. [Google Scholar]
- Lee, S.W.; Lee, S.H.; Park, K.H.; Lan, J.M.; Jang, I.B.; Kim, K.H. Inhibition effect on root rot disease of Panax ginseng by crop cultivation in soil occurring replant failure. Korean J. Med. Crop Sci. 2015, 23, 223–230. [Google Scholar] [CrossRef]
- Jiao, X.L.; Zhang, X.S.; Lu, X.H.; Qin, R.; Bi, Y.M.; Gao, W.W. Effects of maize rotation on the physicochemical properties and microbial communities of American ginseng cultivated soil. Sci. Rep. 2019, 9, 8615. [Google Scholar] [CrossRef]
- Bischoff Nunes, I.; Goodwin, P.H. Interaction of ginseng with Ilyonectria root rot pathogens. Plants 2022, 11, 2152. [Google Scholar] [CrossRef]
- Westerveld, S.M.; Shi, F. 2021. The history, etiology, and management of ginseng replant disease: A Canadian perspective in review. Can. J. Plant Sci. 2021, 101, 886–901. [Google Scholar] [CrossRef]
- He, C.N.; Gao, W.W.; Yang, J.X.; Bi, W.; Zhang, X.S.; Zhao, Y.J. Identification of autotoxic compounds from fibrous roots of Panax quinquefolium L. Plant Soil 2009, 318, 63–72. [Google Scholar] [CrossRef]
- Yousef, L.F.; Bernards, M.A. In vitro metabolism of ginsenosides by the ginseng root pathogen Pythium irregulare. Phytochemistry 2006, 67, 1740–1749. [Google Scholar] [CrossRef]
- OMAFRA. Guide to Ginseng Production; Queen’s Printer for Ontario: Toronto, ON, Canada, 2015. [Google Scholar]
- Kernaghan, G.; Reeleder, R.D.; Hoke, S.M.T. Quantification of Cylindrocarpon destructans f. sp. panacis in soils by real-time PCR. Plant Pathol. 2007, 56, 508–516. [Google Scholar]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- UNITE Community. UNITE QIIME release for Fungi 2; Version 18.11.2018; UNITE Community: Brisbane, QLD, Australia, 2019. [Google Scholar]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool of phylogenetics analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Maechler, M.; Rousseeuw, P.; Struyf, A.; Hubert, M.; Hornik, K. Cluster: Cluster Analysis Basics and Extensions, R package version 2.1.0.; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Ivanov, D.A.; Georgakopoulos, J.R.C.; Bernards, M.A. The chemoattractant potential of ginsenosides in the ginseng—Pythium irregulare pathosystem. Phytochemistry 2016, 122, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Kroetsch, D.B.H.; Wang, C. Particle size distribution. In Soil Sampling and Methods of Analysis, 2nd ed.; Carter, M.R., Gregorich, E.G., Eds.; Lewis Publishers: Boca Raton, FL, USA, 1993; pp. 499–511. [Google Scholar]
- Zhang, Y.L.; Chen, J.B.; Lei, Y.; Zhou, Q.; Sun, S.Q.; Noda, I. Evaluation of different grades of ginseng using Fourier-transform infrared and two-dimensional infrared correlation spectroscopy. J. Mol. Struct. 2010, 974, 94–102. [Google Scholar] [CrossRef]
- Isutsa, D.K.; Merwin, I.A. Malus germplasm varies in resistance or tolerance to apple replant disease in a mixture of New York orchard soils. HortScience 2000, 35, 262–268. [Google Scholar] [CrossRef]
- Farh, M.E.A.; Kim, Y.J.; Kim, Y.J.; Yang, D.C. Cylindrocarpon destructans/Ilyonectria radicicola-species complex: Causative agent of ginseng root-rot disease and rusty symptoms. J. Ginseng Res. 2018, 42, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Shin, K.C.; Oh, D.K. Classification of glycosidases that hydrolyze the specific positions and types of sugar moieties in ginsenosides. Crit. Rev. Biotechnol. 2016, 36, 1036–1049. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yang, H.; You, X.; Li, Y. Diversity of endophytic fungi from roots of Panax ginseng and their saponin yield capacities. SpringerPlus 2013, 2, 107. [Google Scholar] [CrossRef] [PubMed]
- Court, W.A.; Reynolds, L.B.; Hendel, J.G. Influence of root age on the concentration of ginsenosides of American ginseng (Panax quinquefolium). Can. J. Plant Sci. 1996, 76, 853–855. [Google Scholar] [CrossRef]
- Park, C.S.; Yoo, M.H.; Noh, K.H.; Oh, D.K. Biotransformation of ginsenosides by hydrolyzing the sugar moieties of ginsenosides using microbial glycosidases. Appl. Microbiol. Biotechnol. 2010, 87, 9–19. [Google Scholar] [CrossRef]
- Luo, L.F.; Yang, L.; Yan, Z.X.; Jiang, B.B.; Li, S.; Huang, H.C.; Liu, Y.X.; Zhu, S.S.; Yang, M. Ginsenosides in root exudates of Panax notoginseng drive the change of soil microbiota through carbon source different utilization. Plant Soil 2020, 455, 139–153. [Google Scholar] [CrossRef]
- Goodwin, P.H. The rhizosphere microbiome of ginseng. Microorganisms 2022, 10, 1152. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suárez, I.D.S.; Valliani, M.; Hsiang, T.; Goodwin, P.H. Decay of Root Debris after Harvesting American Ginseng (Panax quinquefolius) and Changes in Soil Chemistry and Microbiology. Soil Syst. 2023, 7, 108. https://doi.org/10.3390/soilsystems7040108
Suárez IDS, Valliani M, Hsiang T, Goodwin PH. Decay of Root Debris after Harvesting American Ginseng (Panax quinquefolius) and Changes in Soil Chemistry and Microbiology. Soil Systems. 2023; 7(4):108. https://doi.org/10.3390/soilsystems7040108
Chicago/Turabian StyleSuárez, Iván Darío Samur, Moez Valliani, Tom Hsiang, and Paul H. Goodwin. 2023. "Decay of Root Debris after Harvesting American Ginseng (Panax quinquefolius) and Changes in Soil Chemistry and Microbiology" Soil Systems 7, no. 4: 108. https://doi.org/10.3390/soilsystems7040108
APA StyleSuárez, I. D. S., Valliani, M., Hsiang, T., & Goodwin, P. H. (2023). Decay of Root Debris after Harvesting American Ginseng (Panax quinquefolius) and Changes in Soil Chemistry and Microbiology. Soil Systems, 7(4), 108. https://doi.org/10.3390/soilsystems7040108





