Exploring the Influence of Ecological Niches and Hologenome Dynamics on the Growth of Encephalartos villosus in Scarp Forests
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Sites and Species
2.2. Soil Characteristics
2.3. Coralloid Root Surface Sterilisation
2.4. Bacterial Extraction and Identification from Coralloid Roots and Soils
2.5. Extracellular Enzyme Activities
2.6. Leaf Nutrient Composition
2.7. Percentage N Derived from the Atmosphere (%NDFA)
2.8. Statistical Analysis
3. Results
3.1. Soil Characteristics
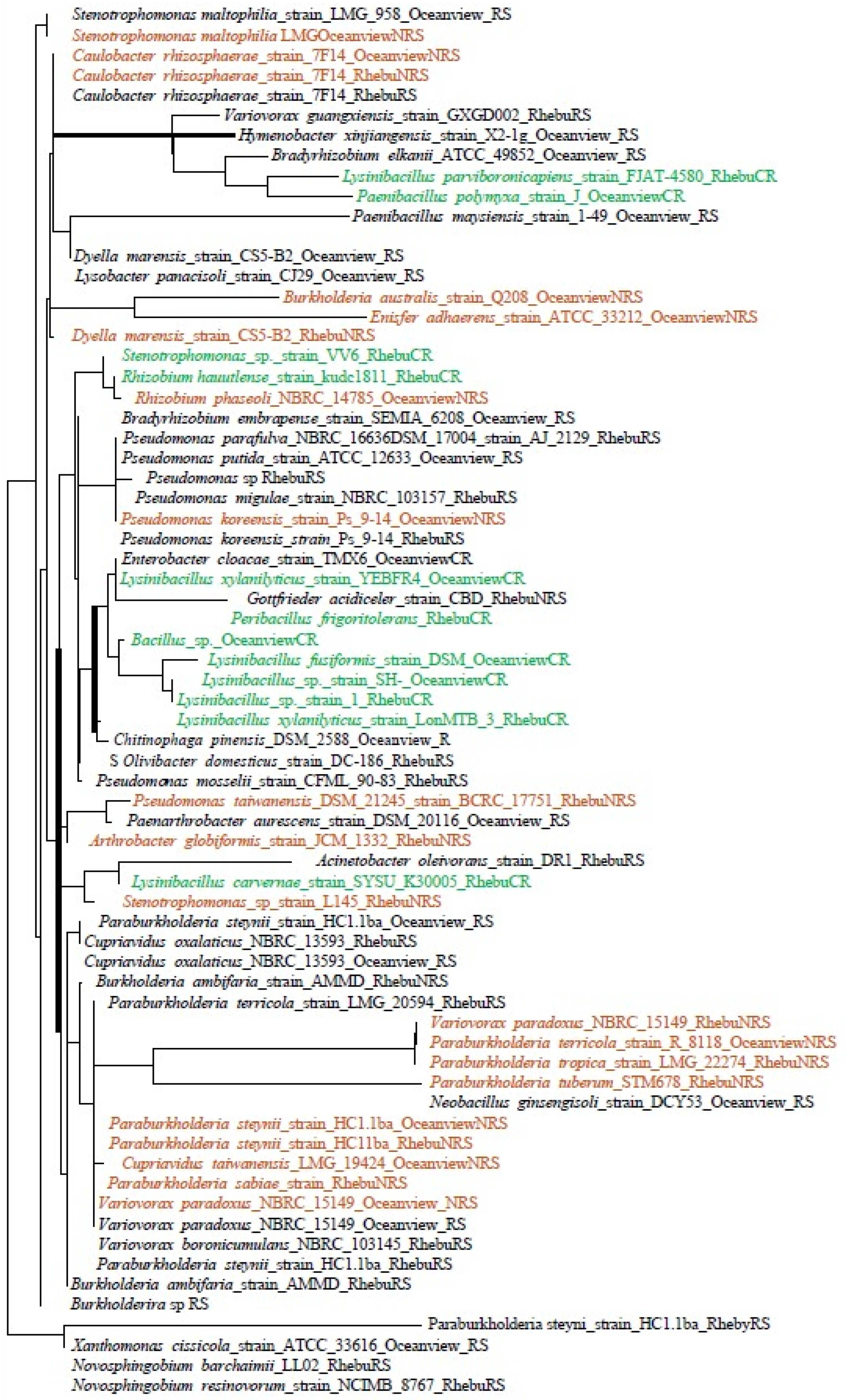
3.2. Bacterial Identification
3.3. Extracellular Enzyme Activities
3.4. Leaf Nutrition and N Source Reliance
3.5. Correlations between Extracellular Enzymes and Soil Nutrition in E. villosus Rhizosphere and Non-Rhizosphere Soils
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brenner, E.D.; Stevenson, D.W.; Twigg, R.W. Cycads: Evolutionary innovations and the role of plant-derived neurotoxins. Trends Plant Sci. 2003, 8, 446–452. [Google Scholar] [CrossRef] [PubMed]
- Marler, T.E.; Krishnapillai, M.V. Cycas micronesica trees alter local soil traits. Forests 2018, 9, 565. [Google Scholar] [CrossRef]
- Cousins, S.R.; Williamson, V.L.; Witkowski, E.T.F. Sifting through cycads: A guide to identifying the stem fragments of six South African medicinal Encephalartos species. S. Afr. J. Bot. 2013, 84, 115–123. [Google Scholar] [CrossRef]
- Donaldson, J.S. Cycads Status Survey and Conservation Action Plan; IUCN/SSC Cycad Specialist Group, IUCN: Gland, Switzerland; Cambridge, UK, 2003; pp. 26–89. [Google Scholar]
- Condamine, F.L.; Nagalingum, N.S.; Marshall, C.R.; Morlon, H. Origin and diversification of living cycads: A cautionary tale on the impact of the branching process prior in Bayesian molecular dating. BMC Evol. Biol. 2015, 15, 65. [Google Scholar] [CrossRef] [PubMed]
- Cousins, S.R.; Williamson, V.L.; Witkowski, E.T.F. Uncovering the cycad taxa (Encephalartos species) traded for traditional medicine in Johannesburg and Durban, South Africa. S. Afr. J. Bot. 2012, 78, 129–138. [Google Scholar] [CrossRef]
- Yessoufou, K.; Bamigboye, S.O.; Daru, B.H.; van der Bank, M. Evidence of constant diversification punctuated by a mass extinction in the African cycads. Ecol. Evol. 2014, 4, 50–58. [Google Scholar] [CrossRef]
- Alvarez-Yepiz, J.C.; Cueva, A.; Dovciak, M.; Teece, M.; Enrico, A.; Yepez, E.A. 2014. Ontogenic resource-use strategies in a rare long-lived cycad along environmental gradients. Conserv. Physiol. 2014, 2, 2–4. [Google Scholar] [CrossRef]
- Mucina, L.; Scott-Shaw, C.R.; Rutherford, M.C.; Camp, K.G.T.; Matthews, W.S.; Powrie, L.W.; Hoare, D.B. Indian Ocean coastal belt. Strelitzia 2006, 19, 569–583. [Google Scholar]
- Marler, T.E.; Calonje, M. Two cycad species affect the Carbon, Nitrogen, and Phosphorus content of soils. Horticulturae 2020, 6, 24. [Google Scholar] [CrossRef]
- Ndlovu, S.; Suinyuy, T.N.; Pérez-Fernández, M.A.; Magadlela, A. Encephalartos natalensis, Their Nutrient-Cycling Microbes and Enzymes: A Story of Successful Trade-Offs. Plants 2023, 12, 1034. [Google Scholar] [CrossRef]
- Ma, Y.; Jiang, H.; Wang, B.; Zhou, G.; Yu, S.; Peng, S.; Hao, Y.; Wei, X.; Liu, J.; Yu, Z. Carbon storage of cycad and other gymnosperm ecosystems in China: Implications to evolutionary trends. Pol. J. Ecol. 2009, 57, 635–646. [Google Scholar]
- Cousins, S.R.; Witkowski, E.T.F. African cycad ecology, ethnobotany and conservation: A synthesis. Bot. Rev. 2017, 83, 152–194. [Google Scholar] [CrossRef]
- García, M.; Hernandez, R.; Lopez, P. The role of cycads in forest restoration efforts. For. Chron. 2019, 95, 12–23. [Google Scholar]
- Brown, T.; Davis, R.; Rodriguez, L. The economic value of cycads in developing countries. Econ. Bot. 2018, 72, 34–45. [Google Scholar]
- Cenciani, K.; dos Santos Freitas, S.; Critter, S.A.M.; Airoldi, C. Microbial enzymatic activity and thermal effect in tropical soil treated with organic material. Soil Sci. Plant Nutr. 2008, 65, 647–680. [Google Scholar] [CrossRef]
- Meena, A.; Rao, K.S. Assessment of soil microbial and enzyme activity in the rhizosphere zone under different land use/cover of a semi-arid region, India. Ecol. Process. 2021, 10, 16. [Google Scholar] [CrossRef]
- Merino, C.; Godoy, R.; Matus, F. Soil enzymes and biological activity at different levels of organic matter stability. J. Soil Sci. Plant Nutr. 2016, 16, 14–30. [Google Scholar]
- Adetunji, A.T.; Lewu, F.B.; Mulidzi, R.; Ncube, B. The biological activities of β-glucosidase, phosphatase, and urease as soil quality indicators: A review. J. Soil Sci. Plant Nutr. 2017, 17, 794–807. [Google Scholar] [CrossRef]
- Gutiérrez-García, K.; Bustos-Díaz, E.D.; Corona-Gómez, J.A.; Ramos-Aboites, H.E.; Sélem-Mojica, N.; Cruz-Morales, P.; Pérez-Farrera, M.A.; Barona-Gómez, F.; Cibrián-Jaramillo, A. Cycad coralloid roots contain bacterial communities including Cyanobacteria and Caulobacter spp. that encode niche-specific biosynthetic gene clusters. Genome Biol. Evol. 2019, 11, 319–334. [Google Scholar] [CrossRef]
- Sasse, J.; Martinoia, E.; Northern, T. Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 2018, 23, 35–41. [Google Scholar] [CrossRef]
- Marler, T.E.; Lindström, A. Leaf nutrient relations of cycads in common garden. Trop. Conserv. Sci. 2021, 14, 19400829211036570. [Google Scholar] [CrossRef]
- Deloso, B.E.; Krishnapillai, M.V.; Ferreras, U.F.; Lindström, A.J.; Calonje, M.; Marler, T.E. Chemical element concentrations of cycad leaves: Do we know enough? Horticulturae 2020, 6, 85. [Google Scholar] [CrossRef]
- Gehringer, M.M.; Pengelly, J.J.L.; Cuddy, W.S.; Fieker, C.; Forster, P.I.; Neilan, B.A. Host selection of symbiotic cyanobacteria in 31 species of the Australian cycad genus: Macrozamia (Zamiaceae). Mol. Plant Microbe Interact. 2010, 23, 811–882. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Moo, P.J.; Vovides, A.P.; Griffith, M.P.; Barona-Gòmez, F.; Cibrián-Jaramillo, A. Unlocking a high bacterial diversity in the coralloid root microbiome from the cycad genus Dioon. PLoS ONE 2019, 14, e0211271. [Google Scholar] [CrossRef] [PubMed]
- Kipp, M.A.; Stüeken, E.E.; Gehringer, M.M.; Sterelny, K.; Scott, J.K.; Forster, P.I.; Strömberg, C.A.E.; Buick, R. Exploring cycad foliage as an archive of the isotopic composition of atmospheric nitrogen. Geobiology 2019, 18, 152–166. [Google Scholar] [CrossRef]
- Marler, T.E.; Lindström, A. Inserting cycads into global climate nutrient relations datasets. Plant Signal. Behav. 2018, 13, e15475781–e15475786. [Google Scholar] [CrossRef]
- Marler, T.E.; Ferreras, U.F. Disruption of leaf nutrient remobilization in coastal Cycas trees by tropical cyclone damage. J. Geogr. Nat. Disasters 2015, 5, 142. [Google Scholar]
- Manson, A.D.; Roberts, V.G. Analytical Methods Used by the Soil Fertility and Analytical Services Section; KZN Agri-Report No. N/A/2001/04; Department of Agriculture and Rural Development: Pietermaritzburg, South Africa, 2000. [Google Scholar]
- Murphy, J.; Riley, J.R. A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta. 1962, 27, 31–36. [Google Scholar] [CrossRef]
- Somasegaran, P.; Hoben, H.J. Handbook for Rhizobia: Methods in Legume-Rhizobium Technology; Springer: New York, NY, USA, 1994. [Google Scholar]
- Ndabankulu, N.; Tsvuura, Z.; Magadlela, A. Soil microbe and associated extracellular enzymes largely impact nutrient bioavailability in acidic and nutrient poor grassland ecosystem soil. Sci. Rep. 2022, 12, 12601. [Google Scholar] [CrossRef]
- Marchesi, J.R.; Sato, T.; Weightman, A.J.; Martin, T.A.; Fry, J.C.; Him, S.J.; Dymock, D.; Wade, W.G. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 1998, 64, 795–799. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.R.; Tyler, H.L.; Millar, J.J. Determination of microbial extracellular enzyme activity in waters, soils, and sediments using high throughput microplate assays. JoVE 2013, 80, e5039. [Google Scholar]
- Kandeler, E. Potential Nitrification. In Methods in Soil Biology; Schinner, F., Öhlinger, R., Kandeler, E., Margesin, R., Eds.; Spinger: Berlin/Heidelberg, Germany, 1995; pp. 146–149. [Google Scholar]
- Marler, T.E.; Krishnapillai, M.V. Incident light and leaf age influence leaflet element concentrations of Cycas micronesica trees. Horticulturae 2019, 5, 58. [Google Scholar] [CrossRef]
- Shearer, G.; Kohl, D.H. N2-fixation in field settings: Estimations based on natural 15N abundance. Funct. Plant Biol. 1986, 13, 699–756. [Google Scholar]
- Johnson, S.L.; Holt, R.D. The evolution of protein function. Science 1989, 246, 1293–1296. [Google Scholar]
- Sparks, D.W.; Marshall, C.R.; Herron, J.C.; Harmon, L.J.; Aiello, L.; Barton, N.H.; Begun, D.R.; Blumenshine, R.J.; Boyer, D.M.; Carlson, S.M.; et al. The evolutionary origins of biodiversity. Science 2017, 358, 692–697. [Google Scholar]
- Hargreaves, S.K.; Willimas, R.J.; Hofmockel, K.S. Environmental filtering of microbial communities in agricultural soil shifts with crop growth. PLoS ONE 2015, 10, e0134345. [Google Scholar] [CrossRef]
- Kropinski, M.; Bordenstein, S.; Bosch, T.; Brown, J.J.; Gilbert, J.A.; Gilbert, W.; Gordon, J.I.; Johnson, D.J.; Ley, R.E.; Moran, N.A.; et al. The hologenome: A novel concept for understanding the organization and dynamics of prokaryotic genomes. PLoS Comput. Biol. 2017, 13, e1005647. [Google Scholar]
- Simonsen, A.K.; Dinnage, R.; Barrett, L.G.; Prober, S.M.; Thrall, P.H. Symbiosis limits establishment of legumes outside their native range at a global scale. Nat. Commun. 2017, 8, 1–9. [Google Scholar] [CrossRef]
- Ulkrike, M. Are legumes different? Origins and consequences of evolving nitrogen fixing symbioses. J. Plant Physiol. Sep. 2022, 76, 153765. [Google Scholar] [CrossRef]
- Motsomane, N.; Suinyuy, T.N.; Pérez-Fernández, M.A.; Magadlela, A. How the right evolved partners in Cycads and Legumes drive enhanced growth in a harsh environment. Symbiosis 2023, 90, 345–353. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Nature 2005, 337, 816–882. [Google Scholar] [CrossRef] [PubMed]
- Lyu, D.; Zajonc, J.; Pagé, A.; Tanney, C.A.S.; Shah, A.; Mongezi, N.; Msimbira, L.A.; Antar, M.; Backer, R.; Smith, D.L. Plant holobiont theory: The phytomicrobiome plays a central role in evolution and success. Microorganisms 2021, 9, 675. [Google Scholar] [CrossRef]
- Roughgarden, J.; Gilbert, S.F.; Rosenberg, E.; Zilber-Rosenberg, I.; Lloyd, E.A. Holobionts as Units of Selection and a Model of Their Population Dynamics and Evolution. Biol. Theory 2018, 13, 44–65. [Google Scholar] [CrossRef]
- Singh, B.K.; Liu, H.; Trivedi, P. Eco-holobiont: A new concept to identify drivers of host-associated microorganisms. Environ. Microbiol. 2020, 22, 564–567. [Google Scholar] [CrossRef]
- Marschner, P.; Neumann, O.; Kania, A.; Weiskopf, I.; Lieberei, R. Spatial and temporal dynamics of the microbial community structure in the rhizosphere of cluster roots of white lupin (Lupinus albus L.). Plant Soil. 2002, 246, 167–174. [Google Scholar] [CrossRef]
- Rosenzweig, N.; Bradeen, J.M.; Tu, Z.J.; McKay, S.J.; Kinkel, L.L. Rhizosphere bacterial communities associated with long lived prairie plants vary in diversity, composition, and structure. Can. J. Microbiol. 2013, 59, 494–502. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef]
- Charpentier, E.; Valgepea, T. The holobiont: A New Paradigm for Life on Earth. Dialogues Hum. Geogr. 2017, 3, 1–14. [Google Scholar]
- Igual, J.M.; Valverde, A.; Cervantes, E.; Velázquez, E. Phosphate-solubilizing bacteria as inoculants for agriculture: Use of updated molecular techniques in their study. Agronomie 2001, 21, 561–568. [Google Scholar] [CrossRef]
- Jorquera, M.A.; Crowley, D.E.; Marschner, P.; Greiner, R.; Fernández, M.T.; Romero, D.; Menezes-Blackburn, D.; De La Luz Mora, M. Identification of b-propeller phytase-encoding genes in culturable Paenibacillus and Bacillus sp. from the rhizosphere of pasture plants on volcanic soils. FEMS Microbiol. Ecol. 2011, 75, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Wouter, J.T.M.; Buijsman, P.J. Secretion of alkaline phosphatase by Bacillus licheniformis 749/C during growth in batch and chemostat cultures. FEMS Microbiol. 1980, 7, 91–95. [Google Scholar] [CrossRef]
- Fujita, K.; Kunito, T.; Moro, H.; Toda, H.; Otsuka, S.; Nagaoka, K. Microbial resource allocation for phosphatase synthesis reflects the availability of inorganic phosphorus across various soils. Biogeochemistry 2017, 136, 325–339. [Google Scholar] [CrossRef]
- Magadlela, A.; Lembede, Z.; Egbewale, S.O.; Olaniran, A.O. The metabolic potential of soil microorganisms and enzymes in phosphorus deficient KwaZulu Natal grassland ecosystem soil. Appl. Soil Ecol. 2023, 181, 104647. [Google Scholar] [CrossRef]
- Grobbelaar, N.; Hattingh, W.; Marshall, J. The occurrence of coralloid roots on the South African species of the Cycadales and their ability to fix nitrogen symbiotically. S. Afr. J. Bot. 1986, 52, 467–471. [Google Scholar] [CrossRef]
- Lindblad, P.; Rai, A.N.; Bergman, B. The Cycas revoluta-Nostoc symbiosis: Enzyme activities of nitrogen and carbon metabolism in the cyanobiont. J. Gen. Microbiol. 1987, 133, 1695–1699. [Google Scholar] [CrossRef]
- Peix, A.; Ramírez-Bahena, M.H.; Velázquez, E.; Bedmar, E.J. Bacterial associations with legumes. Crit. Rev. Pant Sci. 2015, 34, 17–42. [Google Scholar] [CrossRef]
- Martínez-Hidalgo, P.; Hirsch, A.M. The nodule microbiome: N2-fixing rhizobia do not live alone. Phytobiomes J. 2017, 1, 70–82. [Google Scholar] [CrossRef]




| Study Site | Oceanview | Rhebu | ||
|---|---|---|---|---|
| Parameter | Rhizosphere | Non-Rhizosphere Soils | Rhizosphere | Non-Rhizosphere Soils |
| P (mg/kg) | 6.1 ± 2.9 a | 3.86 ± 1.9 a | 22.95 ± 5.9 a | 13.23 ± 3.7 a |
| K (cmolc/kg) | 139.5 ± 33.6 a | 81.9 ± 12.5 a | 330.0 ± 100.9 a | 179.1 ± 54.9 a |
| N (mg/kg) | 3093.7 ± 741.9 a | 2638.0 ± 438.2 a | 5189.3 ± 297.9 a | 3473.3 ± 1396.3 a |
| Ca (cmolc/kg) | 1555.3 ± 274.8 a | 1379.2 ± 153.5 a | 1534.9 ± 974.2 a | 1229.4 ± 1087.2 a |
| Mg (cmolc/kg) | 454.7 ± 46.1 a | 382.12 ± 35.3 b | 838.5 ± 549.3 a | 697.1 ± 548.2 a |
| Mn (mg/kg) | 55.3 ± 21.1 a | 44.8 ± 15.9 a | 89.7 ± 58.0 a | 48.1 ± 20.2 a |
| Zn (mg/kg) | 2.2 ± 0.1 c | 1.3 ± 0.4 d | 4.1 ± 1.6 a | 2.8 ± 1.2 a |
| Cu (mg/kg) | 1.1 ± 0.2 a | 0.6 ± 0.1 a | 0.9 ± 0.1 a | 0.6 ± 0.2 a |
| Exchange acidity (cmol/L) | 0.05 ± 0.01 a | 0.05 ± 0.01 a | 1.46 ± 1.42 a | 1.37 ± 1.19 a |
| Total cations (cmol/L) | 11.53 ± 1.65 a | 11.65 ± 1.62 a | 15.26 ± 8.13 a | 15.10 ± 9.48 a |
| pH | 5.43 ± 0.41 e | 4.97 ± 0.57 f | 4.28 ± 0.61 a | 4.14 ± 0.58 a |
| Oceanview | Rhebu | |||||||
|---|---|---|---|---|---|---|---|---|
| Rhizosphere Soils | Non-Rhizosphere Soils | Rhizosphere Soils | Non-Rhizosphere Soils | |||||
| N | P | N | P | N | P | N | P | |
| Nitrate reductase (µmol/h/g) | 0.89 * | - | 0.98 * | - | 0.94 * | - | 0.70 | - |
| N-acetylglucosaminidase (nmol/h/g) | 0.93 * | - | 0.98 * | - | 0.78 * | - | 0.89 * | - |
| Acid phosphatase (nmol/h/g) | - | 0.68 | - | 0.96 * | - | 0.98 * | - | 0.84 * |
| Alkaline phosphatase (nmol/h/g) | - | 0.98 * | - | 0.97 * | - | 0.97 * | - | 0.76 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Motsomane, N.; Suinyuy, T.N.; Pérez-Fernández, M.A.; Magadlela, A. Exploring the Influence of Ecological Niches and Hologenome Dynamics on the Growth of Encephalartos villosus in Scarp Forests. Soil Syst. 2024, 8, 21. https://doi.org/10.3390/soilsystems8010021
Motsomane N, Suinyuy TN, Pérez-Fernández MA, Magadlela A. Exploring the Influence of Ecological Niches and Hologenome Dynamics on the Growth of Encephalartos villosus in Scarp Forests. Soil Systems. 2024; 8(1):21. https://doi.org/10.3390/soilsystems8010021
Chicago/Turabian StyleMotsomane, Nqobile, Terence N. Suinyuy, María A. Pérez-Fernández, and Anathi Magadlela. 2024. "Exploring the Influence of Ecological Niches and Hologenome Dynamics on the Growth of Encephalartos villosus in Scarp Forests" Soil Systems 8, no. 1: 21. https://doi.org/10.3390/soilsystems8010021
APA StyleMotsomane, N., Suinyuy, T. N., Pérez-Fernández, M. A., & Magadlela, A. (2024). Exploring the Influence of Ecological Niches and Hologenome Dynamics on the Growth of Encephalartos villosus in Scarp Forests. Soil Systems, 8(1), 21. https://doi.org/10.3390/soilsystems8010021





