A Perspective on Information Optimality in a Neural Circuit and Other Biological Systems
Abstract
:1. Introduction
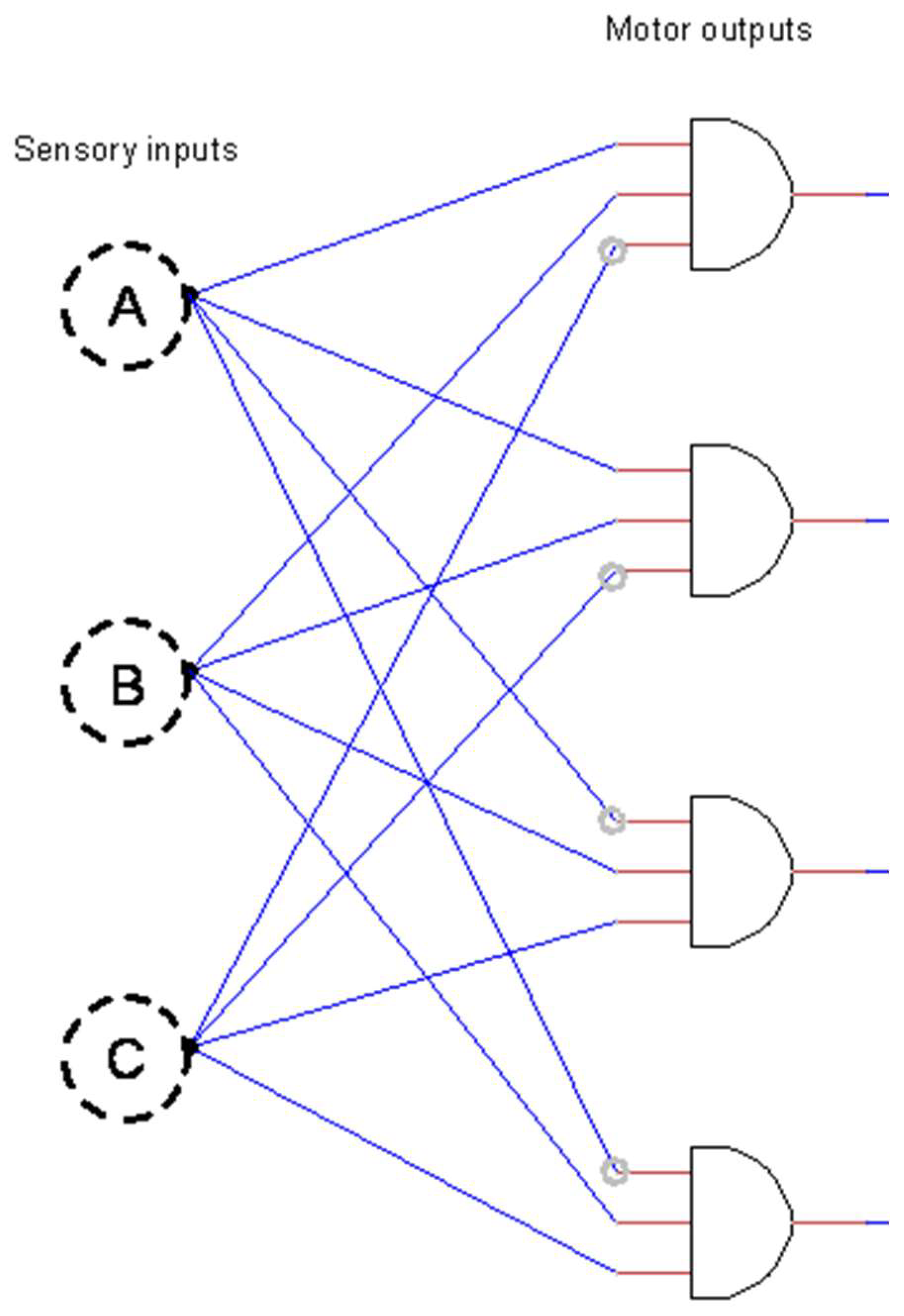
1.1. The Logic Gate Model
1.2. Efficiency of Mathematical Operations
1.3. Optimality of a Neuronal System
1.4. Biological Model of a Neuron
1.5. Approaches of This Study
2. Methods
2.1. Data Retrieval
2.2. Data Processing and Visualization
2.3. Logic Gate Analysis
3. Results
3.1. Optimality of an Idealized Neural Circuit
3.2. Efficiency of Mathematical Operations in a Neural Circuit
4. Discussion
4.1. The Utility of a Logic Gate Model
4.2. Information Processing as an Algorithm in Biological Systems
4.2.1. Overview of Information-Based Systems
4.2.2. Genetic Inheritance
4.2.3. Cellular Immunity in Jawed Vertebrates
4.2.4. Neural Systems in Animals
5. Conclusions
5.1. Information-Based Perspective of Biological Processes
5.2. Future Directions of Study
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McCulloch, W.S.; Pitts, W. A logical calculus of the ideas immanent in nervous activity. Bull. Math. Biophys. 1943, 5, 115–133. [Google Scholar] [CrossRef]
- Boole, G. The Mathematical Analysis of Logic, Being an Essay towards a Calculus of Deductive Reasoning; Macmillan, Barclay, & Macmillan: London, UK, 1847. [Google Scholar]
- Leibniz, G.W.; Gerhardt, C.I. Die Philosophischen Schriften VII; Weidmannsche Buchhandlung: Berlin, Germany, 1890; pp. 236–247. [Google Scholar]
- Malink, M. The logic of Leibniz’s Generales Inquisitiones de Analysi Notionum et Veritatum. Rev. Symb. Log. 2016, 9, 686–751. [Google Scholar] [CrossRef] [Green Version]
- Schmidhuber, J. 1931: Kurt Godel, Founder of THEORETICAL computer Science, Shows Limits of Math, Logic, Computing, and Artificial Intelligence. Available online: people.idsia.ch/~juergen/goedel-1931-founder-theoretical-computer-science-AI.html (accessed on 4 April 2022).
- Leibniz, G.W. De Progressione Dyadica Pars I. 1679; Herrn von Leibniz’ Rechnung mit Null und Einz; Hochstetter, E., Greve, H.-J., Eds.; Siemens Aktiengesellschaft: Berlin, Germany, 1966. [Google Scholar]
- Smith, M.; Pereda, A.E. Chemical synaptic activity modulates nearby electrical synapses. Proc. Natl. Acad. Sci. USA 2003, 100, 4849–4854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reigl, M.; Alon, U.; Chklovskii, D.B. Search for computational modules in the C. elegans brain. BMC Biol. 2004, 2, 25. [Google Scholar] [CrossRef] [Green Version]
- Gray, J.M.; Hill, J.J.; Bargmann, C.I. A circuit for navigation in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2005, 102, 3184–3191. [Google Scholar] [CrossRef] [Green Version]
- Varshney, L.R.; Chen, B.L.; Paniagua, E.; Hall, D.H.; Chklovskii, D.B. Structural properties of the C. elegans neuronal network. PLoS Comput. Biol. 2011, 7, e1001066. [Google Scholar] [CrossRef] [Green Version]
- Bargmann, C.I. Beyond the connectome: How neuromodulators shape neural circuits. Bioessays 2012, 34, 458–465. [Google Scholar] [CrossRef]
- Zhen, M.; Samuel, A.D. C. elegans locomotion: Small circuits, complex functions. Curr. Opin. Neurobiol. 2015, 33, 117–126. [Google Scholar] [CrossRef]
- Niven, J.E.; Chittka, L. Evolving understanding of nervous system evolution. Curr. Biol. 2016, 26, R937–R941. [Google Scholar] [CrossRef] [Green Version]
- Rakowski, F.; Karbowski, J. Optimal synaptic signaling connectome for locomotory behavior in Caenorhabditis elegans: Design minimizing energy cost. PLoS Comput. Biol. 2017, 13, e1005834. [Google Scholar] [CrossRef] [Green Version]
- Jabeen, S.; Thirumalai, V. The interplay between electrical and chemical synaptogenesis. J. Neurophysiol. 2018, 120, 1914–1922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karbowski, J. Deciphering neural circuits for Caenorhabditis elegans behavior by computations and perturbations to genome and connectome. Curr. Opin. Syst. Biol. 2019, 13, 44–51. [Google Scholar] [CrossRef]
- Niebur, E.; Erdos, P. Theory of the locomotion of nematodes: Control of the somatic motor neurons by interneurons. Math. Biosci. 1993, 118, 51–82. [Google Scholar] [CrossRef]
- Kamhi, J.F.; Gronenberg, W.; Robson, S.K.; Traniello, J.F. Social complexity influences brain investment and neural operation costs in ants. Proc. R. Soc. B Biol. Sci. 2016, 283, 20161949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Traniello, I.M.; Chen, Z.; Bagchi, V.A.; Robinson, G.E. Valence of social information is encoded in different subpopulations of mushroom body Kenyon cells in the honeybee brain. Proc. R. Soc. B 2019, 286, 20190901. [Google Scholar] [CrossRef] [Green Version]
- Cover, T.M.; Thomas, J.A. Information Theory and Statistics. In Elements of Information Theory, 1st ed.; John Wiley & Sons: New York, NY, USA, 1991; pp. 279–335. [Google Scholar]
- Borst, A.; Helmstaedter, M. Common circuit design in fly and mammalian motion vision. Nat. Neurosci. 2015, 18, 1067–1076. [Google Scholar] [CrossRef]
- Groschner, L.N.; Malis, J.G.; Zuidinga, B.; Borst, A. A biophysical account of multiplication by a single neuron. Nature 2022, 603, 119–123. [Google Scholar] [CrossRef]
- Schnupp, J.W.; King, A.J. Neural processing: The logic of multiplication in single neurons. Curr. Biol. 2001, 11, R640–R642. [Google Scholar] [CrossRef] [Green Version]
- Laughlin, S.B.; Sejnowski, T.J. Communication in neuronal networks. Science 2003, 301, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.L.; Hall, D.H.; Chklovskii, D.B. Wiring optimization can relate neuronal structure and function. Proc. Natl. Acad. Sci. USA 2006, 103, 4723–4728. [Google Scholar] [CrossRef] [Green Version]
- Bullmore, E.; Sporns, O. The economy of brain network organization. Nat. Rev. Neurosci. 2012, 13, 336–349. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Vertes, P.E.; Towlson, E.K.; Chew, Y.L.; Walker, D.S.; Schafer, W.R.; Barabasi, A.L. Network control principles predict neuron function in the Caenorhabditis elegans connectome. Nature 2017, 550, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Poznanski, R.R. Dendritic integration in a recurrent network. J. Integr. Neurosci. 2002, 1, 69–99. [Google Scholar] [CrossRef]
- Watts, D.J.; Strogatz, S.H. Collective dynamics of ‘small-world’ networks. Nature 1998, 393, 440–442. [Google Scholar] [CrossRef] [PubMed]
- Towlson, E.K.; Vertes, P.E.; Ahnert, S.E.; Schafer, W.R.; Bullmore, E.T. The rich club of the C. elegans neuronal connectome. J. Neurosci. 2013, 33, 6380–6387. [Google Scholar] [CrossRef] [Green Version]
- Milo, R.; Shen-Orr, S.; Itzkovitz, S.; Kashtan, N.; Chklovskii, D.; Alon, U. Network motifs: Simple building blocks of complex networks. Science 2002, 298, 824–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Driscoll, M.; Kaplan, J. Mechanotransduction. In The Nematode C. elegans, II; Cold Spring Harbor Press, Cold Spring Harbor: New York, NY, USA, 1997; pp. 645–677. [Google Scholar]
- Wakabayashi, T.; Kitagawa, I.; Shingai, R. Neurons regulating the duration of forward locomotion in Caenorhabditis elegans. Neurosci. Res. 2004, 50, 103–111. [Google Scholar] [CrossRef]
- Chatterjee, N.; Sinha, S. Understanding the mind of a worm: Hierarchical network structure underlying nervous system function in C. elegans. Prog. Brain Res. 2008, 168, 145–153. [Google Scholar]
- Campbell, J.C.; Chin-Sang, I.D.; Bendena, W.G. Mechanosensation circuitry in Caenorhabditis elegans: A focus on gentle touch. Peptides 2015, 68, 164–174. [Google Scholar] [CrossRef]
- Poznanski, R.R. Biophysical Neural Networks: Foundations of Integrative Neuroscience; Mary Ann Liebert: New York, NY, USA, 2001; pp. 177–214. [Google Scholar]
- Goldental, A.; Guberman, S.; Vardi, R.; Kanter, I. A computational paradigm for dynamic logic-gates in neuronal activity. Front. Comput. Neurosci. 2014, 8, 52. [Google Scholar] [CrossRef] [Green Version]
- Lysiak, A.; Paszkiel, S. A Method to Obtain Parameters of One-Column Jansen–Rit Model Using Genetic Algorithm and Spectral Characteristics. Appl. Sci. 2021, 11, 677. [Google Scholar] [CrossRef]
- Odum, E.P. Energy flow in ecosystems—A historical review. Am. Zool. 1968, 8, 11–18. [Google Scholar] [CrossRef]
- Van Hemmen, J.L.; Sejnowski, T.J. (Eds.) 23 Problems in Systems Neuroscience; Oxford University Press: New York, NY, USA, 2005. [Google Scholar]
- Gerstner, W.; Kistler, W.M.; Naud, R.; Paninski, L. Neuronal Dynamics: From Single Neurons to Networks and Models of Cognition; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar]
- Brette, R. Philosophy of the Spike: Rate-Based vs. Spike-Based Theories of the Brain. Front. Syst. Neurosci. 2015, 9, 151. [Google Scholar] [CrossRef] [PubMed]
- Pregowska, A.; Szczepanski, J.; Wajnryb, E. Temporal code versus rate code for binary Information Sources. Neurocomputing 2016, 216, 756–762. [Google Scholar] [CrossRef] [Green Version]
- Pregowska, A.; Kaplan, E.; Szczepanski, J. How Far can Neural Correlations Reduce Uncertainty? Comparison of Information Transmission Rates for Markov and Bernoulli Processes. Int. J. Neural Syst. 2019, 29, 1950003. [Google Scholar] [CrossRef] [Green Version]
- Di Lorenzo, P.M.; Chen, J.Y.; Victor, J.D. Quality Time: Representation of a Multidimensional Sensory Domain through Temporal Coding. J. Neurosci. 2009, 29, 9227–9238. [Google Scholar] [CrossRef]
- Crumiller, M.; Knight, B.; Kaplan, E. The Measurement of Information Transmitted by a Neural Population: Promises and Challenges. Entropy 2013, 15, 3507–3527. [Google Scholar] [CrossRef] [Green Version]
- Saxena, S.; Cunningham, J.P. Towards the neural population doctrine. Curr. Opin. Neurobiol. 2019, 55, 103–111. [Google Scholar] [CrossRef]
- White, J.G.; Southgate, E.; Thomson, J.N.; Brenner, S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. B Biol. Sci. 1986, 314, 1–340. [Google Scholar]
- Albertson, D.G.; Thomson, J.N. The pharynx of Caenorhabditis elegans. Philos. Trans. R. Soc. B Biol. Sci. 1976, 275, 299–325. [Google Scholar]
- Durbin, R.M. Studies on the Development and Organisation of the Nervous System of Caenorhabditis elegans. Ph.D. Thesis, University of Cambridge, Cambridge, UK, 1987. [Google Scholar]
- Achacoso, T.B.; Yamamoto, W.S. AY’s Neuroanatomy of C. elegans for Computation; CRC Press: Boca Raton, FL, USA, 1992. [Google Scholar]
- Hall, D.H.; Russell, R.L. The posterior nervous system of the nematode Caenorhabditis elegans: Serial reconstruction of identified neurons and complete pattern of synaptic interactions. J. Neurosci. 1991, 11, 1–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hobert, O.; Hall, D.H. Neuroanatomy: A second look with GFP reporters and some comments. Worm Breeder’s Gazette 1999, 16, 24. [Google Scholar]
- Wild, D.J. MINITAB Release 14. J. Chem. Inf. Modeling 2005, 45, 212. [Google Scholar] [CrossRef]
- Pyne, M. TinyCAD Source Code (version 2.90.00). Available online: Sourceforge.net/projects/tinycad (accessed on 4 April 2022).
- Brayton, R.K.; Hachtel, G.D.; McMullen, C.T.; Sangiovanni-Vincentelli, A.L. Logic Minimization Algorithms for VLSI Synthesis; Kluwer Academic: Berlin, Germany, 1985. [Google Scholar]
- Alom, M.Z.; Taha, T.M.; Yakopcic, C.; Westberg, S.; Sidike, P.; Nasrin, M.S.; Hasan, M.; Van Essen, B.C.; Awwal, A.A.S.; Asari, V.K. A State-of-the-Art Survey on Deep Learning Theory and Architectures. Electronics 2019, 8, 292. [Google Scholar] [CrossRef] [Green Version]
- Bengio, Y.; Lecun, Y.; Hinton, G. Deep learning for AI. Commun. ACM 2021, 64, 58–65. [Google Scholar] [CrossRef]
- Friedman, R. Test of robustness of pharyngeal neural networks in Caenorhabditis elegans. NeuroReport 2021, 32, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.; Figueroa, F. Evolution of the major histocompatibility complex. Crit. Rev. Immunol. 1986, 6, 295–386. [Google Scholar]
- Germain, R.N. MHC-dependent antigen processing and peptide presentation: Providing ligands for T lymphocyte activation. Cell 1994, 76, 287–299. [Google Scholar] [CrossRef]
- Davis, M.M.; Bjorkman, P.J. T-cell antigen receptor genes and T-cell recognition. Nature 1988, 334, 395–402. [Google Scholar] [CrossRef]
- Nikolich-Zugich, J.; Slifka, M.K.; Messaoudi, I. The many important facets of T-cell repertoire diversity. Nat. Rev. Immunol. 2004, 4, 123–132. [Google Scholar] [CrossRef]
- Wucherpfennig, K.W. The structural interactions between T cell receptors and MHC-peptide complexes place physical limits on self-nonself discrimination. Curr. Top. Microbiol. Immunol. 2005, 296, 19–37. [Google Scholar] [PubMed]
- Starr, T.K.; Jameson, S.C.; Hogquist, K.A. Positive and negative selection of T cells. Annu. Rev. Immunol. 2003, 21, 139–176. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, T.J.; Rubinsteyn, A.; Laserson, U. MHCflurry 2.0: Improved Pan-Allele Prediction of MHC Class I-Presented Peptides by Incorporating Antigen Processing. Cell Syst. 2020, 11, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Montemurro, A.; Schuster, V.; Povlsen, H.R.; Bentzen, A.K.; Jurtz, V.; Chronister, W.D.; Crinklaw, A.; Hadrup, S.R.; Winther, O.; Peters, B.; et al. NetTCR-2.0 enables accurate prediction of TCR-peptide binding by using paired TCRα and β sequence data. Commun. Biol. 2021, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Clarke, A.; Tyler, L.K. Understanding what we see: How we derive meaning from vision. Trends Cogn. Sci. 2015, 19, 677–687. [Google Scholar] [CrossRef] [Green Version]
- Engel, A.K.; Konig, P.; Singer, W. Direct physiological evidence for scene segmentation by temporal coding. Proc. Natl. Acad. Sci. USA 1991, 88, 9136–9140. [Google Scholar] [CrossRef] [Green Version]
- Lamme, V.A. Why visual attention and awareness are different. Trends Cogn. Sci. 2003, 7, 12–18. [Google Scholar] [CrossRef]
- Spratling, M.W. A review of predictive coding algorithms. Brain Cogn. 2017, 112, 92–97. [Google Scholar] [CrossRef] [Green Version]
- Itti, L.; Koch, C. Computational modelling of visual attention. Nat. Rev. Neurosci. 2001, 2, 194–203. [Google Scholar] [CrossRef] [Green Version]
- Barrett, T.W. Conservation of information. Acta Acust. United Acust. 1972, 27, 44–47. [Google Scholar]
- Chang, L.; Tsao, D.Y. The code for facial identity in the primate brain. Cell 2017, 169, 1013–1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leibniz, G. Dissertatio de Arte Combinatoria, 1666; Akademie Verlag: Berlin, Germany, 1923; Manuscript later published in Smtliche Schriften und Briefe. [Google Scholar]
- Stringer, C.; Pachitariu, M.; Steinmetz, N.; Carandini, M.; Harris, K.D. High-dimensional geometry of population responses in visual cortex. Nature 2019, 571, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Goni, J.; Avena-Koenigsberger, A.; de Mendizabal, N.V.; van den Heuvel, M.P.; Betzel, R.F.; Sporns, O. Exploring the Morphospace of Communication Efficiency in Complex Networks. PLoS ONE 2013, 8, e58070. [Google Scholar] [CrossRef] [PubMed]
- Hinton, G.E. Connectionist learning procedures. Artif. Intell. 1989, 40, 185–234. [Google Scholar] [CrossRef] [Green Version]
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef] [Green Version]
- Silver, D.; Singh, S.; Precup, D.; Sutton, R.S. Reward is enough. Artif. Intell. 2021, 299, 103535. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Hekkelman, M.L.; de Vries, I.; Joosten, R.P.; Perrakis, A. AlphaFill: Enriching the AlphaFold models with ligands and co-factors. bioRxiv 2021. bioRxiv: 2021.11.26.470110. [Google Scholar]
- Gao, M.; Nakajima An, D.; Parks, J.M.; Skolnick, J. Predicting direct physical interactions in multimeric proteins with deep learning. Nat. Commun. 2022, 13, 1744. [Google Scholar] [CrossRef]
- Akdel, M.; Pires, D.E.; Pardo, E.P.; Janes, J.; Zalevsky, A.O.; Mészáros, B.; Bryant, P.; Good, L.L.; Laskowski, R.A.; Pozzati, G.; et al. A structural biology community assessment of AlphaFold 2 applications. bioRxiv 2021. bioRxiv: 2021.09.26.461876. [Google Scholar]
- Mirdita, M.; Schutze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold-Making protein folding accessible to all. bioRxiv 2021. bioRxiv: 2021.08.15.456425. [Google Scholar] [CrossRef] [PubMed]






| OR/NOT | AND/NOT | ||
|---|---|---|---|
| Inputs | Outputs | Inputs | Outputs |
| 0 0 0 | 0 0 0 0 | 0 0 0 | 0 0 0 0 |
| 1 0 0 | 1 1 0 0 | 1 0 0 | 0 0 0 0 |
| 0 1 0 | 0 0 0 0 | 0 1 0 | 0 0 0 0 |
| 0 0 1 | 0 0 1 1 | 0 0 1 | 0 0 0 0 |
| 1 1 0 | 1 1 0 0 | 1 1 0 | 1 1 0 0 |
| 1 0 1 | 0 0 0 0 | 1 0 1 | 0 0 0 0 |
| 0 1 1 | 0 0 1 1 | 0 1 1 | 0 0 1 1 |
| 1 1 1 | 0 0 0 0 | 1 1 1 | 0 0 0 0 |
| Mathematical Operations | |||
|---|---|---|---|
| Input 1 | Input 2 | Output (Multiplication) | Output (Division) |
| A A | B B | AA × BB = | AA/BB = QQ (ERRR) |
| 0 0 | 0 0 | 0 0 0 0 | 0 0 1 0 0 0 |
| 0 0 | 0 1 | 0 0 0 0 | 0 0 0 0 0 0 |
| 0 0 | 1 0 | 0 0 0 0 | 0 0 0 0 0 0 |
| 0 0 | 1 1 | 0 0 0 0 | 0 0 0 0 0 0 |
| 0 1 | 0 0 | 0 0 0 0 | 0 0 1 0 0 0 |
| 0 1 | 0 1 | 0 0 0 1 | 0 1 0 0 0 0 |
| 0 1 | 1 0 | 0 0 1 0 | 0 0 0 1 0 1 |
| 0 1 | 1 1 | 0 0 1 1 | 0 0 0 0 1 1 |
| 1 0 | 0 0 | 0 0 0 0 | 0 0 1 0 0 0 |
| 1 0 | 0 1 | 0 0 1 0 | 1 0 0 0 0 0 |
| 1 0 | 1 0 | 0 1 0 0 | 0 1 0 0 0 0 |
| 1 0 | 1 1 | 0 1 1 0 | 0 0 0 1 1 0 |
| 1 1 | 0 0 | 0 0 0 0 | 0 0 1 0 0 0 |
| 1 1 | 0 1 | 0 0 1 1 | 1 1 0 0 0 0 |
| 1 1 | 1 0 | 0 1 1 0 | 0 1 0 1 0 1 |
| 1 1 | 1 1 | 1 0 0 1 | 0 1 0 0 0 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Friedman, R. A Perspective on Information Optimality in a Neural Circuit and Other Biological Systems. Signals 2022, 3, 410-427. https://doi.org/10.3390/signals3020025
Friedman R. A Perspective on Information Optimality in a Neural Circuit and Other Biological Systems. Signals. 2022; 3(2):410-427. https://doi.org/10.3390/signals3020025
Chicago/Turabian StyleFriedman, Robert. 2022. "A Perspective on Information Optimality in a Neural Circuit and Other Biological Systems" Signals 3, no. 2: 410-427. https://doi.org/10.3390/signals3020025
APA StyleFriedman, R. (2022). A Perspective on Information Optimality in a Neural Circuit and Other Biological Systems. Signals, 3(2), 410-427. https://doi.org/10.3390/signals3020025






