Automatic Identification of Children with ADHD from EEG Brain Waves
Abstract
1. Introduction
- An EEG ML pipeline is presented for ADHD detection, explaining each stage of the pipeline (including signal preprocessing and data preparation) with thorough explanations and rationale.
- Unlike other studies, we employed only the first four sub-bands of EEG, eliminating signals more than 30 Hz and thus reducing the computational load for ML model training while keeping mean accuracy of 93.2%.
- Simple EEG linear features are emphasized in our proposed model development, whereas other works were only based on complex nonlinear features.
- The model was trained on a large dataset of 120 children (the highest of other models was 49) collected from two different sessions at two different places, eliminating the measurement bias in data collection. Also, the experimental setup was child-friendly, easy to reproduce in local settings, and could be employed for future ADHD detection.
- We also performed rigorous validation (unlike other works) to ensure that our model is not impacted by bias and overfitting, which commonly appear in the ML pipeline.
2. Related Works
3. Materials and Methods
4. Preprocessing
5. Feature Extraction and Feature Selection
6. Result
7. Discussion
- To improve the accuracy, we may need to evaluate more features through the use of different machine learning models for comparison of results.
- We will also try different window sizes (0.5 s or 5 s, for example) in future studies.
- We will next work with only two EEG frequency bands (theta and beta), as these two have significant changes in ADHD patients, and investigate more into the sub-band of the EEG signals.
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Attention-Deficit/Hyperactivity Disorder (ADHD) in Children. Available online: https://www.hopkinsmedicine.org/health/conditions-and-diseases/adhdadd (accessed on 18 November 2022).
- Attention-Deficit/Hyperactivity Disorder (ADHD) in Children—Symptoms and Causes. Available online: https://www.mayoclinic.org/diseases-conditions/adhd/symptoms-causes/syc-20350889 (accessed on 18 November 2022).
- Kessler, R.C.; Adler, L.A.; Berglund, P.; Green, J.G.; McLaughlin, K.A.; Fayyad, J.; Russo, L.J.; Sampson, N.A.; Shahly, V.; Zaslavsky, A.M. The Effects of Temporally Secondary Co-Morbid Mental Disorders on the Associations of DSM-IV ADHD with Adverse Outcomes in the US National Comorbidity Survey Replication Adolescent Supplement (NCS-A). Psychol. Med. 2014, 44, 1779–1792. [Google Scholar] [CrossRef] [PubMed]
- Vyvanse® (Lisdexamfetamine Dimesylate). Available online: https://www.vyvanse.com/what-is-adhd (accessed on 18 November 2022).
- Parenting a Child with ADHD (for Parents)—Nemours KidsHealth. Available online: https://kidshealth.org/en/parents/parenting-kid-adhd.html (accessed on 18 November 2022).
- Kooij, S.J.J.; Bejerot, S.; Blackwell, A.; Caci, H.; Casas-Brugué, M.; Carpentier, P.J.; Edvinsson, D.; Fayyad, J.; Foeken, K.; Fitzgerald, M.; et al. European Consensus Statement on Diagnosis and Treatment of Adult ADHD: The European Network Adult ADHD. BMC Psychiatry 2010, 10, 67. [Google Scholar] [CrossRef] [PubMed]
- Diagnostic and Statistical Manual of Mental Disorders. Available online: https://dsm.psychiatryonline.org/doi/book/10.1176/appi.books.9780890425596 (accessed on 18 November 2022).
- Van den Heuvel, M.P.; Sporns, O. Network Hubs in the Human Brain. Trends Cogn. Sci. 2013, 17, 683–696. [Google Scholar] [CrossRef] [PubMed]
- Lubar, J.F. Discourse on the Development of EEG Diagnostics and Biofeedback for Attention-Deficit/Hyperactivity Disorders. Biofeedback Self-Regul. 1991, 16, 201–225. [Google Scholar] [CrossRef] [PubMed]
- Fotuhi, M. Can Neurofeedback Effectively Treat ADHD? Available online: https://neurogrow.com/can-neurofeedback-effectively-treat-adhd/ (accessed on 18 November 2022).
- Ketola, E.C.; Barankovich, M.; Schuckers, S.; Ray-Dowling, A.; Hou, D.; Imtiaz, M.H. Channel Reduction for an EEG-Based Authentication System while Performing Motor Movements. Sensors 2022, 22, 9156. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.; Ahmad, M. Modeling of Human Emotion with Effective Frequency Band during a Test of Sustained Mental Task. In Proceedings of the 2015 IEEE International WIE Conference on Electrical and Computer Engineering (WIECON-ECE), Dhaka, Bangladesh, 19–20 December 2015; pp. 403–406. [Google Scholar] [CrossRef]
- Mazaheri, A.; Coffey-Corina, S.; Mangun, G.R.; Bekker, E.M.; Berry, A.S.; Corbett, B.A. Functional Disconnection of Frontal Cortex and Visual Cortex in Attention-Deficit/Hyperactivity Disorder. Biol. Psychiatry 2010, 67, 617–623. [Google Scholar] [CrossRef]
- Rubia, K.; Cubillo, A.; Smith, A.B.; Woolley, J.; Heyman, I.; Brammer, M.J. Disorder-Specific Dysfunction in Right Inferior Prefrontal Cortex during Two Inhibition Tasks in Boys with Attention-Deficit Hyperactivity Disorder Compared to Boys with Obsessive–Compulsive Disorder. Hum. Brain Mapp. 2010, 31, 287–299. [Google Scholar] [CrossRef]
- Rubia, K.; Halari, R.; Smith, A.B.; Mohammad, M.; Scott, S.; Brammer, M.J. Shared and Disorder-Specific Prefrontal Abnormalities in Boys with Pure Attention-Deficit/Hyperactivity Disorder Compared to Boys with Pure CD during Interference Inhibition and Attention Allocation. J. Child Psychol. Psychiatry 2009, 50, 669–678. [Google Scholar] [CrossRef]
- An Introduction to Machine Learning—GeeksforGeeks. Available online: https://www.geeksforgeeks.org/introduction-machine-learning/ (accessed on 23 December 2022).
- The History of ADHD. Available online: https://www.healthline.com/health/adhd/history (accessed on 19 January 2023).
- Yin, W.; Li, T.; Mucha, P.J.; Cohen, J.R.; Zhu, H.; Zhu, Z.; Lin, W. Altered Neural Flexibility in Children with Attention-Deficit/Hyperactivity Disorder. Mol. Psychiatry 2022, 27, 4673–4679. [Google Scholar] [CrossRef]
- Pulini, A.A.; Kerr, W.T.; Loo, S.K.; Lenartowicz, A. Classification Accuracy of Neuroimaging Biomarkers in Attention-Deficit/Hyperactivity Disorder: Effects of Sample Size and Circular Analysis. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 2019, 4, 108–120. [Google Scholar] [CrossRef]
- Kiiski, H.; Rueda-Delgado, L.M.; Bennett, M.; Knight, R.; Rai, L.; Roddy, D.; Grogan, K.; Bramham, J.; Kelly, C.; Whelan, R. Functional EEG Connectivity Is a Neuromarker for Adult Attention Deficit Hyperactivity Disorder Symptoms. Clin. Neurophysiol. 2020, 131, 330–342. [Google Scholar] [CrossRef]
- Alchalabi, A.E.; Shirmohammadi, S.; Eddin, A.N.; Elsharnouby, M. FOCUS: Detecting ADHD Patients by an EEG-Based Serious Game. IEEE Trans. Instrum. Meas. 2018, 67, 1512–1520. [Google Scholar] [CrossRef]
- Ghassemi, F.; Hassan_Moradi, M.; Tehrani-Doost, M.; Abootalebi, V. Using Non-Linear Features of EEG for ADHD/Normal Participants’ Classification. Procedia Soc. Behav. Sci. 2012, 32, 148–152. [Google Scholar] [CrossRef]
- Mohammadi, M.R.; Khaleghi, A.; Nasrabadi, A.M.; Rafieivand, S.; Begol, M.; Zarafshan, H. EEG Classification of ADHD and Normal Children Using Non-Linear Features and Neural Network. Biomed. Eng. Lett. 2016, 6, 66–73. [Google Scholar] [CrossRef]
- Allahverdy, A.; Khorrami, A.; Mohammadi, M.; Motie Nasrabadi, A. Detecting ADHD Children Using the Attention Continuity as Nonlinear Feature of EEG. Front. Biomed. Technol. 2016, 3, 1–2. [Google Scholar]
- Nasrabadi, A.M. EEG Data for ADHD/Control Children 2020. Available online: https://ieee-dataport.org/open-access/eeg-data-adhd-control-children (accessed on 18 November 2022).
- Stein, D.J.; Phillips, K.A.; Bolton, D.; Fulford, K.W.M.; Sadler, J.Z.; Kendler, K.S. What Is a Mental/Psychiatric Disorder? From DSM-IV to DSM-V. Psychol. Med. 2010, 40, 1759–1765. [Google Scholar] [CrossRef]
- Ritalin Oral: Uses, Side Effects, Interactions, Pictures, Warnings & Dosing—WebMD. Available online: https://www.webmd.com/drugs/2/drug-9475/ritalin-oral/details (accessed on 18 January 2023).
- Direction of Information Flow between Brain Regions in ADHD and Healthy Children Based on EEG by Using Directed Phase Transfer Entropy | SpringerLink. Available online: https://link.springer.com/article/10.1007/s11571-021-09680-3 (accessed on 28 October 2022).
- 10–20 System (EEG). Wikipedia. 2022. Available online: https://en.wikipedia.org/w/index.php?title=10%E2%80%9320_system_(EEG)&oldid=1076814653 (accessed on 18 November 2022).
- Yourtherapysource Auditory and Visual Attention in Children with ADHD. Available online: https://www.yourtherapysource.com/blog1/2017/06/27/auditory-visual-attention-children-adhd/ (accessed on 18 November 2022).
- Feature Extraction. Available online: https://deepai.org/machine-learning-glossary-and-terms/feature-extraction (accessed on 23 December 2022).
- Amin, H.U.; Mumtaz, W.; Subhani, A.R.; Saad, M.N.M.; Malik, A.S. Classification of EEG Signals Based on Pattern Recognition Approach. Front. Comput. Neurosci. 2017, 11, 103. Available online: https://www.frontiersin.org/articles/10.3389/fncom.2017.00103 (accessed on 23 December 2022). [CrossRef] [PubMed]
- Rahman, A.; Chowdhury, M.E.H.; Khandakar, A.; Kiranyaz, S.; Zaman, K.S.; Reaz, M.B.I.; Islam, M.T.; Ezeddin, M.; Kadir, M.A. Multimodal EEG and Keystroke Dynamics Based Biometric System Using Machine Learning Algorithms. IEEE Access 2021, 9, 94625–94643. [Google Scholar] [CrossRef]
- Standard Deviation. Available online: https://www.nlm.nih.gov/nichsr/stats_tutorial/section2/mod8_sd.html (accessed on 9 December 2022).
- Root Mean Square (RMS)—Definition, Formula and RMS Error. Available online: https://byjus.com/maths/root-mean-square/ (accessed on 9 December 2022).
- Measures of Skewness and Kurtosis. Available online: https://www.itl.nist.gov/div898/handbook/eda/section3/eda35b.htm (accessed on 9 December 2022).
- Hjorth Parameters. Wikipedia. 2021. Available online: https://en.wikipedia.org/w/index.php?title=Hjorth_parameters&oldid=1022917018 (accessed on 9 December 2022).
- Cocconcelli, M.; Strozzi, M.; Cavalaglio Camargo Molano, J.; Rubini, R. Detectivity: A Combination of Hjorth’s Parameters for Condition Monitoring of Ball Bearings. Mech. Syst. Signal Process. 2022, 164, 108247. [Google Scholar] [CrossRef]
- Stephanie Shannon Entropy. Available online: https://www.statisticshowto.com/shannon-entropy/ (accessed on 9 December 2022).
- Spectral Density—Wikipedia. Available online: https://en.wikipedia.org/wiki/Spectral_density (accessed on 9 December 2022).
- What Is the Band Power Measurement? Technical Support Knowledge Center Open. Available online: https://edadocs.software.keysight.com/kkbopen/what-is-the-band-power-measurement-589310400.html (accessed on 9 December 2022).
- What Is Analysis of Variance (ANOVA)? Available online: https://www.tibco.com/reference-center/what-is-analysis-of-variance-anova (accessed on 27 January 2023).
- Feature Selection and Feature Transformation Using Classification Learner App—MATLAB & Simulink. Available online: https://www.mathworks.com/help/stats/feature-selection-and-feature-transformation.html (accessed on 28 December 2022).
- Darshan, M. Using ANOVA for Feature Selection in Python. Available online: https://machinehack.com/story/using-anova-for-feature-selection-in-python (accessed on 23 January 2023).
- Kumar, A. PCA Explained Variance Concepts with Python Example. Data Anal. 2022. [Google Scholar]
- What Is a Support Vector Machine (SVM)? Definition from Techopedia. Available online: http://www.techopedia.com/definition/30364/support-vector-machine-svm (accessed on 29 December 2022).
- Team, D. Kernel Functions-Introduction to SVM Kernel & Examples. Available online: https://data-flair.training/blogs/svm-kernel-functions/ (accessed on 29 December 2022).
- Kernel Method. Available online: https://www.engati.com/glossary/kernel-method (accessed on 29 January 2023).
- Awasthi, S. Seven Most Popular SVM Kernels. Dataaspirant. 2020. Available online: https://dataaspirant.com/svm-kernels/ (accessed on 29 December 2022).
- Major Kernel Functions in Support Vector Machine (SVM). GeeksforGeeks. 2020. Available online: https://www.geeksforgeeks.org/major-kernel-functions-in-support-vector-machine-svm/ (accessed on 25 January 2023).
- Brownlee, J. A Gentle Introduction to k-fold Cross-Validation. MachineLearningMastery.com. 2018. Available online: https://machinelearningmastery.com/k-fold-cross-validation/ (accessed on 30 December 2022).
- Allibhai, E. Holdout vs. Cross-Validation in Machine Learning Medium. 2018. Available online: https://medium.com/@eijaz/holdout-vs-cross-validation-in-machine-learning-7637112d3f8f (accessed on 9 December 2022).
- Cross Validation. Available online: https://www.cs.cmu.edu/~schneide/tut5/node42.html (accessed on 9 December 2022).
- Why Use K-Fold Cross Validation? KDnuggets. Available online: https://www.kdnuggets.com/2022/07/kfold-cross-validation.html (accessed on 17 December 2022).
- Introduction to K-Fold Cross-Validation in Python—SQLRelease. Available online: https://sqlrelease.com/introduction-to-k-fold-cross-validation-in-python (accessed on 25 January 2023).
- Stancin, I.; Cifrek, M.; Jovic, A. A Review of EEG Signal Features and Their Application in Driver Drowsiness Detection Systems. Sensors 2021, 21, 3786. [Google Scholar] [CrossRef] [PubMed]
- Turney, S. What Is Kurtosis? | Definition, Examples & Formula. Available online: https://www.scribbr.com/statistics/kurtosis/ (accessed on 25 January 2023).
- Wang, R.; Wang, J.; Yu, H.; Wei, X.; Yang, C.; Deng, B. Power Spectral Density and Coherence Analysis of Alzheimer’s EEG. Cogn. Neurodyn. 2015, 9, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Principal Component Analysis (PCA) Explained | Built In. Available online: https://builtin.com/data-science/step-step-explanation-principal-component-analysis (accessed on 30 December 2022).
- Joby, A. What Is Cross-Validation? Comparing Machine Learning Models. Available online: https://learn.g2.com/cross-validation (accessed on 29 December 2022).
- Richhariya, B.; Tanveer, M. EEG Signal Classification Using Universum Support Vector Machine. Expert Syst. Appl. 2018, 106, 169–182. [Google Scholar] [CrossRef]
- Raj, A. Everything about Support Vector Classification—Above and beyond. Available online: https://towardsdatascience.com/everything-about-svm-classification-above-and-beyond-cc665bfd993e (accessed on 31 December 2022).
- Blekh, A. Answer to ‘What Makes the Gaussian Kernel so Magical for PCA, and Also in General?’. Cross Validated. 2015. Available online: https://stats.stackexchange.com/questions/131138/what-makes-the-gaussian-kernel-so-magical-for-pca-and-also-in-general (accessed on 29 January 2023).
- Keelawat, P.; Thammasan, N.; Numao, M.; Kijsirikul, B. A Comparative Study of Window Size and Channel Arrangement on EEG-Emotion Recognition Using Deep CNN. Sensors 2021, 21, 1678. [Google Scholar] [CrossRef] [PubMed]
- Implementation of a Mind-Controlled Wheelchair. Available online: https://www.researchgate.net/publication/362075774_Implementation_of_a_Mind-Controlled_Wheelchair (accessed on 2 February 2023).
- Ketola, E.; Lloyd, C.; Shuhart, D.; Schmidt, J.; Morenz, R.; Khondker, A.; Imtiaz, M. Lessons Learned from the Initial Development of a Brain Controlled Assistive Device. In Proceedings of the 2022 IEEE 12th Annual Computing and Communication Workshop and Conference (CCWC), Las Vegas, NV, USA, 26–29 January 2022; pp. 0580–0585. [Google Scholar] [CrossRef]
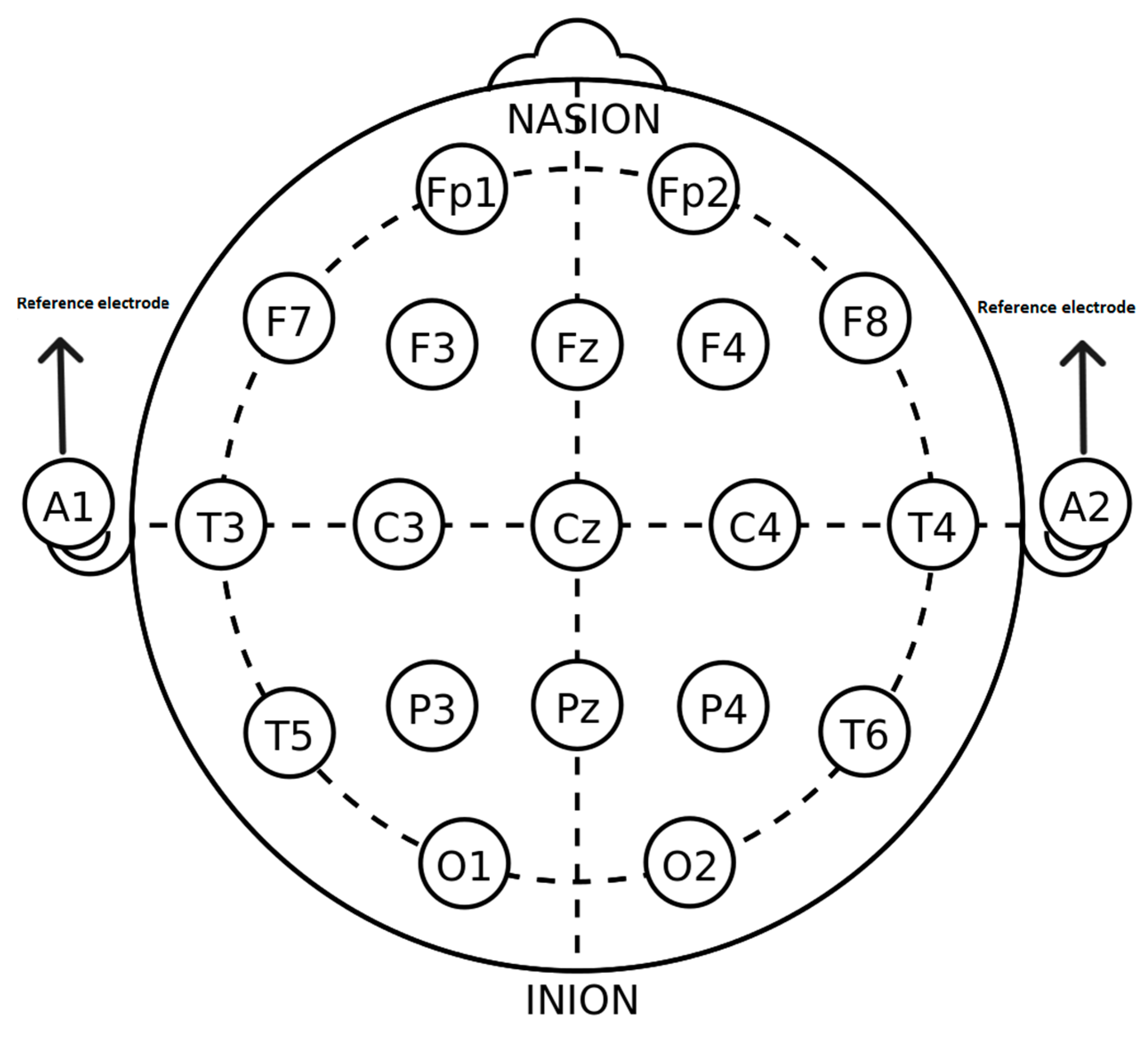
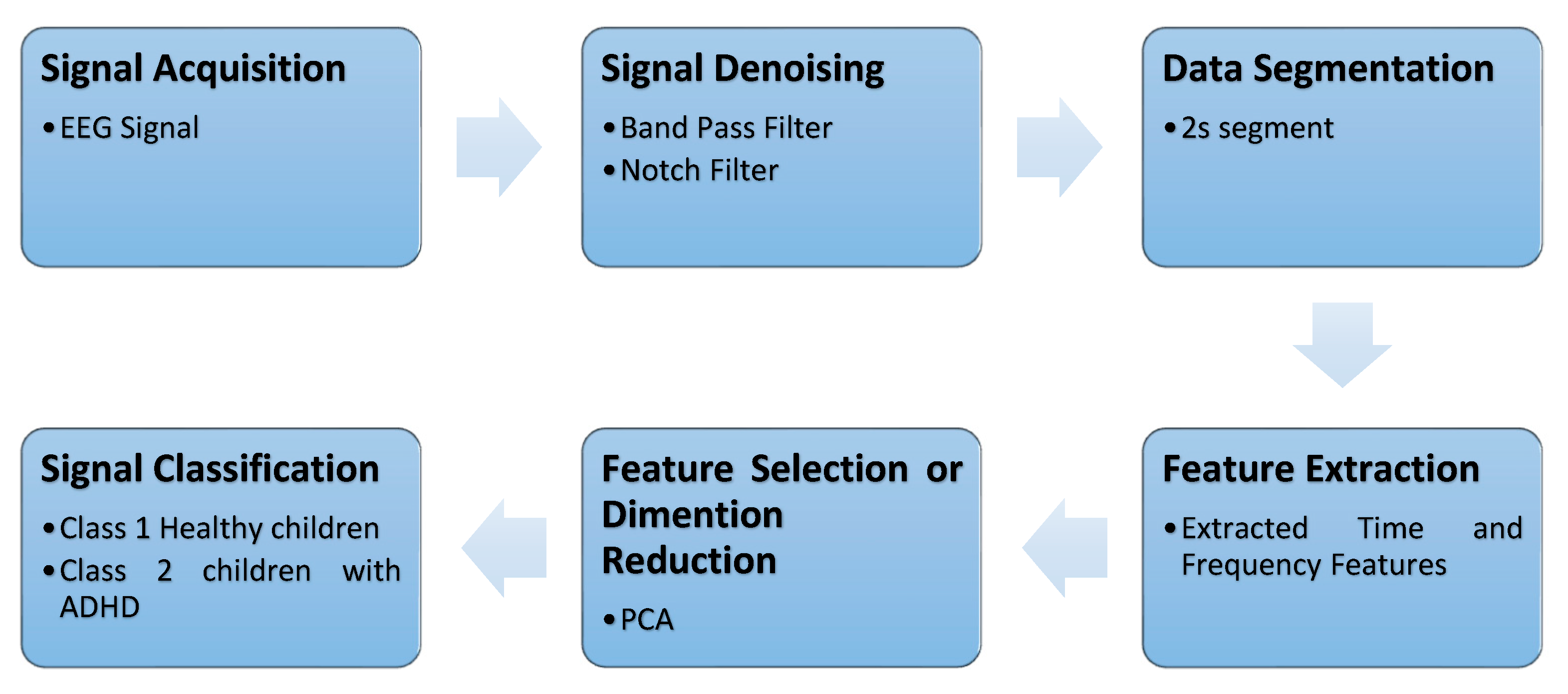
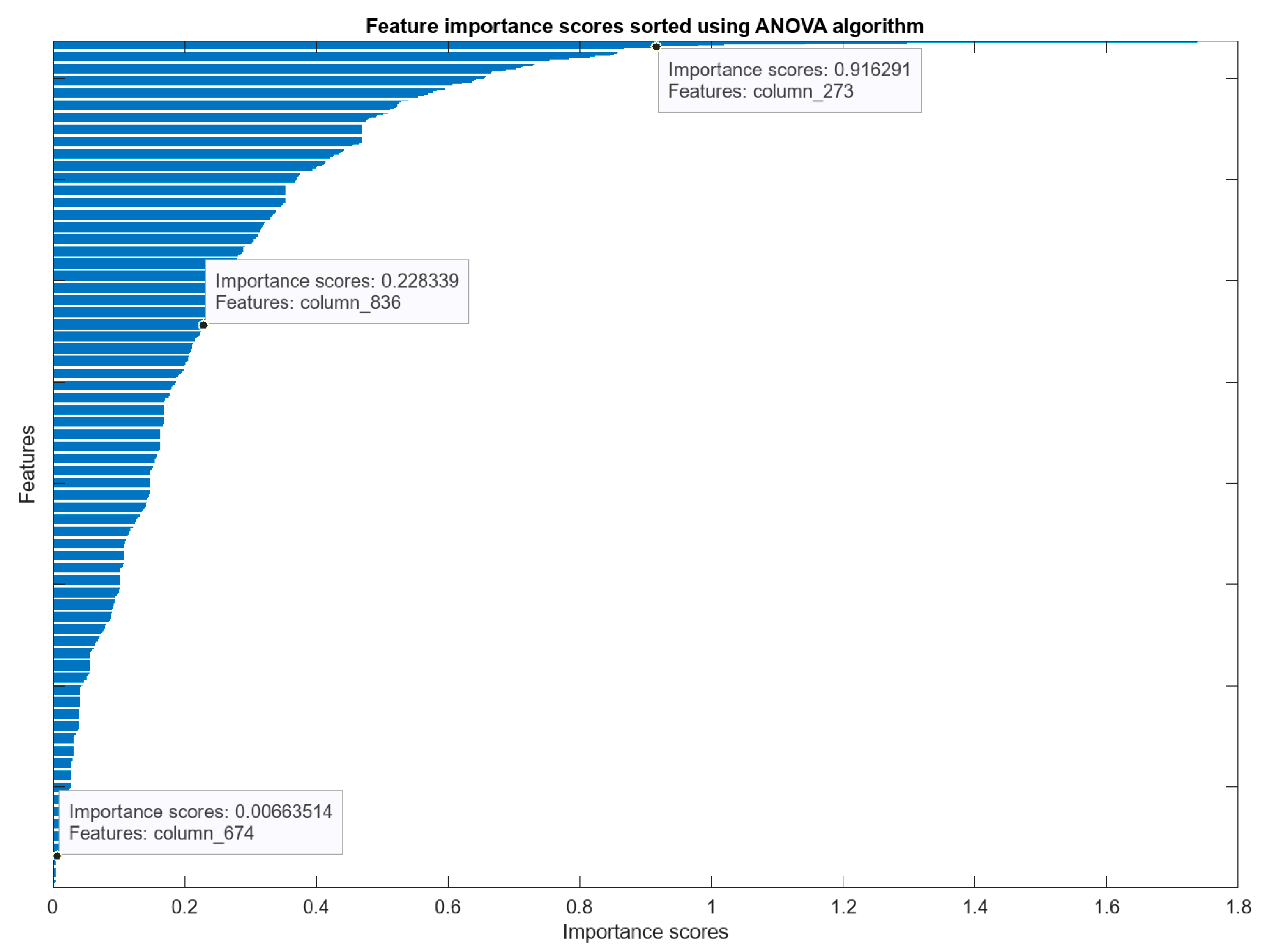

| Sub-Band | Frequency Range | Associated Brain Function |
|---|---|---|
| Delta | 0.5–4 Hz | Deep sleep or unconsciousness |
| Theta | 4–8 Hz | Sleep or drowsiness and recall |
| Alpha | 8–13 Hz | Eye closed and visual stimuli are limited |
| Beta | 13–30 Hz | Attentive to stimuli or problem-solving |
| Gamma | 30 Hz and above | Movement, emotional processing, and high-level mental activity |
| Boys | Girls | Age | Mean Age | Dominant Hand | |
|---|---|---|---|---|---|
| Healthy Children | 50 | 10 | 7–12 | 9.85 ± 1.77 | Right-Handed |
| ADHD | 48 | 12 | 7–12 | 9.62 ± 1.75 | Right-Handed |
| Feature Name | Definition | Mathematical Description |
|---|---|---|
| Standard Deviation | It is a statistical feature that is a measure of how spread out the data is to the mean. | xn = n-th data sample, N = Total No. of samples, µ = mean [33,34] |
| RMS | RMS is the square root-mean-square value of a signal | xn = n-th data sample, N = Total No. of samples [35] |
| Skewness | Skewness is the measure of the lack of symmetry from the mean of the dataset. | xn = n-th data sample, N = Total No. of samples, µ = mean [33] |
| Kurtosis | Kurtosis is a measure of whether the data are heavy-tailed or light-tailed relative to a normal distribution. | xn = n-th data sample, N = Total No. of samples, µ = mean [36] |
| Hjorth Activity | It is the variance of the amplitude of the signal in a time function. Represent the signal power. | Ha = var(x(t)) x(t) = amplitude of time-varying signal [37,38] |
| Hjorth Mobility | The mobility is the square root of the activity of the first derivative of the signal divided by the activity of the signal. Represents the mean frequency. | x′(t) = 1st derivation of the amplitude of the signal [37,38] |
| Hjorth Complexity | It represents the change in frequency. It is defined as the ratio between the mobility of the first derivative of the signal and the mobility of the signal. | [37,38] |
| Shannon’s Entropy | Shannon’s entropy measures the uncertainty/randomness in a dataset | Pn = probability of occurrence xn [39] |
| Spectral Entropy (SEN) | SEN is the normalized Shannon’s entropy | P = spectral of normalized frequency, N = number of frequencies in binary. [28] |
| Power Spectral Density (PSD) | PSD of the signal describes the power present in the signal as a function of frequency [40] | |
| Band Power | It measures both power and power spectral density in a specified channel bandwidth [41] |
| % Variance | Accuracy (Validation) | Accuracy (Test) | Confusion Matrix (Test) | |||
|---|---|---|---|---|---|---|
| TP | TN | FP | FN | |||
| 80% | 88.49% | 88.06% | 967 | 1263 | 166 | 124 |
| 85% | 92.18% | 92.51% | 1011 | 1312 | 122 | 75 |
| 90% | 93.17% | 93.24% | 994 | 1354 | 139 | 33 |
| 95% | 88.29% | 88.36% | 867 | 1358 | 266 | 29 |
| 97% | 85.0% | 85.5% | 769 | 1344 | 342 | 16 |
| Number | Accuracy (Validation) | Accuracy (Test) | Confusion Matrix (Test) | |||
|---|---|---|---|---|---|---|
| TP | TN | FP | FN | |||
| 1 | 93.4% | 93.9% | 990 | 1331 | 120 | 30 |
| 2 | 93.1% | 92.7% | 969 | 1322 | 141 | 39 |
| 3 | 93.0% | 93.8% | 992 | 1326 | 118 | 35 |
| 4 | 92.7% | 93.0% | 959 | 1339 | 152 | 21 |
| 5 | 92.3% | 93.6% | 982 | 1330 | 128 | 31 |
| 6 | 91.3% | 92.5% | 953 | 1332 | 158 | 28 |
| 7 | 93.3% | 93.6% | 987 | 1325 | 124 | 35 |
| 8 | 91.6% | 93.0% | 976 | 1321 | 135 | 39 |
| 9 | 93.1% | 93.2% | 981 | 1321 | 129 | 40 |
| 10 | 93.17% | 93.24% | 994 | 1354 | 139 | 33 |
| Mean | 93.2% | |||||
| STD | 0.44 | |||||
| % Variance | Accuracy (Validation) | Accuracy (Test) | Confusion Matrix (Test) | |||
|---|---|---|---|---|---|---|
| TP | TN | FP | FN | |||
| 80% | 89.3% | 89.7% | 650 | 828 | 90 | 79 |
| 85% | 92.7% | 93.9% | 681 | 866 | 59 | 41 |
| 90% | 93.4% | 94.2% | 665 | 886 | 75 | 21 |
| 95% | 88.5% | 88.3% | 560 | 895 | 180 | 12 |
| 97% | 84.8% | 84.4% | 493 | 897 | 247 | 10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alim, A.; Imtiaz, M.H. Automatic Identification of Children with ADHD from EEG Brain Waves. Signals 2023, 4, 193-205. https://doi.org/10.3390/signals4010010
Alim A, Imtiaz MH. Automatic Identification of Children with ADHD from EEG Brain Waves. Signals. 2023; 4(1):193-205. https://doi.org/10.3390/signals4010010
Chicago/Turabian StyleAlim, Anika, and Masudul H. Imtiaz. 2023. "Automatic Identification of Children with ADHD from EEG Brain Waves" Signals 4, no. 1: 193-205. https://doi.org/10.3390/signals4010010
APA StyleAlim, A., & Imtiaz, M. H. (2023). Automatic Identification of Children with ADHD from EEG Brain Waves. Signals, 4(1), 193-205. https://doi.org/10.3390/signals4010010





