Engineering the 2-Oxoglutarate Dehydrogenase Complex to Understand Catalysis and Alter Substrate Recognition
Abstract
:1. Introduction
1.1. Strategies for Protein Engineering
1.2. The E. coli 2-Oxoglutarate Dehydrogenase Complex Is a Promising Multienzyme Complex for Production of α-Hydroxyketones and for Synthesis of Acyl CoA Analogues
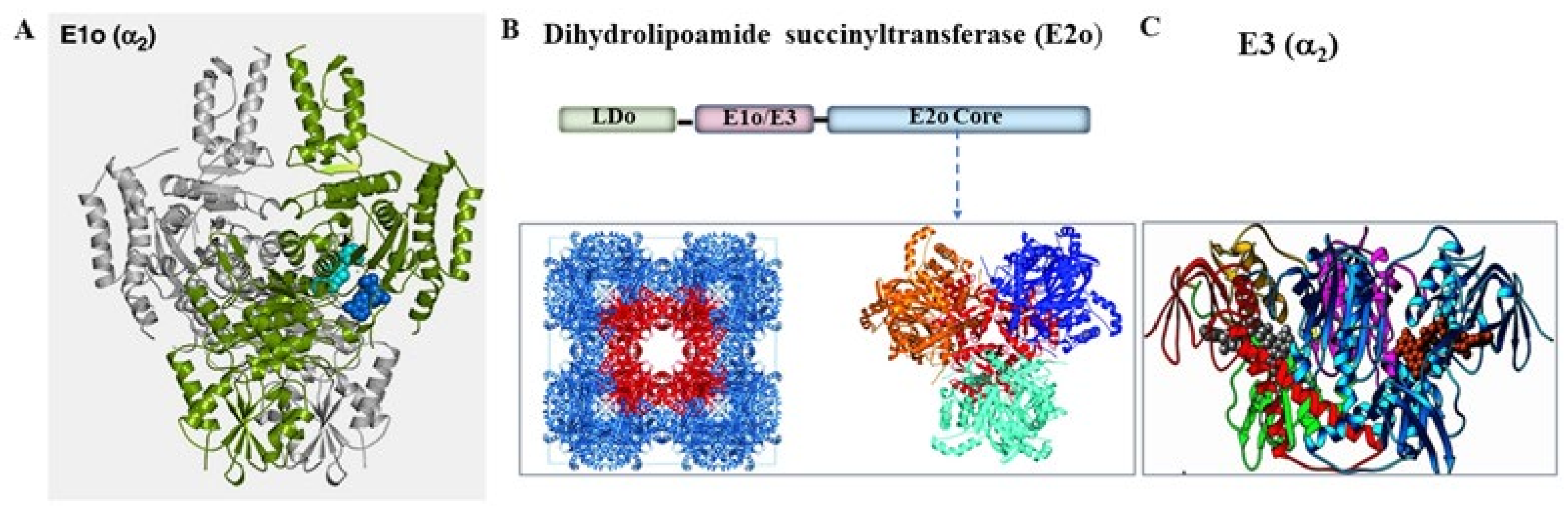
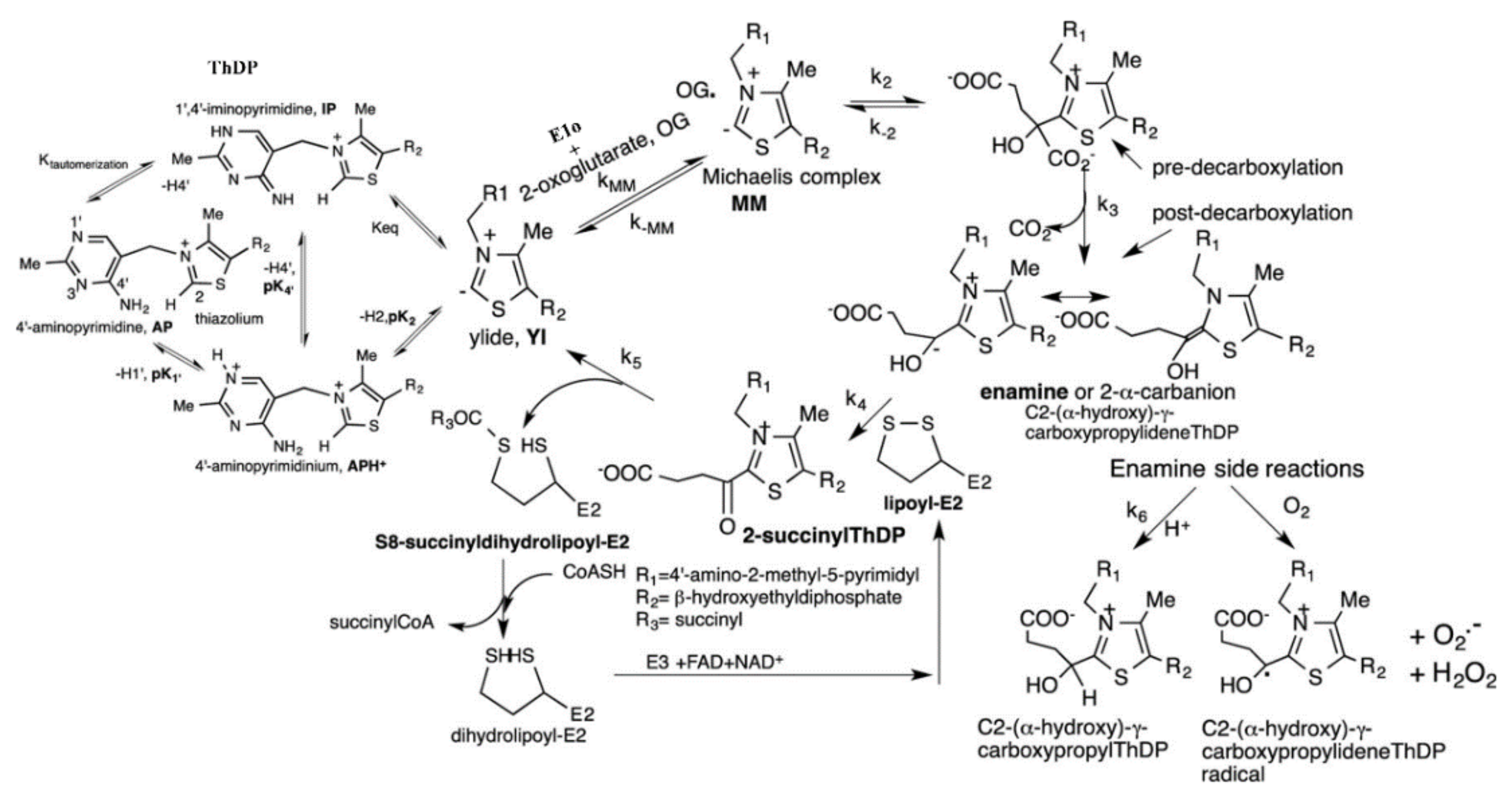
2. Engineering of the E1o Component of the E. coli OGDHc that Leads to Acceptance of Substrate Analogues Lacking the 5-Carboxyl Group
3. E. coli E1o Catalyzes the Formation of Acetoin-like Products in a Carboligase Condensation Reaction
4. Site-Saturation Mutagenesis Studies Clarified Catalysis of the Transthioesterification Reaction on the E. coli E2o and Corrected a Long-Misunderstood Mechanism Relevant to All E2 Components
5. Expansion of E2o Substrate Specificity and Creation of the 2-oxo Aliphatic Acid Dehydrogenase Complex with Goals of Acyl CoA Analogue Synthesis
6. Conclusions and Perspectives
- (a)
- The E1o component of the E. coli OGDHc was engineered to accept substrates lacking the 5-carboxylate group by subjecting His260 and His298 in the active center to site saturation mutagenesis. The results rule out an acid–base or hydrogen-bonding role for His298; however, they confirmed a hydrogen-bonding role for His260. It was suggested that the E2o component is a ‘gatekeeper’ for 2-OG channeling through the pathway.
- (b)
- It was demonstrated that the E. coli E1o can accept 2-oxovalerate and 2-oxoisovalerate in addition to its natural substrate 2-oxoglutarate in the carboligation reaction and that glyoxylate, ethyl glyoxylate and methylglyoxal can serve as aldehyde acceptors, producing acetoin-like compounds with good enantioselectivity. The novel products formed by the carboligation reaction include chiral 2-hydroxy-1,3-diketones and 2-hydroxy-β-ketoesters. The E. coli E1o offers a good starting point for protein engineering and optimization to synthesize stable chiral intermediates for fine chemical synthesis.
- (c)
- The fundamental mechanism of the transthioesterification reaction carried out by the E2o component of the E. coli OGDHc was elucidated, leading to the identification of Asp374 and His375 residues in the E2o catalytic center that are important for succinyl transfer in both the physiological and the reverse directions. The evidence provided ruled out a role of acid–base catalysis by His375. It was suggested that His375 and Asp 374 are involved in the stabilization of the tetrahedral oxyanionic intermediate through hydrogen bond donation. It was concluded that a rate-limiting step on E. coli OGDHc is succinyl transfer to CoA, thus providing crucial information for producing a variety of acyl-CoA analogues.
- (d)
- 6.4. The engineering of two consecutive components, E1o and E2o, was accomplished. The promiscuous variant components assembled into OGDHc were able to catalyze reactions with both natural and unnatural substrates, leaving significant room for optimization. Similar to natural evolution, the new pathway could be engineered to suit particular user-defined goals. These designed routes can function together with the existing pathways and could be utilized for chemical and synthetic purposes [118].
Author Contributions
Funding
Conflicts of Interest
References
- Hammer, S.C.; Knight, A.M.; Arnold, F.H. Design and evolution of enzymes for non-natural chemistry. Curr. Opin. Green Sustain. Chem. 2017, 7, 23–30. [Google Scholar] [CrossRef]
- Tracewell, C.A.; Arnold, F.H. Directed enzyme evolution: Climbing fitness peaks one amino acid at a time. Curr. Opin. Chem. Biol. 2009, 13, 3–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carter, P.; Nilsson, B.; Burnier, J.P.; Burdick, D.; Wells, J.A. Engineering subtilisin BPN′ for site-specific proteolysis. Proteins Struct. Funct. Genet. 1989, 6, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Nigel, S.; Berry, A.; Perham, R. Redesing of the coenzyme specificity of a dehygrogenase by protein engineering. Nature 1990, 343, 38–43. [Google Scholar]
- Dalby, P.A. Optimising enzyme function by directed evolution. Curr. Opin. Struct. Biol. 2003, 13, 500–505. [Google Scholar] [CrossRef]
- Park, S.; Morley, K.L.; Horsman, G.P.; Holmquist, M.; Hult, K.; Kazlauskas, R.J. Focusing Mutations into the P. fluorescens Esterase Binding Site Increases Enantioselectivity More Effectively than Distant Mutations. Chem. Biol. 2005, 12, 45–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strausberg, S.L.; Ruan, B.; Fisher, K.E.; Alexander, P.A.; Bryan, P.N. Directed Coevolution of Stability and Catalytic Activity in Calcium-free Subtilisin. Biochemistry 2005, 44, 3272–3279. [Google Scholar] [CrossRef]
- Morley, K.L.; Kazlauskas, R.J. Improving enzyme properties: When are closer mutations better? Trends Biotechnol. 2005, 23, 231–237. [Google Scholar] [CrossRef]
- Chica, R.A.; Doucet, N.; Pelletier, J.N. Semi-rational approaches to engineering enzyme activity: Combining the benefits of directed evolution and rational design. Curr. Opin. Biotechnol. 2005, 16, 378–384. [Google Scholar] [CrossRef]
- Arnold, F.H.; Georgiou, G. Directed Enzyme Evolution: Screening and Selection Methods; Humana Press: Totowa, NJ, USA, 2003; 383p. [Google Scholar]
- Gupta, N.; Farinas, E.T. Directed evolution of CotA laccase for increased substrate specificity using Bacillus subtilis spores. Protein. Eng. Des. Sel. 2010, 23, 679–682. [Google Scholar] [CrossRef] [Green Version]
- Gupta, N.; Farinas, E.T. Narrowing laccase substrate specificity using active site saturation mutagenesis. Comb. Chem. High. Throughput Screen. 2009, 12, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Glieder, A.; Farinas, E.T.; Arnold, F.H. Laboratory evolution of a soluble, self-sufficient, highly active alkane hydroxylase. Nat. Biotechnol. 2002, 20, 1135–1139. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Arnold, F.H. Tuning the activity of an enzyme for unusual environments: Sequential random mutagenesis of subtilisin E for catalysis in dimethylformamide. Proc. Natl. Acad. Sci. USA 1993, 90, 5618–5622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnold, F.H. Protein engineering for unusual environments. Curr. Opin. Biotechnol. 1993, 4, 450–455. [Google Scholar] [CrossRef]
- You, L.; Arnold, F.H. Directed evolution of subtilisin E in Bacillus subtilis to enhance total activity in aqueous dimethylformamide. Protein. Eng. Des. Sel. 1996, 9, 77–83. [Google Scholar] [CrossRef]
- Kan, S.B.J.; Lewis, R.D.; Chen, K.; Arnold, F.H. Directed evolution of cytochrome c for carbon–silicon bond formation: Bringing silicon to life. Science 2014, 522, 1048–1051. [Google Scholar] [CrossRef] [Green Version]
- Andrews, F.H.; McLeish, M.J. Using site-saturation mutagenesis to explore mechanism and substrate specificity in thiamin diphosphate-dependent enzymes. FEBS J. 2013, 280, 6395–6411. [Google Scholar] [CrossRef]
- Zhou, C.; Saravanan, T.; Lorillière, M.; Wei, D.; Charmantray, F.; Hecquet, L.; Fessner, W.; Yi, D. Second-Generation Engineering of a Thermostable Transketolase (TKGst) for Aliphatic Aldehyde Acceptors with Either Improved or Reversed Stereoselectivity. ChemBioChem. 2017, 18, 455–459. [Google Scholar] [CrossRef]
- Yu, H.; Yan, Y.; Zhang, C.; Dalby, P.A. Two strategies to engineer flexible loops for improved enzyme thermostability. Sci. Rep. 2017, 7, 41212. [Google Scholar] [CrossRef] [Green Version]
- Morris, P.; García-Arrazola, R.; Rios-Solis, L.; Dalby, P.A. Biophysical characterization of the inactivation of E. coli transketolase by aqueous co-solvents. Sci. Rep. 2021, 11, 23584. [Google Scholar] [CrossRef]
- Müller, M.; Sprenger, G.A.; Pohl, M. CC bond formation using ThDP-dependent lyases. Curr. Opin. Chem. Biol. 2013, 17, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Ricca, E.; Brucher, B.; Schrittwieser, J.H. Multi-Enzymatic Cascade Reactions: Overview and Perspectives. Adv. Synth. Catal. 2011, 353, 2239–2262. [Google Scholar] [CrossRef]
- Schrittwieser, J.H.; Velikogne, S.; Hall, M.; Kroutil, W. Artificial Biocatalytic Linear Cascades for Preparation of Organic Molecules. Chem. Rev. Am. Chem. Soc. 2018, 118, 270–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Junceda, E.; Lavandera, I.; Rother, D.; Schrittwieser, J.H. (Chemo)enzymatic cascades—Nature’s synthetic strategy transferred to the laboratory. J. Mol. Catal. B Enzym. 2015, 114, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Bugada, L.F.; Smith, M.R.; Wen, F. Engineering Spatially Organized Multienzyme Assemblies for Complex Chemical Transformation. ACS Catal. 2018, 8, 7898–7906. [Google Scholar] [CrossRef] [Green Version]
- Wilner, O.I.; Weizmann, Y.; Gill, R.; Lioubashevski, O.; Freeman, R.; Willner, I. Enzyme cascades activated on topologically programmed DNA scaffolds. Nat. Nanotechnol. 2009, 4, 249–254. [Google Scholar] [CrossRef]
- Dueber, J.E.; Wu, G.C.; Malmirchegini, G.R.; Moon, T.S.; Petzold, C.J.; Ullal, A.V.; Prather, K.L.J.; Keasling, J.D. Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 2009, 27, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Grotzky, A.; Nauser, T.; Erdogan, H.; Schlüter, A.D.; Walde, P. A fluorescently labeled dendronized polymer-enzyme conjugate carrying multiple copies of two different types of active enzymes. J. Am. Chem. Soc. 2012, 134, 11392–11395. [Google Scholar] [CrossRef]
- Chen, S.; Wen, L.; Svec, F.; Tan, T.; Lv, Y. Magnetic metal-organic frameworks as scaffolds for spatial co-location and positional assembly of multi-enzyme systems enabling enhanced cascade biocatalysis. RSC Adv. 2017, 7, 21205–21213. [Google Scholar] [CrossRef] [Green Version]
- Sheldon, R.A. Characteristic features and biotechnological applications of cross-linked enzyme aggregates (CLEAs). Appl. Microbiol. Biotechnol. 2011, 92, 467–477. [Google Scholar] [CrossRef] [Green Version]
- Wei, Q.; He, S.; Qu, J.; Xia, J. Synthetic Multienzyme Complexes Assembled on Virus-like Particles for Cascade Biosynthesis in Cellulo. Bioconjug. Chem. 2020, 31, 2413–2420. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.J.; Schmidt-Dannert, C. Organizing Multi-Enzyme Systems into Programmable Materials for Biocatalysis. Catalysts 2021, 11, 409. [Google Scholar] [CrossRef]
- Abdallah, W.; Hong, X.; Banta, S.; Wheeldon, I. Microenvironmental effects can masquerade as substrate channelling in cascade biocatalysis. Curr. Opin. Biotechnol. 2022, 73, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tsitkov, S.; Hess, H. Proximity does not contribute to activity enhancement in the glucose oxidase-horseradish peroxidase cascade. Nat. Commun. 2016, 7, 13982. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Li, X.; Xiong, J.; Wang, L.; Yan, L.T.; Ge, J. Investigating the origin of high efficiency in confined multienzyme catalysis. Nanoscale 2019, 11, 22108–22117. [Google Scholar] [CrossRef]
- Tretter, L.; Adam-Vizi, V. Alpha-ketoglutarate dehydrogenase: A target and generator of oxidative stress. Phil. Trans. R Soc. B. 2005, 360, 2335–2345. [Google Scholar] [CrossRef] [Green Version]
- Garrett, R.H.; Grisham, C.M. Biochemistry; Cengage Learning: Belmont, CA, USA, 2008. [Google Scholar]
- Frank, R.A.W.; Price, A.J.; Northrop, F.D.; Perham, R.N.; Luisi, B.F. Crystal Structure of the E1 Component of the Escherichia coli 2-Oxoglutarate Dehydrogenase Multienzyme Complex. J. Mol. Biol. 2007, 368, 639–651. [Google Scholar] [CrossRef]
- Perham, R.N. Domains, motifs, and linkers in 2-oxo acid dehydrogenase multienzyme complexes: A paradigm in the design of a multifunctional protein. Biochemistry 1991, 30, 8501–8512. [Google Scholar] [CrossRef]
- Perham, R.N. Swinging Arms and Swinging Domains in Multifunctional Enzymes: Catalytic Machines for Multistep Reactions. Annu. Rev. Biochem. 2000, 69, 961–1004. [Google Scholar] [CrossRef]
- Reed, L.J. A Trail of Research from Lipoic Acid to α-Keto Acid Dehydrogenase Complexes. J. Biol. Chem. 2001, 276, 38329–38336. [Google Scholar] [CrossRef] [Green Version]
- Murphy, G.E.; Jensen, G.J. Electron Cryotomography of the E. coli Pyruvate and 2-Oxoglutarate Dehydrogenase Complexes. Structure 2005, 13, 1765–1773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, M.; Wynn, R.M.; Chuang, J.L.; Brautigam, C.A.; Custorio, M.; Chuang, D.T. A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain a-ketoacid dehydrogenase complex. Eur. Mol. Biol. Organ. 2006, 25, 5983–5994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Izard, T.; Aevarsson, A.; Allen, M.D.; Westphal, A.H.; Perham, R.N.; de Kok, A.; Hol, W.G. Principles of quasi-equivalence and Euclidean geometry govern the assembly of cubic and dodecahedral cores of pyruvate dehydrogenase complexes. Proc. Natl. Acad. Sci. USA 1999, 96, 1240–1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattevi, A.; Obmolova, G.; Kalk, K.H.; Teplyakov, A.; Hol, W.G. Crystallographic analysis of substrate binding and catalysis in dihydrolipoyl transacetylase (E2p). Biochemistry 1993, 32, 3887–3901. [Google Scholar] [CrossRef]
- Milne, J.L.S.; Wu, X.; Borgnia, M.J.; Lengyel, J.S.; Brooks, B.R.; Shi, D.; Perham, R.N.; Subramaniam, S. Molecular structure of a 9-MDa icosahedral pyruvate dehydrogenase subcomplex containing the E2 and E3 enzymes using cryoelectron microscopy. J. Biol. Chem. 2006, 281, 4364–4370. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekhar, K.; Wang, J.; Arjunan, P.; Sax, M.; Park, Y.H.; Nemeria, N.S.; Kumaran, S.; Song, J.; Jordan, F.; Furey, W. Insight to the interaction of the dihydrolipoamide acetyltransferase (E2) core with the peripheral components in the Escherichia coli pyruvate dehydrogenase complex via multifaceted structural approaches. J. Biol. Chem. 2013, 288, 15402–15417. [Google Scholar] [CrossRef] [Green Version]
- Andi, B.; Soares, A.S.; Shi, W.; Fuchs, M.R.; McSweeney, S.; Liu, Q. Structure of the dihydrolipoamide succinyltransferase catalytic domain from Escherichia coli in a novel crystal form: A tale of a common protein crystallization contaminant. Acta Cryst. Sect. F Struct. Biol. Commun. 2019, 75, 616–624. [Google Scholar] [CrossRef] [Green Version]
- Wagenknecht, T.; Grassucci, R.; Schaak, D. Cryoelectron microscopy of frozen-hydrated α-ketoacid dehydrogenase complexes from Escherichia coli. J. Biol. Chem. 1990, 265, 22402–22408. [Google Scholar] [CrossRef]
- Wagenknecht, T.; Grassucci, R.; Berkowitz, J.; Forneris, C. Configuration of interdomain linkers in pyruvate dehydrogenase complex of Escherichia coli as determined by cryoelectron microscopy. J. Struct. Biol. 1992, 109, 70–77. [Google Scholar] [CrossRef]
- Škerlová, J.; Berndtsson, J.; Nolte, H.; Ott, M.; Stenmark, P. Structure of the native pyruvate dehydrogenase complex reveals the mechanism of substrate insertion. Nat. Commun. 2021, 12, 5277. [Google Scholar] [CrossRef]
- Bezerra, G.A.; Foster, W.R.; Bailey, H.J.; Hicks, K.G.; Sauer, S.W.; Dimitrov, B.; McCorvie, T.J.; Okun, J.G.; Rutter, J.; Kölker, S.; et al. Crystal structure and interaction studies of human DHTKD1 provide insight into a mitochondrial megacomplex in lysine catabolism. IUCrJ. 2020, 7, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Nagy, B.; Polak, M.; Ozohanics, O.; Zambo, Z.; Szabo, E.; Hubert, A.; Jordan, F.; Novaček, J.; Adam-Vizi, V.; Ambrus, A. Structure of the Dihydrolipoamide Succinyltransferase (E2) Component of the Human α-ketoglutarate dehydrogenase complex (hKGDHc) revealed by cryo-EM and Cross-linking mass spectrometry: Implications for the overall hKGDHc structure. Free Radic. Biol. Med. 2020, 159, S29–S30. [Google Scholar] [CrossRef]
- Reed, L.J.; Koike, M.; Levitch, M.E.; Leach, F.R. Studies on the nature and reactions of protein-bound Lipoic acid. J. Biol. Chem. 1958, 232, 143–158. [Google Scholar] [CrossRef]
- Reed, L.J. Multienzyme complexes. Acc. Chem. Res. 1974, 7, 40–46. [Google Scholar] [CrossRef]
- Miles, J.S.; Guest, J.R.; Radford, S.E.; Perham, R.N. Investigation of the mechanism of active site coupling in the pyruvate dehydrogenase multienzyme complex of Escherichia coli by protein engineering. J. Mol. Biol. 1988, 202, 97–106. [Google Scholar] [CrossRef]
- Texter, F.L.; Radford, S.E.; Laue, E.D.; Perham, R.N.; Miles, J.S.; Guest, J.R. Site-directed mutagenesis and proton NMR spectroscopy of an interdomain segment in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Biochemistry 2002, 27, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Prajapati, S.; Haselbach, D.; Wittig, S.; Patel, M.S.; Chari, A.; Schmidt, C.; Stark, H.; Tittmann, K. Structural and Functional Analyses of the Human PDH Complex Suggest a “Division-of-Labor” Mechanism by Local E1 and E3 Clusters. Structure 2019, 27, 1124–1136.e4. [Google Scholar] [CrossRef] [PubMed]
- Niebisch, A.; Kabus, A.; Schultz, C.; Weil, B.; Bott, M. Corynebacterial protein kinase G controls 2-oxoglutarate dehydrogenase activity via the phosphorylation status of the OdhI protein. J. Biol. Chem. 2006, 281, 12300–12307. [Google Scholar] [CrossRef] [Green Version]
- Wagner, T.; Bellinzoni, M.; Wehenkel, A.; O’Hare, H.M.; Alzari, P.M. Functional plasticity and allosteric regulation of α-ketoglutarate decarboxylase in central mycobacterial metabolism. Chem. Biol. 2011, 18, 1011–1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffelder, M.; Raasch, K.; Van Ooyen, J.; Eggeling, L. The E2 domain of OdhA of Corynebacterium glutamicum has succinyltransferase activity dependent on lipoyl residues of the acetyltransferase AceF. J. Bacteriol. 2010, 192, 5203–5211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nemeria, N.S.; Gerfen, G.; Nareddy, P.; Yang, L.; Zhang, X.; Szostak, M.; Jordan, F. The mitochondrial 2-oxoadipate and 2-oxoglutarate dehydrogenase complexes share their E2 and E3 components for their function and both generate reactive oxygen species. Free Radic. Biol. Med. 2018, 115, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Nemeria, N.S.; Gerfen, G.; Guevara, E.; Nareddy, P.R.; Szostak, M.; Jordan, F. The human Krebs cycle 2-oxoglutarate dehydrogenase complex creates an additional source of superoxide/hydrogen peroxide from 2-oxoadipate as alternative substrate. Free Radic. Biol. Med. 2017, 108, 644–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leandro, J.; Khamrui, S.; Wang, H.; Suebsuwong, C.; Nemeria, N.S.; Huynh, K.; Moustakim, M.; Secor, C.; Wang, M.; Dodatko, T.; et al. Inhibition and Crystal Structure of the Human DHTKD1-Thiamin Diphosphate Complex. ACS Chem. Biol. 2020, 15, 2041–2047. [Google Scholar] [CrossRef] [PubMed]
- Knapp, J.E.; Carroll, D.; Lawson, J.E.; Ernst, S.R.; Reed, L.J.; Hackert, M.L. Expression, purification, and structural analysis of the trimeric form of the catalytic domain of the Escherichia coli dihydrolipoamide succinyltransferase. Protein. Sci. 2000, 9, 37–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nemeria, N.S.; Ambrus, A.; Patel, H.; Gerfen, G.; Adam-Vizi, V.; Tretter, L.; Zhou, J.; Wang, J.; Jordan, F. Human 2-oxoglutarate dehydrogenase complex E1 component forms a thiamin-derived radical by aerobic oxidation of the enamine intermediate. J. Biol. Chem. 2014, 289, 29859–29873. [Google Scholar] [CrossRef] [Green Version]
- Shim, D.J.; Nemeria, N.S.; Balakrishnan, A.; Patel, H.; Song, J.; Wang, J.; Jordan, F.; Farinas, E.T. Assignment of function to histidines 260 and 298 by engineering the E1 component of the Escherichia coli 2-oxoglutarate dehydrogenase complex; Substitutions that lead to acceptance of substrates lacking the 5-carboxyl group. Biochemistry 2011, 50, 7705–7709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guevara, E.L. Different Modes of Activation of the Four Regulatory Pyruvate Dehydrogenase Kinases by the E2 and E3 Binding Protein Components of the Human Pyurvate Dehydrogenase Complex. Ph.D. Thesis, Rutgers University, Newark, NJ, USA, 2017. [Google Scholar]
- Chakraborty, J. Engineering of Escherichia coli 2-Oxoglutarate Dehydrogenase Complex with Mechanistic and Synthetic Goals. Ph.D. Thesis, New Jersey Institute of Technology, Newark, NJ, USA, 2019. [Google Scholar]
- Baykal, A.; Chakraborty, S.; Dodoo, A.; Jordan, F. Synthesis with good enantiomeric excess of both enantiomers of alpha-ketols and acetolactates by two thiamin diphosphate-dependent decarboxylases. Bioorg. Chem. 2006, 34, 380–393. [Google Scholar] [CrossRef] [Green Version]
- Nemeria, N.; Korotchkina, L.; McLeish, M.J.; Kenyon, G.L.; Patel, M.S.; Jordan, F. Elucidation of the Chemistry of Enzyme-Bound Thiamin Diphosphate Prior to Substrate Binding: Defining Internal Equilibria among Tautomeric and Ionization States. Biochemistry 2007, 46, 10739–10744. [Google Scholar] [CrossRef]
- Polovnikova, E.S.; McLeish, M.J.; Sergienko, E.A.; Burgner, J.T.; Anderson, N.L.; Bera, A.K.; Jordan, F.; Kenyon, A.G.L.; Hasson, M.S. Structural and Kinetic Analysis of Catalysis by a Thiamin Diphosphate-Dependent Enzyme, Benzoylformate Decarboxylase. Biochemistry 2003, 42, 1820–1830. [Google Scholar] [CrossRef] [PubMed]
- Meyer, D.; Walter, L.; Kolter, G.; Pohl, M.; Müller, M.; Tittmann, K. Conversion of Pyruvate Decarboxylase into an Enantioselective Carboligase with Biosynthetic Potential. J. Am. Chem. Soc. 2011, 133, 3609–3616. [Google Scholar] [CrossRef]
- Beigi, M.; Waltzer, S.; Fries, A.; Eggeling, L.; Sprenger, G.A.; Müller, M. TCA cycle involved enzymes SucA and Kgd, as well as MenD: Efficient biocatalysts for asymmetric C-C bond formation. Org. Lett. 2013, 15, 452–455. [Google Scholar] [CrossRef] [PubMed]
- Kasparyan, E.; Richter, M.; Dresen, C.; Walter, L.S.; Fuchs, G.; Leeper, F.J.; Wacker, T.; Andrade, S.L.A.; Kolter, G.; Pohl, M.; et al. Asymmetric Stetter reactions catalyzed by thiamine diphosphate-dependent enzymes. Appl. Microbiol. Biotechnol. 2014, 98, 9681–9690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jordan, F. Current mechanistic understanding of thiamin diphosphate-dependent enzymatic reactions. Nat. Prod. Rep. 2003, 20, 184–201. [Google Scholar] [CrossRef] [PubMed]
- Brandt, G.S.; Kneen, M.M.; Petsko, G.A.; Ringe, D.; McLeish, M.J. Active-site engineering of benzaldehyde lyase shows that a point mutation can confer both new reactivity and susceptibility to mechanism-based inhibition. J. Am. Chem. Soc. 2010, 132, 438–439. [Google Scholar] [CrossRef] [PubMed]
- Hailes, H.C.; Rother, D.; Müller, M.; Westphal, R.; Ward, J.M.; Pleiss, J.; Vogel, C.; Pohl, M. Engineering stereoselectivity of ThDP-dependent enzymes. FEBS J. 2013, 280, 6374–6394. [Google Scholar] [CrossRef] [PubMed]
- Baraibar, Á.G.; Von Lieres, E.; Wiechert, W.; Pohl, M.; Rother, D. Effective production of (S)-α-hydroxy ketones: An reaction engineering approach. Top. Catal. 2014, 57, 401–411. [Google Scholar] [CrossRef]
- Marsden, S.R.; McMillan, D.G.G.; Hanefeld, U. Assessing the Thiamine Diphosphate Dependent Pyruvate Dehydrogenase E1 Subunit for Carboligation Reactions with Aliphatic Ketoacids. Int. J. Mol. Sci. 2020, 21, 8641. [Google Scholar] [CrossRef]
- Schapfl, M.; Baier, S.; Fries, A.; Ferlaino, S.; Waltzer, S.; Müller, M.; Sprenger, G.A. Extended substrate range of thiamine diphosphate-dependent MenD enzyme by coupling of two C–C-bonding reactions. Appl. Microbiol. Biotechnol. 2018, 102, 8359–8372. [Google Scholar] [CrossRef]
- Kulig, J.; Sehl, T.; Mackfeld, U.; Wiechert, W.; Pohl, M.; Rother, D. An Enzymatic 2-Step Cofactor and Co-Product Recycling Cascade towards a Chiral 1,2-Diol. Part I: Cascade Design. Adv. Synth. Catal. 2019, 361, 2607–2615. [Google Scholar] [CrossRef]
- Nilsson, U.; Meshalkina, L.; Lindqvist, Y.; Schneidere, G. Examination of substrate binding in thiamin diphosphate-dependent transketolase by protein crystallography and site-directed mutagenesis. J. Biol. Chem. 1997, 272, 1864–1869. [Google Scholar] [CrossRef] [Green Version]
- Hibbert, E.G.; Senussi, T.; Costelloe, S.J.; Lei, W.; Smith, M.E.; Ward, J.M.; Hailes, H.C.; Dalby, P.A. Directed evolution of transketolase activity on non-phosphorylated substrates. J. Biotechnol. 2007, 131, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.E.B.; Hibbert, E.G.; Jones, A.B.; Dalby, P.A.; Hailesa, H.C. Enhancing and Reversing the Stereoselectivity of Escherichia coli Transketolase via Single-Point Mutations. Adv. Synth. Catal. 2008, 350, 2631–2638. [Google Scholar] [CrossRef]
- Engel, S.; Vyazmensky, M.; Geresh, S.; Barak, Z.; Chipman, D.M. Acetohydroxyacid synthase: A new enzyme for chiral synthesis of R-phenylacetylcarbinol. Biotechnol. Bioeng. 2003, 83, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Chipman, D.M.; Duggleby, R.G.; Tittmann, K. Mechanisms of acetohydroxyacid synthases. Curr. Opin. Chem. Biol. 2005, 9, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Vyazmensky, M.; Steinmetz, A.; Meyer, D.; Golbik, R.; Barak, E.; Tittmann, K.; Chipman, D.M. Significant Catalytic Roles for Glu47 and Gln 110 in All Four of the CÀC Bond-Making and-Breaking Steps of the Reactions of Acetohydroxyacid Synthase II. Biochemistry 2011, 50, 3250–3260. [Google Scholar] [CrossRef]
- Lingen, B.; Kolter-Jung, D.; Dünkelmann, P.; Feldmann, R.; Grötzinger, J.; Pohl, M.; Müller, M. Alteration of the substrate specificity of benzoylformate decarboxylase from Pseudomonas putida by directed evolution. ChemBioChem. 2003, 4, 721–726. [Google Scholar] [CrossRef]
- Siegert, P.; McLeish, M.; Baumann, M.; Iding, H.; Kneen, M.M.; Kenyon, G.L.; Pohl, M. Exchanging the substrate specificities of pyruvate decarboxylase from Zymomonas mobilis and benzoylformate decarboxylase from Pseudomonas putida. Protein. Eng. Des. Sel. 2005, 18, 345–357. [Google Scholar] [CrossRef] [Green Version]
- De María, P.D.; Pohl, M.; Gocke, D.; Gröger, H.; Trauthwein, H.; Stillger, T.; Walter, L.; Müller, M. Asymmetric synthesis of aliphatic 2-hydroxy ketones by enzymatic carboligation of aldehydes. Eur. J. Org. Chem. 2007, 2009, 2940–2944. [Google Scholar] [CrossRef]
- Kurutsch, A.; Richter, M.; Brecht, V.; Sprenger, G.A.; Müller, M. MenD as a versatile catalyst for asymmetric synthesis. J. Mol. Catal. B Enzym. 2009, 61, 56–66. [Google Scholar] [CrossRef]
- Beigi, M.; Loschonsky, S.; Lehwald, P.; Brecht, V.; Andrade, S.L.A.; Leeper, F.J.; Hummeld, W.; Müller, M. Organic & Biomolecular Chemistry α-Hydroxy-β-keto acid rearrangement-decarboxylation: Impact on thiamine diphosphate-dependent enzymatic transformation. Org. Biomol. Chem. 2013, 11, 252. [Google Scholar]
- Sergienko, E.A.; Jordan, F. Catalytic acid-base groups in yeast pyruvate decarboxylase. 3. A steady-state kinetic model consistent with the behavior of both wild-type and variant enzymes at all relevant pH values. Biochemistry 2001, 40, 7382–7403. [Google Scholar] [CrossRef]
- Nemeria, N.; Tittmann, K.; Joseph, E.; Zhou, L.; Vazquez-Coll, M.B.; Arjunan, P.; Hübner, G.; Furey, W.; Jordan, F. Glutamate 636 of the Escherichia coli Pyruvate Dehydrogenase-E1 Participates in Active Center Communication and Behaves as an Engineered Acetolactate Synthase with Unusual Stereoselectivity. J. Biol. Chem. 2005, 280, 21473–21482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, H.; Shim, D.J.; Farinas, E.T.; Jordan, F. Investigation of the donor and acceptor range for chiral carboligation catalyzed by the E1 component of the 2-oxoglutarate dehydrogenase complex. J. Mol. Catal. B Enzym. 2013, 98, 42–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jordan, F.; Nemeria, N.S.; Balakrishnan, A.; Chakraborty, J.; Guevara, E.; Nareddy, P.; Patel, H.; Shim, D.J.; Wang, J.; Yang, L.; et al. An Update on Developments in the Field of Thiamin Diphosphate-Dependent Enzymes. Compr. Nat. Prod. III 2020, 4, 58–110. [Google Scholar]
- Knapp, J.E.; Mitchell, D.T.; Yazdi, M.A.; Ernst, S.R.; Reed, L.J.; Hackert, M.L. Crystal structure of the truncated cubic core component of the Escherichia coli 2-oxoglutarate dehydrogenase multienzyme complex. J. Mol. Biol. 1998, 280, 655–668. [Google Scholar] [CrossRef] [PubMed]
- Hendle, J.; Mattevi, A.; Westphal, A.H.; Spee, J.; De Kok, A.; Teplyakov, A.; Hol, W.G. Crystallographic and Enzymatic Investigations on the Role of Ser558, His610, and Asn614 in the Catalytic Mechanism of Azotobacter vinelandii Dihydrolipoamide Acetyltransferase (Elp)1. Biochemistry 1995, 34, 4287–4298. [Google Scholar] [CrossRef]
- Yu, X.; Hiromasa, Y.; Tsen, H.; Stoops, J.K.; Roche, T.E.; Zhou, Z.H. Structures of the Human Pyruvate Dehydrogenase Complex Cores: A Highly Conserved Catalytic Center with Flexible N-Terminal Domains. Structure 2008, 16, 104–114. [Google Scholar] [CrossRef] [Green Version]
- Kyrilis, F.L.; Semchonok, D.A.; Skalidis, I.; Tüting, C.; Hamdi, F.; O’Reilly, F.J.; Rappsilber, J.; Kastritis, P.L. Integrative structure of a 10-megadalton eukaryotic pyruvate dehydrogenase complex from native cell extracts. Cell Rep. 2021, 34, 108727. [Google Scholar] [CrossRef]
- Guest, J.R. Functional implications of structural homologies between chloramphenicol acetyltransferase and dihydrolipoamide acetyltransferase. FEMS Microbiol. Lett. 1987, 44, 417–422. [Google Scholar] [CrossRef]
- Shaw, W.V.; Leslie, A.G.W. Chloramphenicol Acetyltransferase. Annu. Rev. Biophys. Biophys. Chem. 1991, 20, 363–386. [Google Scholar] [CrossRef]
- Carter, P.; Wells, J.A. Dissecting the catalytic triad of a serine protease. Nature 1988, 332, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, J.; Nemeria, N.S.; Farinas, E.; Jordan, F. Catalysis of transthiolacylation in the active centers of dihydrolipoamide acyltransacetylase components of 2-oxo acid dehydrogenase complexes. FEBS Open Bio. 2018, 8, 880–896. [Google Scholar] [CrossRef] [PubMed]
- Lewendon, A.; Murray, I.A.; Shaw, W.V.; Gibbs, M.R.; Leslie, A.G.W. Replacement of catalytic histidine-195 of chloramphenicol acetyltransferase: Evidence for a general base role for glutamate. Biochemistry 1994, 33, 1944–1950. [Google Scholar] [CrossRef] [PubMed]
- Russell, G.C.; Guest, J.R. Site-directed mutagenesis of the lipoate acetyltransferase of Escherichia coli. Proc. R Soc. B Biol. Sci. 1991, 243, 155–160. [Google Scholar]
- Russell, G.C.; Guest, J.R. Sequence similarities within the family of dihydrolipoamide acyltransferases and discovery of a previously unidentified fungal enzyme. Biochim. Biophys. Acta BBA Protein. Struct. Mol. 1991, 1076, 225–232. [Google Scholar] [CrossRef]
- Chakraborty, J.; Nemeria, N.S.; Zhang, X.; Nareddy, P.R.; Szostak, M.; Farinas, E.; Jordan, F. Engineering 2- oxoglutarate dehydrogenase to a 2-oxo aliphatic dehydrogenase complex by optimizing consecutive components. AIChE J. 2020, 66, e16769. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Choi, J.-M.; Han, S.-S.; Kim, H.-S. Industrial applications of enzyme biocatalysis: Current status and future aspects. Biotechnol. Adv. 2015, 33, 1443–1454. [Google Scholar] [CrossRef]
- Guan, L.-J.; Ohtsuka, J.; Okai, M.; Miyakawa, T.; Mase, T.; Zhi, Y.; Hou, F.; Ito, N.; Iwasaki, A.; Yasohara, Y.; et al. A new target region for changing the substrate specificity of amine transaminases. Sci. Rep. 2015, 5, 10753. [Google Scholar] [CrossRef] [Green Version]
- Cobb, R.E.; Chao, R.; Zhao, H. Directed Evolution: Past, Present and Future. AIChE J. 2013, 59, 1432–1440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desai, A.A. Sitagliptin Manufacture: A Compelling Tale of Green Chemistry, Process Intensification, and Industrial Asymmetric Catalysis. Angew. Chem. Int. Ed. 2011, 50, 1974–1976. [Google Scholar] [CrossRef] [PubMed]
- Kazemi, M.; Shiri, L. Thioesters synthesis: Recent adventures in the esterification of thiols. J. Sulfur. Chem. 2015, 36, 613–623. [Google Scholar] [CrossRef]
- Hutchison, C.A.; Chuang, R.-Y.; Noskov, V.N.; Assad-Garcia, N.; Deerinck, T.J.; Ellisman, M.H.; Gill, J.; Kannan, K.; Karas, B.J.; Ma, L.; et al. Design and synthesis of a minimal bacterial genome. Science 2016, 351, aad6253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leveson-Gower, R.B.; Mayer, C.; Roelfes, G. The importance of catalytic promiscuity for enzyme design and evolution. Nat. Rev. Chem. Nat. Res. 2019, 3, 687–705. [Google Scholar] [CrossRef]
- Currin, A.; Swainston, N.; Day, P.J.; Kell, D.B. Synthetic biology for the directed evolution of protein biocatalysts: Navigating sequence space intelligently. Chem. Soc. Rev. R. Soc. Chem. 2015, 44, 1172–1239. [Google Scholar] [CrossRef] [Green Version]
- Yuen, C.M.; Liu, D.R. Dissecting protein structure and function using directed evolution. Nat. Methods 2007, 4, 995–997. [Google Scholar] [CrossRef]
- Newton, M.S.; Arcus, V.L.; Gerth, M.L.; Patrick, W.M. Enzyme evolution: Innovation is easy, optimization is complicated. Curr. Opin. Struct. Biol. 2018, 48, 110–116. [Google Scholar] [CrossRef]
- Arnold, F.H. Directed Evolution: Bringing New Chemistry to Life. Angew. Chem. Int. Ed. 2018, 57, 4143–4148. [Google Scholar] [CrossRef] [Green Version]
- Khosla, C.; Harbury, P.B. Detection of modular enzymes. Nature 2001, 409, 247–252. [Google Scholar] [CrossRef]






| E1o Variant | DCPIP Activity a | |
|---|---|---|
| kcat/Km, 2-OG (s−1mM−1) | kcat/Km, 2-OV (s−1mM−1) | |
| Wild type | 824 | 0.0047 |
| His298Leu | 122 | 0.015 |
| His298Thr | 1.5 | 0.0020 |
| His298Asp | Nd | 0.18 |
| His298Val | 0.6 | 0.062 |
| His260Glu/His298Asn | 49.1 | Nd |
| His260Glu | 0.0084 | Nd |
| Substrates | Acceptors | Product | Enantiomeric Excess |
|---|---|---|---|
| 2-OG 2-OV 2-OiV | Glyoxylate | 1 2 3 | 90% (R) 96% (S) 67% (S) |
| 2-OG 2-OV 2-OiV | Ethyl-glyoxylate | 4 5 6 | 81% (R) 52% (S) 81% (S) |
| 2-OG 2-OV 2-OiV | Methyl-glyoxal | 7 8 9 | 83% (S) 81% (R) >90% (S) |
| E2o Substitution | kcat/Km, 2-OG (s−1mM−1) | kcat/Km, 2-OV (s−1mM−1) | kcat/Km, 2-OHe (s−1mM−1) |
|---|---|---|---|
| None b | 875 | No activity | No Activity |
| None c | 703 | No activity | No activity |
| His348Phe d | 244 | 81 | 1.98 |
| His348Gln d | 149 | 6.8 | 0.59 |
| His348Tyr d | 338 | 57 | 1.17 |
| Ser333Met d | 210 | 4.7 | 15.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chakraborty, J.; Nemeria, N.; Shim, Y.; Zhang, X.; Guevara, E.L.; Patel, H.; Farinas, E.T.; Jordan, F. Engineering the 2-Oxoglutarate Dehydrogenase Complex to Understand Catalysis and Alter Substrate Recognition. Reactions 2022, 3, 139-159. https://doi.org/10.3390/reactions3010011
Chakraborty J, Nemeria N, Shim Y, Zhang X, Guevara EL, Patel H, Farinas ET, Jordan F. Engineering the 2-Oxoglutarate Dehydrogenase Complex to Understand Catalysis and Alter Substrate Recognition. Reactions. 2022; 3(1):139-159. https://doi.org/10.3390/reactions3010011
Chicago/Turabian StyleChakraborty, Joydeep, Natalia Nemeria, Yujeong Shim, Xu Zhang, Elena L. Guevara, Hetal Patel, Edgardo T. Farinas, and Frank Jordan. 2022. "Engineering the 2-Oxoglutarate Dehydrogenase Complex to Understand Catalysis and Alter Substrate Recognition" Reactions 3, no. 1: 139-159. https://doi.org/10.3390/reactions3010011






