DNA-Based Mechanical Sensors for Cell Applications
Abstract
:1. Introduction
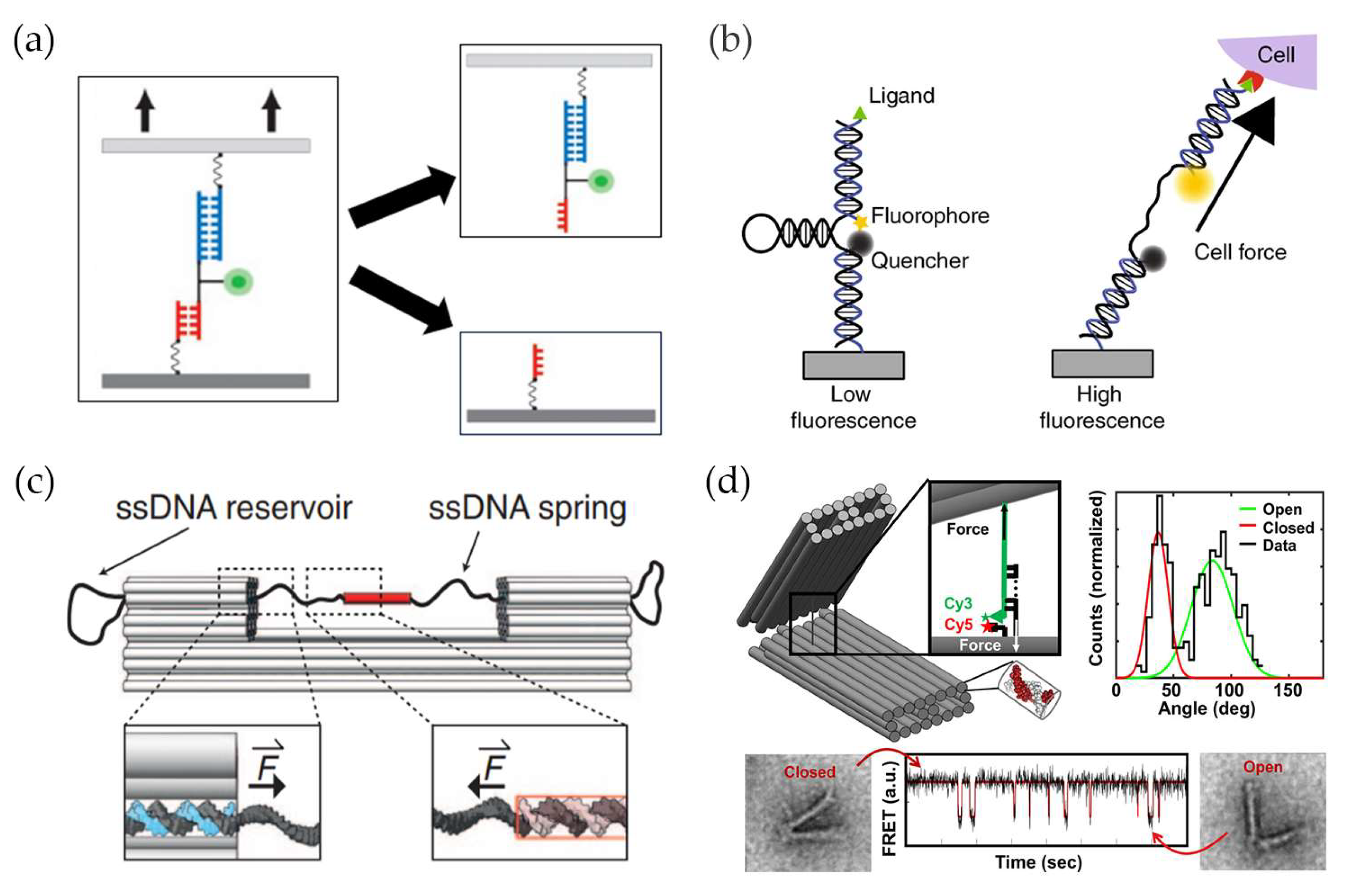
2. DNA-Based Tension Probes
3. Force Measurement at Cell–Extracellular Matrix
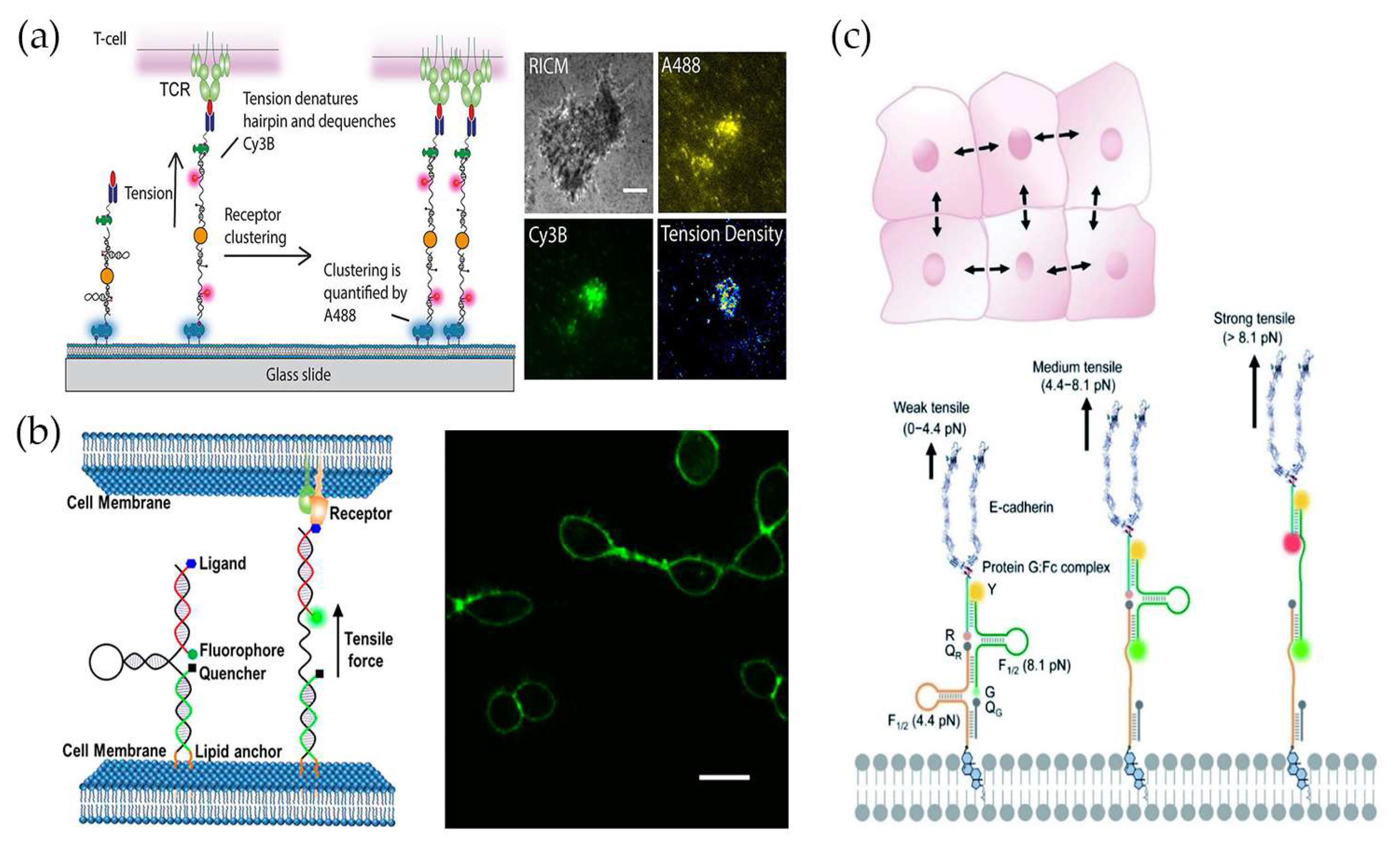
4. Force Measurement at Cell–Cell Junctions
4.1. Measuring Intercellular Forces at the Single-Cell Level
4.2. Measuring Intercellular Forces at the Collective-Cell Level
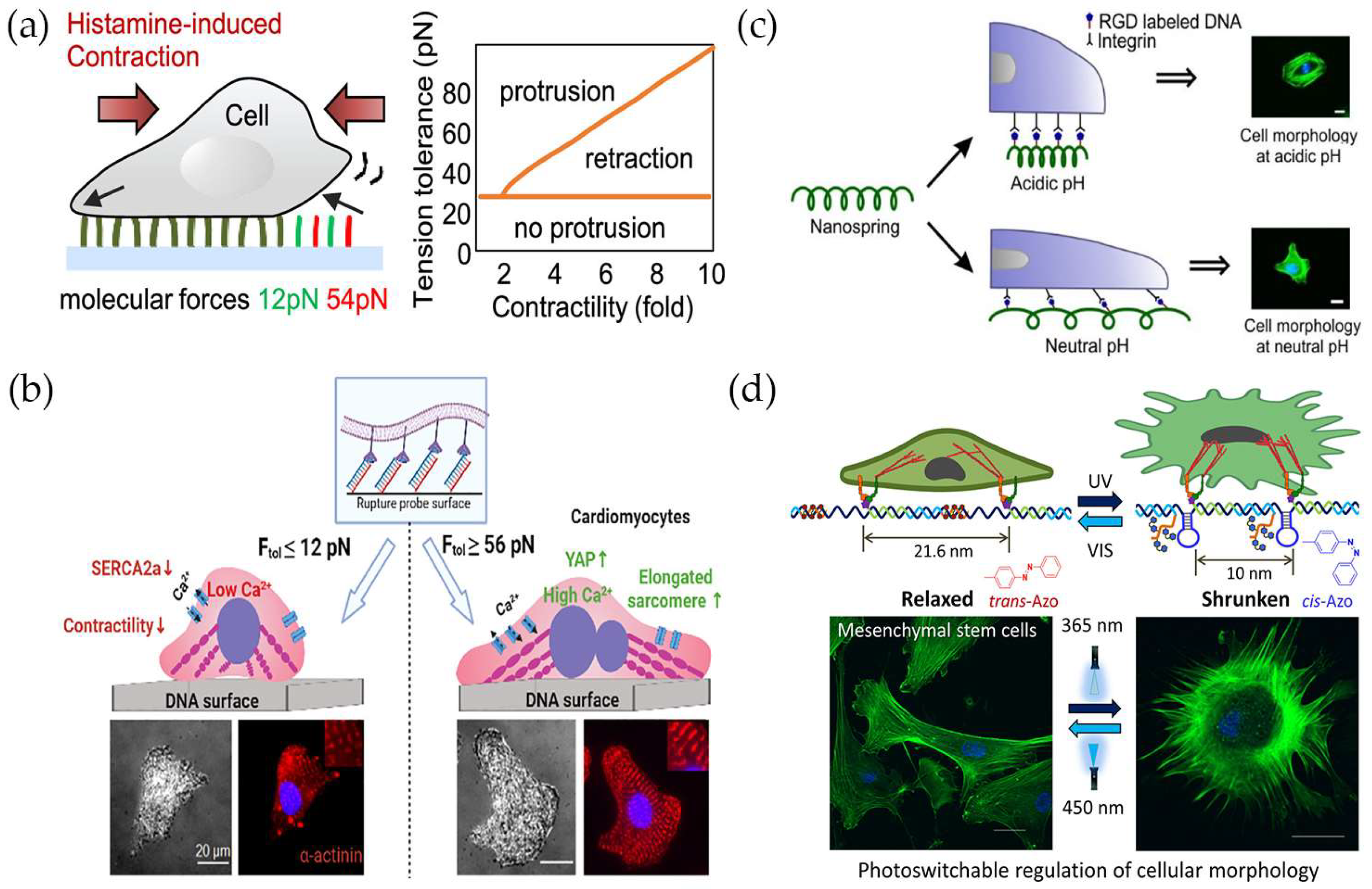
5. Monitoring and Regulating Cellular Mechanical Functions
6. Summary
Author Contributions
Funding
Conflicts of Interest
References
- Splitt, R.L.; DeMali, K.A. Metabolic reprogramming in response to cell mechanics. Biol. Cell 2023, 115, e202200108. [Google Scholar] [CrossRef] [PubMed]
- Clarke, D.N.; Martin, A.C. Actin-based force generation and cell adhesion in tissue morphogenesis. Curr. Biol. 2021, 31, R667–R680. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.S.; Tan, J.; Tien, J. Mechanotransduction at Cell-Matrix and Cell-Cell Contacts. Annu. Rev. Biomed. Eng. 2004, 6, 275–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortes, P.; Riser, B.L. Mechanical strain of glomerular mesangial cells in the pathogenesis of glomerulosclerosis: Clinical implications. Nephrol. Dial. Transplant. 1999, 14, 1351–1354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrick, S.K.; Greenberg, M.J. Cardiac myosin contraction and mechanotransduction in health and disease. J. Biol. Chem. 2021, 297, 101297. [Google Scholar] [CrossRef]
- Libring, S.; Solorio, L. 16—Cancer mechanobiology: Interaction of biomaterials with cancer cells. In Biomaterials for Cancer Therapeutics, 2nd ed.; Park, K., Ed.; Woodhead Publishing: Swaston, UK, 2020; pp. 445–470. [Google Scholar]
- Wang, A.; Vijayraghavan, K.; Solgaard, O.; Butte, M.J. Fast Stiffness Mapping of Cells Using High-Bandwidth Atomic Force Microscopy. ACS Nano 2016, 10, 257–264. [Google Scholar] [CrossRef] [Green Version]
- Coceano, G.; Yousafzai, M.S.; Ma, W.; Ndoye, F.; Venturelli, L.; Hussain, I.; Bonin, S.; Niemela, J.; Scoles, G.; Cojoc, D.; et al. Investigation into local cell mechanics by atomic force microscopy mapping and optical tweezer vertical indentation. Nanotechnology 2016, 27, 065102. [Google Scholar] [CrossRef]
- Tapia-Rojo, R.; Eckels, E.C.; Fernández, J.M. Ephemeral states in protein folding under force captured with a magnetic tweezers design. Proc. Natl. Acad. Sci. USA 2019, 116, 7873–7878. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Gao, X.; Lei, H.; Wang, W.; Cao, Y. Biophysical Approaches for Applying and Measuring Biological Forces. Adv. Sci. 2022, 9, e2105254. [Google Scholar] [CrossRef]
- Roca-Cusachs, P.; Conte, V.; Trepat, X. Quantifying forces in cell biology. Nat. Cell. Biol. 2017, 19, 742–751. [Google Scholar] [CrossRef]
- Hang, X.; He, S.; Dong, Z.; Minnick, G.; Rosenbohm, J.; Chen, Z.; Yang, R.; Chang, L. Nanosensors for single cell mechanical interrogation. Biosens. Bioelectron. 2021, 179, 113086. [Google Scholar] [CrossRef]
- Stabley, D.R.; Jurchenko, C.; Marshall, S.S.; Salaita, K.S. Visualizing mechanical tension across membrane receptors with a fluorescent sensor. Nat. Methods 2011, 9, 64–67. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ge, C.; Zhu, C.; Salaita, K. DNA-based digital tension probes reveal integrin forces during early cell adhesion. Nat. Commun. 2014, 5, 5167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morimatsu, M.; Mekhdjian, A.H.; Chang, A.C.; Tan, S.J.; Dunn, A.R. Visualizing the interior architecture of focal adhesions with high-resolution traction maps. Nano Lett. 2015, 15, 2220–2228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galior, K.; Liu, Y.; Yehl, K.; Vivek, S.; Salaita, K. Titin-Based Nanoparticle Tension Sensors Map High-Magnitude Integrin Forces within Focal Adhesions. Nano Lett. 2016, 16, 341–348. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Tian, C.; Li, X.; Qian, H.; Hao, C.; Jiang, W.; Mao, C. Reversibly switching the surface porosity of a DNA tetrahedron. J. Am. Chem. Soc. 2012, 134, 11998–12001. [Google Scholar] [CrossRef]
- Yang, D.; Hartman, M.R.; Derrien, T.L.; Hamada, S.; An, D.; Yancey, K.G.; Cheng, R.; Ma, M.; Luo, D. DNA materials: Bridging nanotechnology and biotechnology. Acc. Chem. Res. 2014, 47, 1902–1911. [Google Scholar] [CrossRef]
- Seeman, N.C. Nucleic acid junctions and lattices. J. Theor. Biol. 1982, 99, 237–247. [Google Scholar] [CrossRef]
- Neuman, K.C.; Nagy, A. Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 2008, 5, 491–505. [Google Scholar] [CrossRef]
- Albrecht, C.; Blank, K.; Lalic-Mülthaler, M.; Hirler, S.; Mai, T.; Gilbert, I.; Schiffmann, S.; Bayer, T.; Clausen-Schaumann, H.; Gaub, H.E. DNA: A programmable force sensor. Science 2003, 301, 367–370. [Google Scholar] [CrossRef] [Green Version]
- Wan, Z.; Chen, X.; Chen, H.; Ji, Q.; Chen, Y.; Wang, J.; Cao, Y.; Wang, F.; Lou, J.; Tang, Z.; et al. The activation of IgM- or isotype-switched IgG- and IgE-BCR exhibits distinct mechanical force sensitivity and threshold. elife 2015, 4, e06925. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Liu, Z.; Liu, Y.; Ma, V.P.-Y.; Blanchard, A.; Zhao, J.; Galior, K.; Dyer, R.B.; Salaita, K. Light-Responsive Polymer Particles as Force Clamps for the Mechanical Unfolding of Target Molecules. Nano Lett. 2018, 18, 2630–2636. [Google Scholar] [CrossRef] [PubMed]
- Woodside, M.T.; Behnke-Parks, W.M.; Larizadeh, K.; Travers, K.; Herschlag, D.; Block, S.M. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc. Natl. Acad. Sci. USA 2006, 103, 6190–6195. [Google Scholar] [CrossRef] [PubMed]
- Brockman, J.; Blanchard, A.T.; Ma, V.P.-Y.; Derricotte, W.D.; Zhang, Y.; Fay, M.E.; Lam, W.A.; Evangelista, F.A.; Mattheyses, A.L.; Salaita, K. Mapping the 3D orientation of piconewton integrin traction forces. Nat. Methods 2018, 15, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Nickels, P.C.; Wünsch, B.; Holzmeister, P.; Bae, W.; Kneer, L.M.; Grohmann, D.; Tinnefeld, P.; Liedl, T. Molecular force spectroscopy with a DNA origami-based nanoscopic force clamp. Science 2016, 354, 305–307. [Google Scholar] [CrossRef]
- Wang, Y.; Le, J.V.; Crocker, K.; Darcy, M.A.; Halley, P.D.; Zhao, D.; Andrioff, N.; Croy, C.; Poirier, M.G.; Bundschuh, R.; et al. A nanoscale DNA force spectrometer capable of applying tension and compression on biomolecules. Nucleic Acids Res. 2021, 49, 8987–8999. [Google Scholar] [CrossRef]
- Darcy, M.; Crocker, K.; Wang, Y.; Le, J.V.; Mohammadiroozbahani, G.; Abdelhamid, M.A.S.; Craggs, T.D.; Castro, C.E.; Bundschuh, R.; Poirier, M.G. High-Force Application by a Nanoscale DNA Force Spectrometer. ACS Nano 2022, 16, 5682–5695. [Google Scholar] [CrossRef]
- Humphrey, J.D.; Dufresne, E.R.; Schwartz, M.A. Mechanotransduction and extracellular matrix homeostasis. Nat. Rev. Mol. Cell Biol. 2014, 15, 802–812. [Google Scholar] [CrossRef] [Green Version]
- Berrier, A.L.; Yamada, K.M. Cell-matrix adhesion. J. Cell. Physiol. 2007, 213, 565–573. [Google Scholar] [CrossRef]
- Conway, J.R.W.; Jacquemet, G. Cell matrix adhesion in cell migration. Essays Biochem. 2019, 63, 535–551. [Google Scholar] [CrossRef]
- Poujade, M.; Grasland-Mongrain, E.; Hertzog, A.; Jouanneau, J.; Chavrier, P.; Ladoux, B.; Buguin, A.; Silberzan, P. Collective migration of an epithelial monolayer in response to a model wound. Proc. Natl. Acad. Sci. USA 2007, 104, 15988–15993. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, L.; Wan, D.; Zhou, L.; Zheng, S.; Lin, S.; Qiao, Y. Extracellular matrix and its therapeutic potential for cancer treatment. Signal Transduct. Target Ther. 2021, 6, 153. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ha, T. Defining single molecular forces required to activate integrin and notch signaling. Science 2013, 340, 991–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, R.; Kellner, A.V.; Ma, V.P.-Y.; Su, H.; Deal, B.R.; Brockman, J.M.; Salaita, K. DNA probes that store mechanical information reveal transient piconewton forces applied by T cells. Proc. Natl. Acad. Sci. USA 2019, 116, 16949–16954. [Google Scholar] [CrossRef] [Green Version]
- Ma, V.P.; Liu, Y.; Yehl, K.; Galior, K.; Zhang, Y.; Salaita, K. Mechanically Induced Catalytic Amplification Reaction for Readout of Receptor-Mediated Cellular Forces. Angew. Chem. Int. Ed. Engl. 2016, 55, 5488–5492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, Y.; Glazier, R.; Bazrafshan, A.; Hu, Y.; Rashid, S.A.; Petrich, B.G.; Ke, Y.; Salaita, K. Mechanically Triggered Hybridization Chain Reaction. Angew. Chem. Int. Ed. Engl. 2021, 60, 19974–19981. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Sarkar, A.; Wang, X. Peptide nucleic acid based tension sensor for cellular force imaging with strong DNase resistance. Biosens. Bioelectron. 2020, 150, 111959. [Google Scholar] [CrossRef]
- Dutta, P.K.; Zhang, Y.; Blanchard, A.T.; Ge, C.; Rushdi, M.; Weiss, K.; Zhu, C.; Ke, Y.; Salaita, K. Programmable Multivalent DNA-Origami Tension Probes for Reporting Cellular Traction Forces. Nano Lett. 2018, 18, 4803–4811. [Google Scholar] [CrossRef]
- Hang, X.; He, S.; Dong, Z.; Li, Y.; Huang, Z.; Zhang, Y.; Sun, H.; Lin, L.; Li, H.; Wang, Y.; et al. High-Throughput DNA Tensioner Platform for Interrogating Mechanical Heterogeneity of Single Living Cells. Small 2022, 18, e2106196. [Google Scholar] [CrossRef]
- Pawlak, M.R.; Smiley, A.T.; Ramirez, M.P.; Kelly, M.D.; Shamsan, G.A.; Anderson, S.M.; Smeester, B.A.; Largaespada, D.A.; Odde, D.J.; Gordon, W.R. RAD-TGTs: High-throughput measurement of cellular mechanotype via rupture and delivery of DNA tension probes. Nat. Commun. 2023, 14, 2468. [Google Scholar] [CrossRef]
- Wang, X.; Sun, J.; Xu, Q.; Chowdhury, F.; Roein-Peikar, M.; Wang, Y.; Ha, T. Integrin Molecular Tension within Motile Focal Adhesions. Biophys. J. 2015, 109, 2259–2267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Champion, J.A.; Mitragotri, S. Role of target geometry in phagocytosis. Proc. Natl. Acad. Sci. USA 2006, 103, 4930–4934. [Google Scholar] [CrossRef] [PubMed]
- Dickopf, S.; Georges, G.J.; Brinkmann, U. Format and geometries matter: Structure-based design defines the functionality of bispecific antibodies. Comput. Struct. Biotechnol. J. 2020, 18, 1221–1227. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Ma, V.P.; Ma, R.; Chen, W.; Duan, Y.; Glazier, R.; Petrich, B.G.; Li, R.; Salaita, K. DNA-Based Microparticle Tension Sensors (μTS) for Measuring Cell Mechanics in Non-planar Geometries and for High-Throughput Quantification. Angew. Chem. Int. Ed. Engl. 2021, 60, 18044–18050. [Google Scholar] [CrossRef]
- Acharya, B.R.; Wu, S.K.; Lieu, Z.Z.; Parton, R.G.; Grill, S.W.; Bershadsky, A.D.; Gomez, G.A.; Yap, A.S. Mammalian Diaphanous 1 Mediates a Pathway for E-cadherin to Stabilize Epithelial Barriers through Junctional Contractility. Cell Rep. 2017, 18, 2854–2867. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Heemskerk, I.; Ibar, C.; Shraiman, B.I.; Irvine, K.D. Differential growth triggers mechanical feedback that elevates Hippo signaling. Proc. Natl. Acad. Sci. USA 2016, 113, E6974–E6983. [Google Scholar] [CrossRef]
- Charras, G.; Yap, A.S. Tensile Forces and Mechanotransduction at Cell-Cell Junctions. Curr. Biol. 2018, 28, R445–R457. [Google Scholar] [CrossRef] [Green Version]
- Manz, B.N.; Groves, J.T. Spatial organization and signal transduction at intercellular junctions. Nat. Rev. Mol. Cell Biol. 2010, 11, 342–352. [Google Scholar] [CrossRef] [Green Version]
- Ma, V.P.-Y.; Liu, Y.; Blanchfield, L.; Su, H.; Evavold, B.D.; Salaita, K. Ratiometric Tension Probes for Mapping Receptor Forces and Clustering at Intermembrane Junctions. Nano Lett. 2016, 16, 4552–4559. [Google Scholar] [CrossRef]
- Glazier, R.; Salaita, K. Supported lipid bilayer platforms to probe cell mechanobiology. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1465–1482. [Google Scholar] [CrossRef]
- Zhao, B.; O’brien, C.; Mudiyanselage, A.P.K.K.K.; Li, N.; Bagheri, Y.; Wu, R.; Sun, Y.; You, M. Visualizing Intercellular Tensile Forces by DNA-Based Membrane Molecular Probes. J. Am. Chem. Soc. 2017, 139, 18182–18185. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Li, N.; Xie, T.; Bagheri, Y.; Liang, C.; Keshri, P.; Sun, Y.; You, M. Quantifying tensile forces at cell-cell junctions with a DNA-based fluorescent probe. Chem. Sci. 2020, 11, 8558–8566. [Google Scholar] [CrossRef] [PubMed]
- Friedl, P.; Gilmour, D. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 2009, 10, 445–457. [Google Scholar] [CrossRef]
- Wang, X.-H.; Yang, F.; Pan, J.-B.; Kang, B.; Xu, J.-J.; Chen, H.-Y. Quantitative Imaging of pN Intercellular Force and Energetic Costs during Collective Cell Migration in Epithelial Wound Healing. Anal. Chem. 2020, 92, 16180–16187. [Google Scholar] [CrossRef]
- Wang, X.-H.; Liu, Y.; Kang, B.; Xu, J.-J.; Chen, H.-Y. Cell mechanics and energetic costs of collective cell migration under confined microchannels. Chin. Chem. Lett. 2023, 34, 107789. [Google Scholar] [CrossRef]
- Zhang, W.; Gunst, S.J. Interactions of airway smooth muscle cells with their tissue matrix: Implications for contraction. Proc. Am. Thorac. Soc. 2008, 5, 32–39. [Google Scholar] [CrossRef] [Green Version]
- Webb, R.C. Smooth muscle contraction and relaxation. Adv. Physiol. Educ. 2003, 27, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Jo, M.H.; Kim, B.C.; Sung, K.; Panettieri, R.A.; An, S.S.; Liu, J.; Ha, T. Molecular Nanomechanical Mapping of Histamine-Induced Smooth Muscle Cell Contraction and Shortening. ACS Nano 2021, 15, 11585–11596. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Pu, W.T. Cardiomyocyte Maturation: New Phase in Development. Circ. Res. 2020, 126, 1086–1106. [Google Scholar] [CrossRef]
- Rashid, S.A.; Blanchard, A.T. DNA Tension Probes Show that Cardiomyocyte Maturation Is Sensitive to the Piconewton Traction Forces Transmitted by Integrins. ACS Nano 2022, 16, 5335–5348. [Google Scholar] [CrossRef]
- Zhang, K.; Ma, Y.; Wang, D.; Liu, J.; An, J.; Li, Y.; Ma, C.; Pei, Y.; Zhang, Z.; Liu, J.; et al. In Vivo Activation of T-Cell Proliferation by Regulating Cell Surface Receptor Clustering Using a pH-Driven Interlocked DNA Nano-Spring. Nano Lett. 2022, 22, 1937–1945. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Deng, R.; Sun, Y.; Zhang, L.; Li, J. Reversible control of cell membrane receptor function using DNA nano-spring multivalent ligands. Chem. Sci. 2017, 8, 7098–7105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karna, D.; Stilgenbauer, M.; Jonchhe, S.; Ankai, K.; Kawamata, I.; Cui, Y.; Zheng, Y.-R.; Suzuki, Y.; Mao, H. Chemo-Mechanical Modulation of Cell Motions Using DNA Nanosprings. Bioconjug. Chem. 2021, 32, 311–317. [Google Scholar] [CrossRef]
- Li, H.; Zhang, C.; Hu, Y.; Liu, P.; Sun, F.; Chen, W.; Zhang, X.; Ma, J.; Wang, W.; Wang, L.; et al. A reversible shearing DNA probe for visualizing mechanically strong receptors in living cells. Nat. Cell Biol. 2021, 23, 642–651. [Google Scholar] [CrossRef]
- Sethi, S.; Hidaka, K.; Sugiyama, H.; Endo, M. Non-invasive Regulation of Cellular Morphology Using a Photoswitchable Mechanical DNA Polymer. Angew. Chem. Int. Ed. Engl. 2021, 60, 20342–20349. [Google Scholar] [CrossRef]
- Yu, Z.; Schonhoft, J.D.; Dhakal, S.; Bajracharya, R.; Hegde, R.; Basu, S.; Mao, H. ILPR G-quadruplexes formed in seconds demonstrate high mechanical stabilities. J. Am. Chem. Soc. 2009, 131, 1876–1882. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, S.; Schonhoft, J.D.; Koirala, D.; Yu, Z.; Basu, S.; Mao, H. Coexistence of an ILPR i-motif and a partially folded structure with comparable mechanical stability revealed at the single-molecule level. J. Am. Chem. Soc. 2010, 132, 8991–8997. [Google Scholar] [CrossRef] [Green Version]
- Shrestha, P.; Emura, T.; Koirala, D.; Cui, Y.; Hidaka, K.; Maximuck, W.; Endo, M.; Sugiyama, H.; Mao, H. Mechanical properties of DNA origami nanoassemblies are determined by Holliday junction mechanophores. Nucleic Acids Res. 2016, 44, 6574–6582. [Google Scholar] [CrossRef] [Green Version]
- Borisenko, G.G.; Zaitseva, M.A.; Chuvilin, A.N.; Pozmogova, G.E. DNA modification of live cell surface. Nucleic Acids Res. 2009, 37, e28. [Google Scholar] [CrossRef] [Green Version]
- Bagheri, Y.; Chedid, S.; Shafiei, F.; Zhao, B.; You, M. A quantitative assessment of the dynamic modification of lipid-DNA probes on live cell membranes. Chem. Sci. 2019, 10, 11030–11040. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, X.; Hao, P.; Wu, N. DNA-Based Mechanical Sensors for Cell Applications. Chemistry 2023, 5, 1546-1559. https://doi.org/10.3390/chemistry5030106
Sun X, Hao P, Wu N. DNA-Based Mechanical Sensors for Cell Applications. Chemistry. 2023; 5(3):1546-1559. https://doi.org/10.3390/chemistry5030106
Chicago/Turabian StyleSun, Xiaoya, Pengyan Hao, and Na Wu. 2023. "DNA-Based Mechanical Sensors for Cell Applications" Chemistry 5, no. 3: 1546-1559. https://doi.org/10.3390/chemistry5030106
APA StyleSun, X., Hao, P., & Wu, N. (2023). DNA-Based Mechanical Sensors for Cell Applications. Chemistry, 5(3), 1546-1559. https://doi.org/10.3390/chemistry5030106




