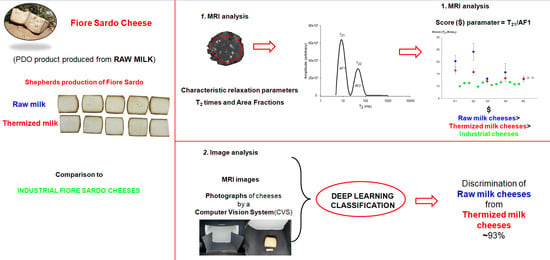
Quality Control in Fiore Sardo PDO Cheese: Detection of Heat Treatment Application and Production Chain by MRI Relaxometry and Image Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Production of Fiore Sardo (FS) Cheese
2.1.1. Dataset 1
2.1.2. Dataset 2
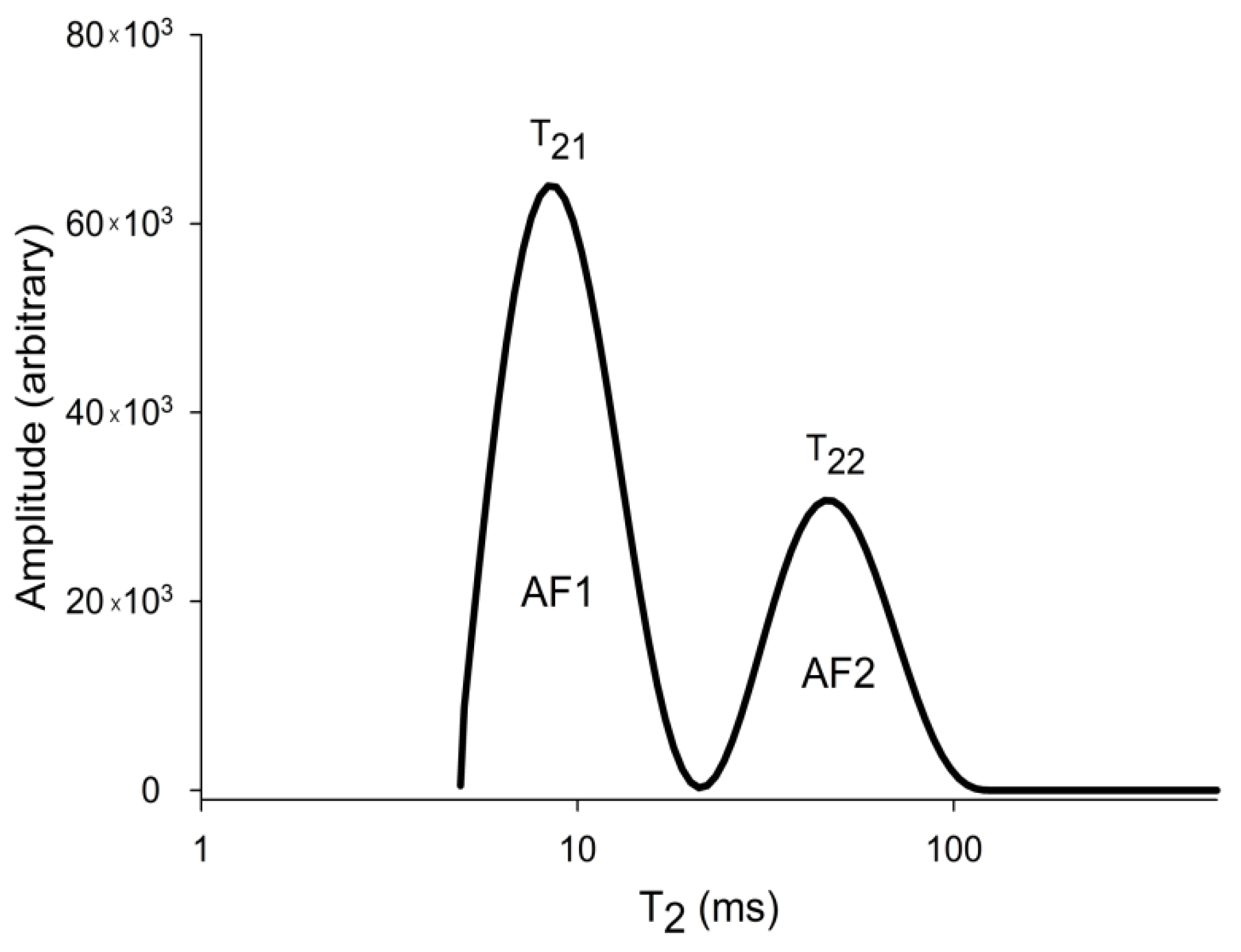
2.2. Magnetic Resonance Imaging (MRI) Analysis
2.3. Moisture Content
2.4. Statistical Analysis
2.5. MRI Data Conversion
2.6. Photographic Acquisition and Processing of Cheese Paste Surfaces
2.7. Deep Transfer Learning Based Classification
3. Results and Discussion
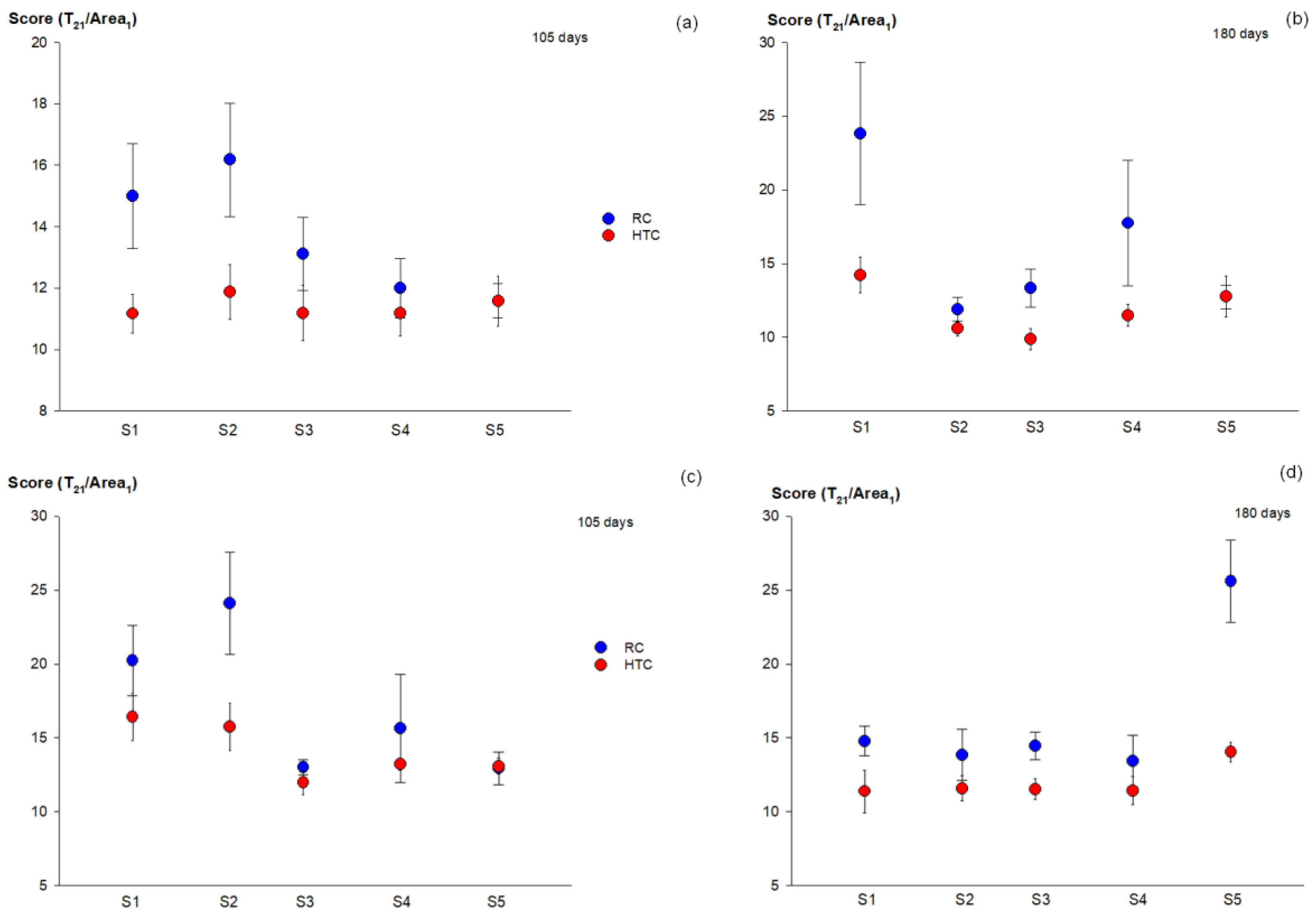
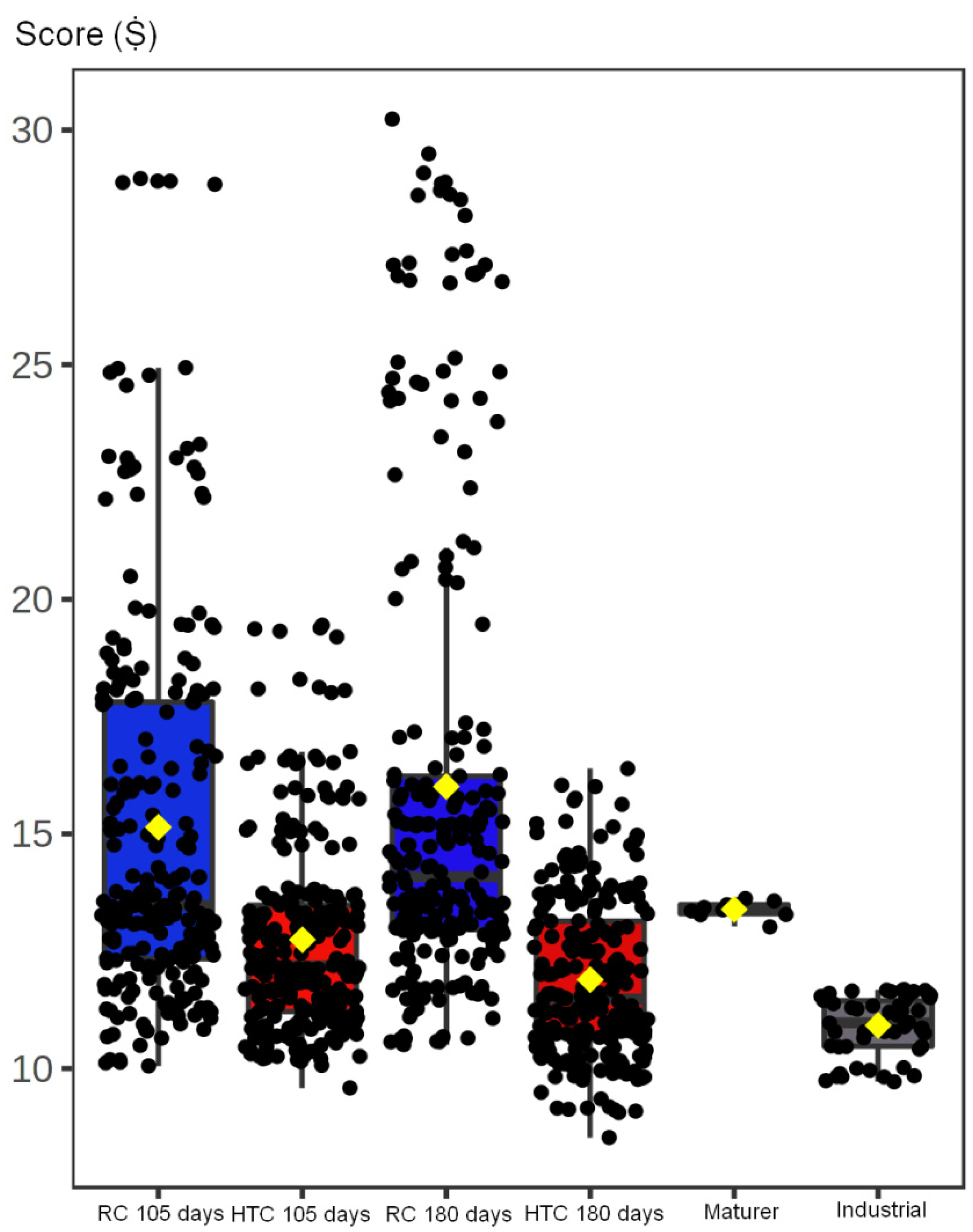
3.1. MRI Analysis of Dataset 1
3.2. MRI Analysis of Dataset 2
3.3. Moisture Content
3.4. Image Analysis of Fiore Sardo Pictures and MRI Images
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- European Commission. Quality Schemes Explained. Available online: https://ec.europa.eu/info/food-farming-fisheries/food-safety-and-quality/certification/quality-labels/quality-schemes-explained (accessed on 16 February 2021).
- European Commission eAmbrosia—The EU Geographical Indications Register. Available online: https://ec.europa.eu/info/food-farming-fisheries/food-safety-and-quality/certification/quality-labels/geographical-indications-register/# (accessed on 11 April 2021).
- Sardo, P.; Barletta, M. European Designations between Identity Values and Market. 2019. Available online: https://www.slowfood.com/wp-content/uploads/2019/09/European-designations-between-identity-values-and-market_ENG.pdf (accessed on 26 February 2021).
- Fox, P.; McSweeney, P.; Cogan, T.; Guinee, T. Cheese—Chemistry, Physics and Microbiology, 3rd ed.; Guinee, P., Fox, P., McSweeney, T., Cogan, T., Eds.; Academic Press: Cambridge, MA, USA, 2004; ISBN 978-1-4613-6138-1. [Google Scholar]
- Fox, P.; Uniacke-Lowe, T.; McSweeney, P.; O’Mahony, J. (Eds.) Heat-Induced Changes in Milk. In Dairy Chemistry and Biochemistry; Springer International Publishing: Basel, Switzerland, 2015; pp. 345–375. ISBN 978-3-319-14891-5. [Google Scholar]
- Pirisi, A.; Pinna, G.; Papoff, C.M. Effect of milk thermisation on Fiore Sardo PDO cheese: 1. Physicochemical characteristics. Sci. Tecn. Latt. Cas. 1999, 50, 353–366. [Google Scholar]
- EUR-Lex—31996R1236—EN—EUR-Lex. Available online: https://eur-lex.europa.eu/legal-content/IT/TXT/?uri=CELEX%3A31996R1236 (accessed on 26 February 2021).
- Anedda, R. Magnetic Resonance Analysis of Dairy Processing Suitable Tools for the Dairy Industry. In Magnetic Resonance in Food Science: Defining Food by Magnetic Resonance; The Royal Society of Chemistry: London, UK, 2015; pp. 49–64. ISBN 978-1-78262-031-0. [Google Scholar]
- Anedda, R.; Pala, M.; Curti, E. Cheeses That Made History in Italian Dairy Tradition. In Cheeses around the World: Types, Production, Properties and Cultural and Nutritional Relevance; Ferrão, A.C., Guiné, R.P.F., Correia, P.R., Eds.; Nova Science Publishers: New York City, NY, USA, 2019; pp. 203–228. ISBN 978-1-53615-419-1. [Google Scholar]
- Mendia, C.; Ibañez, F.C.; Torre, P.; Barcina, Y. Effect of pasteurization on the sensory characteristics of a ewe’s-milk cheese. J. Sens. Stud. 1999, 14, 415–424. [Google Scholar] [CrossRef]
- Scintu, M.F.; Del Caro, A.; Urgeghe, P.P.; Piga, C.; Di Salvo, R. Sensory profile development for an Italian PDO ewe’s milk cheese at two different ripening times. J. Sens. Stud. 2010, 25, 577–590. [Google Scholar] [CrossRef]
- Siniscalchi, V. Environment, regulation and the moral economy of food in the Slow Food movement. J. Polit. Ecol. 2013, 20, 295–305. [Google Scholar] [CrossRef] [Green Version]
- Pittalis, P.; Cappellacci, U.; Contu, M.I.; Fasolino, G.; Peru, A.; Tedde, M.; Tocco, E.; Tunis, S.; Zedda, A. XV Legislature—Regional Question N.1380. Available online: http://www3.consregsardegna.it/XVlegislatura/interrogazioni/1380 (accessed on 26 February 2021).
- Ledda A XV Legislature—Regional Question N.1706/A. Available online: http://www3.consregsardegna.it/XVlegislatura/interrogazioni/1706 (accessed on 26 February 2021).
- Siniscalchi, V. Food, Slow Food and Middle Class Activism. In Food Activism: Agency, Democracy and Economy; Bloomsbury Academic: London, UK, 2014; pp. 9–24. [Google Scholar]
- Anedda, R.; Pardu, A.; Korb, J.P.; Curti, E. Effect of the manufacturing process on Fiore Sardo PDO cheese microstructure by multi-frequency NMR relaxometry. Food Res. Int. 2021, 140, 110079. [Google Scholar] [CrossRef]
- Caboni, P.; Maxia, D.; Scano, P.; Addis, M.; Dedola, A.; Pes, M.; Murgia, A.; Casula, M.; Profumo, A.; Pirisi, A. A gas chromatography-mass spectrometry untargeted metabolomics approach to discriminate Fiore Sardo cheese produced from raw or thermized ovine milk. J. Dairy Sci. 2019, 102, 5005–5018. [Google Scholar] [CrossRef]
- Dedola, A.S.; Piras, L.; Addis, M.; Pirisi, A.; Piredda, G.; Mara, A.; Sanna, G. New analytical tools for unmasking frauds in raw milk-based dairy products: Assessment, validation and application to fiore sardo PDO cheese of a RP-HPLC method for the evaluation of the α-l-fucosidase activity. Separations 2020, 7, 40. [Google Scholar] [CrossRef]
- Mulas, G.; Roggio, T.; Uzzau, S.; Anedda, R. A new magnetic resonance imaging approach for discriminating Sardinian sheep milk cheese made from heat-treated or raw milk. J. Dairy Sci. 2013, 96, 7393–7403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazza, M.; Guglielmetti, C.; Brusadore, S.; Sciuto, S.; Esposito, G.; Caramelli, M.; Peletto, S.; Acutis, P.L.; Marengo, E.; Manfredi, M.; et al. A proteomic approach to the safeguard of a typical agri-food product: Fiore sardo PDO. Adv. Dairy Res. 2019, 7. [Google Scholar] [CrossRef]
- Piga, C.; Urgeghe, P.P.; Piredda, G.; Scintu, M.F.; Di Salvo, R.; Sanna, G. Thermal inactivation and variability of γ-glutamyltransferase and α-l-fucosidase enzymatic activity in sheep milk. LWT Food Sci. Technol. 2013, 54, 152–156. [Google Scholar] [CrossRef]
- Lambelet, P.; Berrocal, R.; Ducret, F. Low resolution NMR spectroscopy: A tool to study protein denaturation: I. Application to diamagnetic whey proteins. J. Dairy Res. 1989, 56, 211–222. [Google Scholar] [CrossRef] [Green Version]
- Lambelet, P.; Berrocal, R.; Renevey, F. Low-field nuclear magnetic resonance relaxation study of thermal effects on milk proteins. J. Dairy Res. 1992, 59, 517–526. [Google Scholar] [CrossRef]
- Curti, E.; Pardu, A.; Melis, R.; Addis, M.; Pes, M.; Pirisi, A.; Anedda, R. Molecular mobility changes after high-temperature, short-time pasteurization: An extended time-domain nuclear magnetic resonance screening of ewe milk. J. Dairy Sci. 2020, 103, 9881–9892. [Google Scholar] [CrossRef]
- Curti, E.; Pardu, A.; Del Vigo, S.; Sanna, R.; Anedda, R. Non-invasive monitoring of curd syneresis upon renneting of raw and heat-treated cow’s and goat’s milk. Int. Dairy J. 2019, 90. [Google Scholar] [CrossRef]
- Curti, E.; Pardu, A.; Del Vigo, S.; Sanna, R.; Anedda, R. A low-field Nuclear Magnetic Resonance dataset of whole milk during coagulation and syneresis. Data Br. 2019, 26. [Google Scholar] [CrossRef] [PubMed]
- Bjarnason, T.A.; Mitchell, J.R. AnalyzeNNLS: Magnetic resonance multiexponential decay image analysis. J. Magn. Reson. 2010, 206, 200–204. [Google Scholar] [CrossRef] [PubMed]
- AOAC International. AOAC 948.12-2002, Moisture in Cheese. Method II (Rapid Screening Method): AOAC Official Method. Available online: http://www.aoacofficialmethod.org/index.php?main_page=product_info&products_id=1145 (accessed on 26 February 2021).
- Pang, Z.; Chong, J.; Li, S.; Xia, J. MetaboAnalystR 3.0: Toward an optimized workflow for global metabolomics. Metabolites 2020, 10, 186. [Google Scholar] [CrossRef]
- Laurence, A. NIfTI Image Converter (nii2png) for Python and Matlab|NIfTI-Image-Converter. Available online: https://alexlaurence.github.io/NIfTI-Image-Converter/ (accessed on 26 February 2021).
- Shrestha, A.; Mahmood, A. Review of deep learning algorithms and architectures. IEEE Access 2019, 7, 53040–53065. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. In Proceedings of the 25th International Conference on Neural Information Processing Systems, Siem Reap, Cambodia, 13–16 December 2018; Curran Associates Inc.: Red Hook, NY, USA, 2012; Volume 1, pp. 1097–1105. [Google Scholar]
- Liu, S.; Deng, W. Very deep convolutional neural network based image classification using small training sample size. In Proceedings of the 2015 3rd IAPR Asian Conference on Pattern Recognition (ACPR), Kuala Lumpur, Malaysia, 3–6 November 2015; pp. 730–734. [Google Scholar]
- Iandola, F.N.; Moskewicz, M.W.; Ashraf, K.; Han, S.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <1MB model size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Google Google Colaboratory. Available online: https://colab.research.google.com/notebooks/intro.ipynb (accessed on 26 February 2021).
- Hills, B.P.; Takacs, S.F.; Belton, P.S. A new interpretation of proton NMR relaxation time measurements of water in food. Food Chem. 1990, 37, 95–111. [Google Scholar] [CrossRef]
- Mariette, F.; Tellier, C.; Brule, G.; Marchal, P. Multinuclear Nmr-Study of the Ph dependent water state in skim milk and caseinate solutions. J. Dairy Res. 1993, 60, 175–188. [Google Scholar] [CrossRef]
- Mariette, F. NMR Relaxation of Dairy Products. In Modern Magnetic Resonance; Webb, G.A., Ed.; Springer: Dordrecht, The Netherlands; Berlin, Germany, 2006; pp. 1697–1701. ISBN 978-1-4020-3894-5. [Google Scholar]
- Gianferri, R.; D’Aiuto, V.; Curini, R.; Delfini, M.; Brosio, E. Proton NMR transverse relaxation measurements to study water dynamic states and age-related changes in Mozzarella di Bufala Campana cheese. Food Chem. 2007, 105, 720–726. [Google Scholar] [CrossRef]
- Gianferri, R.; Maioli, M.; Delfini, M.; Brosio, E. A low-resolution and high-resolution nuclear magnetic resonance integrated approach to investigate the physical structure and metabolic profile of Mozzarella di Bufala Campana cheese. Int. Dairy J. 2007, 17, 167–176. [Google Scholar] [CrossRef]
- Chaland, B.; Mariette, F.; Marchal, P.; De Certaines, J. 1H nuclear magnetic resonance relaxometric characterization of fat and water states in soft and hard cheese. J. Dairy Res. 2000, 64, 609–618. [Google Scholar] [CrossRef]
- Godefroy, S.; Korb, J.-P.; Creamer, L.K.; Watkinson, P.J.; Callaghan, P.T. Probing protein hydration and aging of food materials by the magnetic field dependence of proton spin-lattice relaxation times. J. Colloid Interface Sci. 2003, 267, 337–342. [Google Scholar] [CrossRef]
- Venu, K.; Denisov, V.P.; Halle, B. Water 1H magnetic relaxation dispersion in protein solutions. A quantitative assessment of internal hydration, proton exchange, and cross-relaxation. J. Am. Chem. Soc. 1997, 119, 3122–3134. [Google Scholar] [CrossRef]
- Conte, P.; Cinquanta, L.; Lo Meo, P.; Mazza, F.; Micalizzi, A.; Corona, O. Fast field cycling NMR relaxometry as a tool to monitor Parmigiano Reggiano cheese ripening. Food Res. Int. 2021, 139, 109845. [Google Scholar] [CrossRef]
- Boiani, M.; Fenelon, M.; FitzGerald, R.J.; Kelly, P.M. Use of 31P NMR and FTIR to investigate key milk mineral equilibria and their interactions with micellar casein during heat treatment. Int. Dairy J. 2018, 81, 12–18. [Google Scholar] [CrossRef] [Green Version]
- Wahlgren, N.M.; Dejmek, P.; Drakenberg, T. A 43Ca and 31P NMR study of the calcium and phosphate equilibria in heated milk solutions. J. Dairy Res. 1990, 57, 355–364. [Google Scholar] [CrossRef]
- Pisanu, S.; Pagnozzi, D.; Pes, M.; Pirisi, A.; Roggio, T.; Uzzau, S.; Addis, M.F. Differences in the peptide profile of raw and pasteurised ovine milk cheese and implications for its bioactive potential. Int. Dairy J. 2015, 42, 26–33. [Google Scholar] [CrossRef]
- Grappin, R.; Beuvier, E. Possible implications of milk pasteurization on the manufacture and sensory quality of ripened cheese. Int. Dairy J. 1997, 7, 751–761. [Google Scholar] [CrossRef]
- Singh, H.; Waungana, A. Influence of heat treatment of milk on cheesemaking properties. Int. Dairy J. 2001, 11, 543–551. [Google Scholar] [CrossRef]
- Slade, L.; Levine, H. Beyond water activity: Recent advances based on an alternative approach to the assessment of food quality and safety. Crit. Rev. Food Sci. Nutr. 1991, 30, 115–360. [Google Scholar] [CrossRef]
- Brosnan, T.; Sun, D.-W. Inspection and grading of agricultural and food products by computer vision systems—A review. Comput. Electron. Agric. 2002, 36, 193–213. [Google Scholar] [CrossRef]
- Narendra, V.G.; Hareesha, K.S. Quality inspection and grading of agricultural and food products by computer vision—A review. Int. J. Comput. Appl. 2010, 2. [Google Scholar] [CrossRef]
- Wang, H.-H.; Sun, D.-W. Melting characteristics of cheese: Analysis of effect of cheese dimensions using computer vision techniques. J. Food Eng. 2002, 52, 279–284. [Google Scholar] [CrossRef]
- Caccamo, M.; Melilli, C.; Barbano, D.M.; Portelli, G.; Marino, G.; Licitra, G. Measurement of gas holes and mechanical openness in cheese by image analysis. J. Dairy Sci. 2004, 87, 739–748. [Google Scholar] [CrossRef] [Green Version]
- Ni, H.; Guansekaran, S. Image processing algorithm for cheese shred evaluation. J. Food Eng. 2004, 61, 37–45. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, C.; Liu, F.; Qiu, Z.; He, Y. Application of deep learning in food: A review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1793–1811. [Google Scholar] [CrossRef] [Green Version]
- Lundervold, A.S.; Lundervold, A. An overview of deep learning in medical imaging focusing on MRI. Z. Med. Phys. 2019, 29, 102–127. [Google Scholar] [CrossRef]
- Weiss, K.; Khoshgoftaar, T.M.; Wang, D. A survey of transfer learning. J. Big Data 2016, 3, 9. [Google Scholar] [CrossRef] [Green Version]

| T21 | T22 | AF1 | AF2 | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RC | HTC | RC | HTC | RC | HTC | RC | HTC | |||||||||||||||
| M | SD | M | SD | M | SD | M | SD | M | SD | M | SD | M | SD | M | SD | |||||||
| Season 1 | 105 days | S1 | 11.16 | 0.47 | 9.48 | 0.40 | * | 52.58 | 3.70 | 56.01 | 1.56 | * | 0.75 | 0.07 | 0.85 | 0.02 | * | 0.24 | 0.07 | 0.14 | 0.02 | * |
| S2 | 11.28 | 0.94 | 9.24 | 0.42 | * | 49.05 | 3.84 | 52.15 | 1.85 | * | 0.70 | 0.03 | 0.78 | 0.03 | * | 0.32 | 0.05 | 0.21 | 0.03 | * | ||
| S3 | 10.74 | 0.78 | 9.13 | 0.58 | * | 52.37 | 3.54 | 47.51 | 0.77 | * | 0.82 | 0.02 | 0.82 | 0.02 | 0.17 | 0.02 | 0.18 | 0.02 | ||||
| S4 | 8.46 | 0.31 | 8.64 | 0.51 | 48.33 | 4.15 | 52.67 | 3.12 | * | 0.71 | 0.05 | 0.77 | 0.03 | * | 0.28 | 0.05 | 0.21 | 0.03 | * | |||
| S5 | 8.86 | 0.31 | 9.33 | 0.52 | * | 51.73 | 1.48 | 49.69 | 1.46 | * | 0.77 | 0.02 | 0.81 | 0.02 | * | 0.23 | 0.02 | 0.19 | 0.02 | * | ||
| 180 days | S1 | 11.94 | 1.17 | 8.76 | 0.24 | * | 47.83 | 3.91 | 48.31 | 2.23 | 0.52 | 0.09 | 0.62 | 0.03 | * | 0.44 | 0.06 | 0.37 | 0.03 | * | ||
| S2 | 8.83 | 0.14 | 8.18 | 0.23 | * | 44.92 | 1.38 | 53.34 | 1.05 | * | 0.74 | 0.02 | 0.77 | 0.02 | * | 0.25 | 0.02 | 0.22 | 0.02 | * | ||
| S3 | 10.52 | 0.91 | 7.79 | 0.47 | * | 51.98 | 1.92 | 44.31 | 1.81 | * | 0.79 | 0.01 | 0.79 | 0.03 | 0.20 | 0.01 | 0.20 | 0.02 | ||||
| S4 | 9.74 | 1.64 | 7.98 | 0.28 | * | 40.67 | 3.29 | 47.27 | 2.17 | * | 0.56 | 0.03 | 0.70 | 0.03 | * | 0.42 | 0.08 | 0.29 | 0.03 | * | ||
| S5 | 8.50 | 0.31 | 8.80 | 0.74 | * | 49.42 | 1.49 | 46.66 | 1.93 | * | 0.67 | 0.03 | 0.69 | 0.05 | * | 0.32 | 0.03 | 0.30 | 0.05 | * | ||
| Season 2 | 105 days | S1 | 12.70 | 1.60 | 12.70 | 0.69 | 52.47 | 2.85 | 57.72 | 2.80 | * | 0.68 | 0.05 | 0.78 | 0.04 | * | 0.31 | 0.05 | 0.22 | 0.04 | * | |
| S2 | 14.11 | 1.45 | 12.23 | 0.93 | * | 54.40 | 1.59 | 53.04 | 2.98 | * | 0.63 | 0.06 | 0.78 | 0.04 | * | 0.36 | 0.06 | 0.21 | 0.04 | * | ||
| S3 | 10.26 | 0.26 | 10.02 | 0.39 | * | 45.93 | 1.05 | 52.22 | 1.46 | * | 0.79 | 0.01 | 0.84 | 0.03 | * | 0.20 | 0.02 | 0.16 | 0.03 | * | ||
| S4 | 10.20 | 1.48 | 10.49 | 0.31 | 46.95 | 3.28 | 50.16 | 2.18 | * | 0.67 | 0.07 | 0.79 | 0.01 | * | 0.32 | 0.07 | 0.20 | 0.01 | * | |||
| S5 | 9.29 | 1.08 | 10.74 | 0.42 | * | 43.55 | 3.08 | 53.18 | 1.05 | * | 0.68 | 0.02 | 0.82 | 0.01 | * | 0.31 | 0.02 | 0.17 | 0.01 | * | ||
| 180 days | S1 | 9.08 | 0.18 | 8.82 | 0.50 | * | 49.26 | 2.67 | 52.37 | 4.38 | * | 0.62 | 0.04 | 0.78 | 0.06 | * | 0.37 | 0.04 | 0.21 | 0.06 | * | |
| S2 | 9.94 | 0.66 | 9.21 | 0.44 | * | 53.10 | 2.35 | 52.33 | 1.93 | 0.72 | 0.05 | 0.80 | 0.03 | * | 0.27 | 0.05 | 0.19 | 0.03 | * | |||
| S3 | 10.39 | 0.50 | 8.78 | 0.35 | * | 50.97 | 1.01 | 52.90 | 1.61 | * | 0.72 | 0.03 | 0.76 | 0.03 | * | 0.27 | 0.03 | 0.23 | 0.03 | * | ||
| S4 | 9.09 | 0.74 | 8.31 | 0.51 | * | 50.87 | 3.87 | 48.03 | 3.28 | * | 0.68 | 0.05 | 0.73 | 0.03 | * | 0.31 | 0.05 | 0.26 | 0.03 | * | ||
| S5 | 14.55 | 2.24 | 10.07 | 0.38 | * | 44.94 | 3.34 | 53.00 | 1.14 | * | 0.57 | 0.03 | 0.72 | 0.02 | * | 0.42 | 0.03 | 0.27 | 0.02 | * | ||
| T21 | T22 | Area 1 | Area 2 | |||||
|---|---|---|---|---|---|---|---|---|
| M | SD | M | SD | M | SD | M | SD | |
| I1 | 8.301 | 0.055 | 40.337 | 0.100 | 0.835 | 0.002 | 0.157 | 0.002 |
| I2 | 10.032 | 0.056 | 52.046 | 0.141 | 0.891 | 0.001 | 0.102 | 0.001 |
| I3 | 9.214 | 0.038 | 45.078 | 0.035 | 0.814 | 0.001 | 0.178 | 0.001 |
| I4 | 8.523 | 0.030 | 29.646 | 0.043 | 0.871 | 0.002 | 0.120 | 0.002 |
| I5 | 8.018 | 0.024 | 51.936 | 0.047 | 0.697 | 0.002 | 0.294 | 0.002 |
| I6 | 8.742 | 0.020 | 44.319 | 0.042 | 0.836 | 0.001 | 0.156 | 0.001 |
| I7 | 9.318 | 0.023 | 51.352 | 0.133 | 0.858 | 0.012 | 0.130 | 0.000 |
| I8 | 9.462 | 0.022 | 51.054 | 0.058 | 0.813 | 0.001 | 0.180 | 0.000 |
| I9 | 9.463 | 0.018 | 51.058 | 0.037 | 0.813 | 0.001 | 0.180 | 0.001 |
| I10 | 9.466 | 0.114 | 48.470 | 0.292 | 0.876 | 0.002 | 0.117 | 0.002 |
| I11 | 9.188 | 0.047 | 46.314 | 0.131 | 0.843 | 0.001 | 0.150 | 0.001 |
| I12 | 7.702 | 0.098 | 34.887 | 0.084 | 0.575 | 0.004 | 0.415 | 0.004 |
| I13 | 7.301 | 0.029 | 34.772 | 0.013 | 0.545 | 0.002 | 0.442 | 0.002 |
| RC | HTC | Maturer | Industrial | |||
|---|---|---|---|---|---|---|
| S1 | Season 1 | 105 days | 14.99 ± 1.70 a | 11.17 ± 0.62 c | 13.39 ± 0.18 b | 10.92 ± 0.62 c |
| 180 days | 23.83 ± 4.83 a | 14.22 ± 1.22 b | 13.39 ± 0.18 b | 10.92 ± 0.62 c | ||
| Season 2 | 105 days | 20.25 ± 2.38 a | 16.43 ± 1.60 b | 13.39 ± 0.18 c | 10.92 ± 0.62 d | |
| 180 days | 14.78 ± 1.00 a | 11.38 ± 1.45 c | 13.39 ± 0.18 b | 10.92 ± 0.62 c | ||
| S2 | Season 1 | 105 days | 16.18 ± 1.85 a | 11.87 ± 0.90 c | 13.39 ± 0.18 b | 10.92 ± 0.62 d |
| 180 days | 11.90 ± 0.81 b | 10.57 ± 0.48 c | 13.39 ± 0.18 a | 10.92 ± 0.62 c | ||
| Season 2 | 105 days | 24.10 ± 3.44 a | 15.74 ± 1.59 b | 13.39 ± 0.18 c | 10.92 ± 0.62 d | |
| 180 days | 13.86 ± 1.74 a | 11.58 ± 0.86 b | 13.39 ± 0.18 a | 10.92 ± 0.62 c | ||
| S3 | Season 1 | 105 days | 13.12 ± 1.19 a | 11.19 ± 0.90 b | 13.39 ± 0.18 a | 10.92 ± 0.62 b |
| 180 days | 13.33 ± 1.29 a | 9.86 ± 0.73 c | 13.39 ± 0.18 a | 10.92 ± 0.62 b | ||
| Season 2 | 105 days | 13.02 ± 0.51 a | 11.98 ± 0.82 b | 13.39 ± 0.18 a | 10.92 ± 0.62 c | |
| 180 days | 14.47 ± 0.94 a | 11.53 ± 0.70 c | 13.39 ± 0.18 b | 10.92 ± 0.62 d | ||
| S4 | Season 1 | 105 days | 12.00 ± 0.97 b | 11.18 ± 0.75 c | 13.39 ± 0.18 a | 10.92 ± 0.62 c |
| 180 days | 17.74 ± 4.27 a | 11.48 ± 0.75 bc | 13.39 ± 0.18 b | 10.92 ± 0.62 c | ||
| Season 2 | 105 days | 15.65 ± 3.68 a | 13.23 ± 0.40 b | 13.39 ± 0.18 b | 10.92 ± 0.62 c | |
| 180 days | 13.41 ± 1.77 a | 11.42 ± 0.96 b | 13.39 ± 0.18 a | 10.92 ± 0.62 b | ||
| S5 | Season 1 | 105 days | 11.58 ± 0.56 b | 11.57 ± 0.81 b | 13.39 ± 0.18 a | 10.92 ± 0.62 c |
| 180 days | 12.74 ± 0.81 a | 12.76 ± 1.39 a | 13.39 ± 0.18 a | 10.92 ± 0.62 b | ||
| Season 2 | 105 days | 12.93 ± 1.10 a | 13.08 ± 0.53 a | 13.39 ± 0.18 a | 10.92 ± 0.62 b | |
| 180 days | 25.61 ± 2.79 a | 14.05 ± 0.69 b | 13.39 ± 0.18 b | 10.92 ± 0.62 c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anedda, R.; Melis, R.; Curti, E. Quality Control in Fiore Sardo PDO Cheese: Detection of Heat Treatment Application and Production Chain by MRI Relaxometry and Image Analysis. Dairy 2021, 2, 270-287. https://doi.org/10.3390/dairy2020023
Anedda R, Melis R, Curti E. Quality Control in Fiore Sardo PDO Cheese: Detection of Heat Treatment Application and Production Chain by MRI Relaxometry and Image Analysis. Dairy. 2021; 2(2):270-287. https://doi.org/10.3390/dairy2020023
Chicago/Turabian StyleAnedda, Roberto, Riccardo Melis, and Elena Curti. 2021. "Quality Control in Fiore Sardo PDO Cheese: Detection of Heat Treatment Application and Production Chain by MRI Relaxometry and Image Analysis" Dairy 2, no. 2: 270-287. https://doi.org/10.3390/dairy2020023
APA StyleAnedda, R., Melis, R., & Curti, E. (2021). Quality Control in Fiore Sardo PDO Cheese: Detection of Heat Treatment Application and Production Chain by MRI Relaxometry and Image Analysis. Dairy, 2(2), 270-287. https://doi.org/10.3390/dairy2020023