Glycoproteomic and Lipidomic Characterization of Industrially Produced Whey Protein Phospholipid Concentrate with Emphasis on Antimicrobial Xanthine Oxidase, Oxylipins and Small Milk Fat Globules
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and Chemicals
2.2. Selection of Samples for Specific Analyses
2.3. Compositional Analysis of Whey Protein Phospholipid Concentrate (WPPC) and Bovine Milk
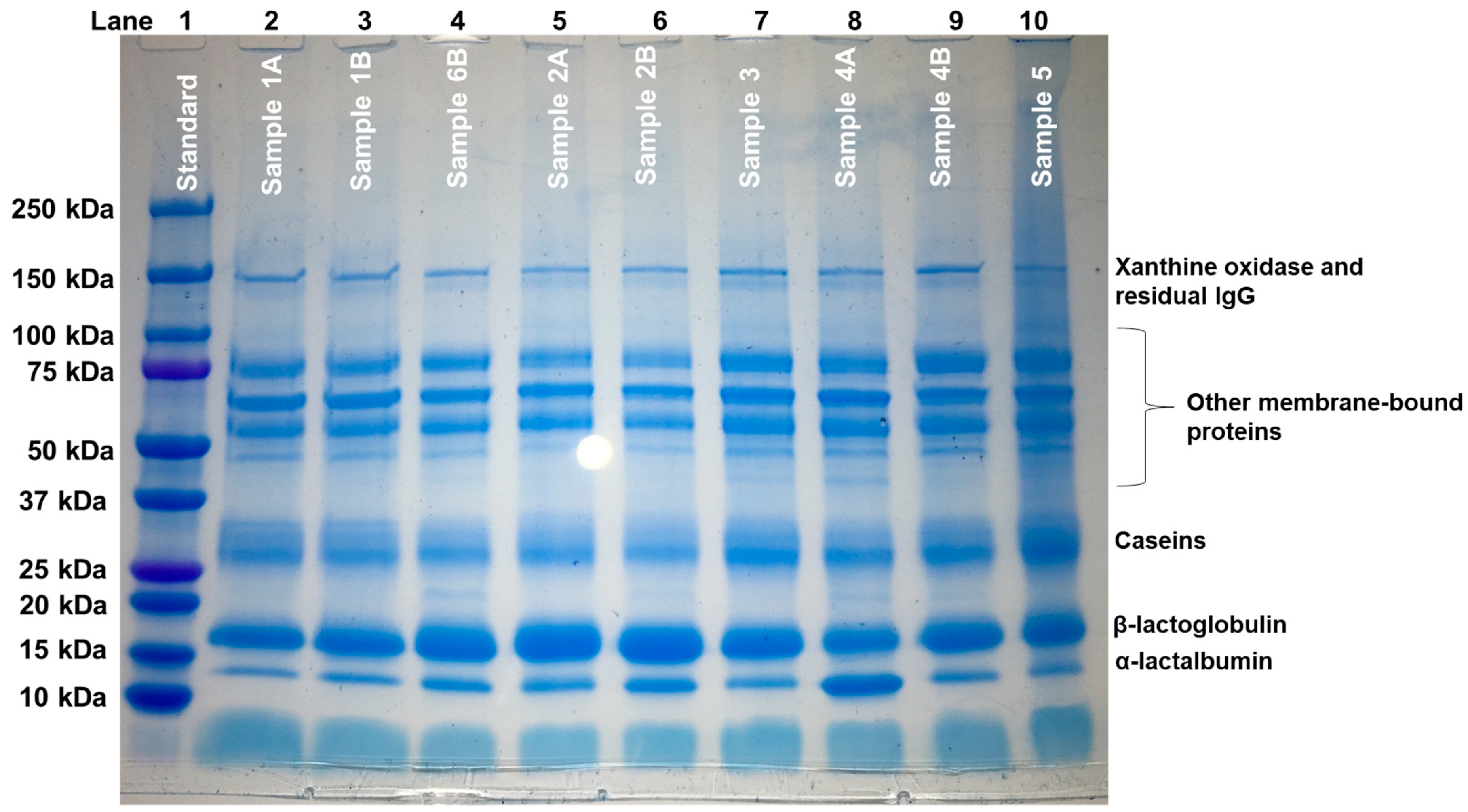
2.4. Identification of Proteins in WPPC and Comparison with WPI by SDS-PAGE
2.5. In-Gel Digestion and Proteomics
2.6. Proteomics Data Analysis
2.7. N-linked Glycans: Release from Glycoproteins and Analysis by Mass Spectrometry
2.8. Sialic Acid Quantification
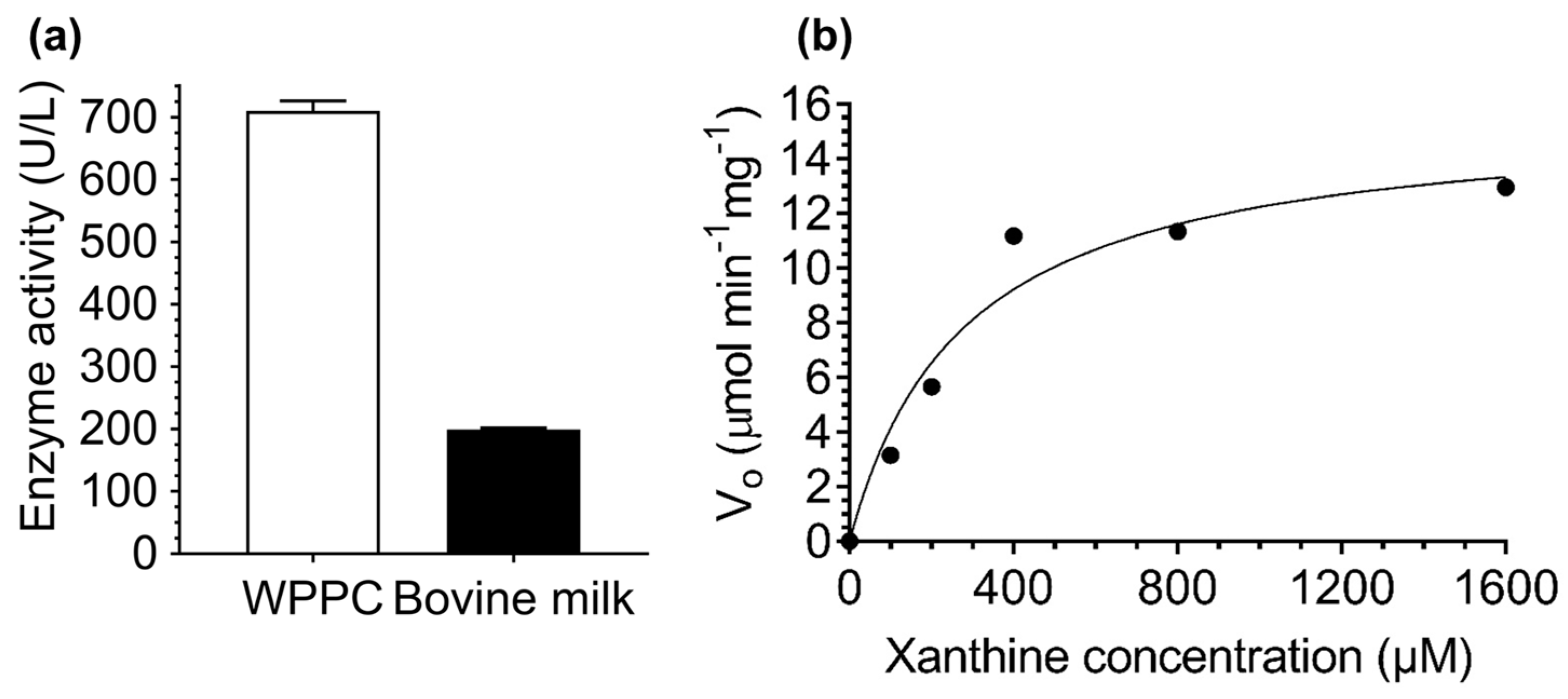
2.9. Determination of Xanthine Oxidase Activity in WPPC and Kinetic Parameters
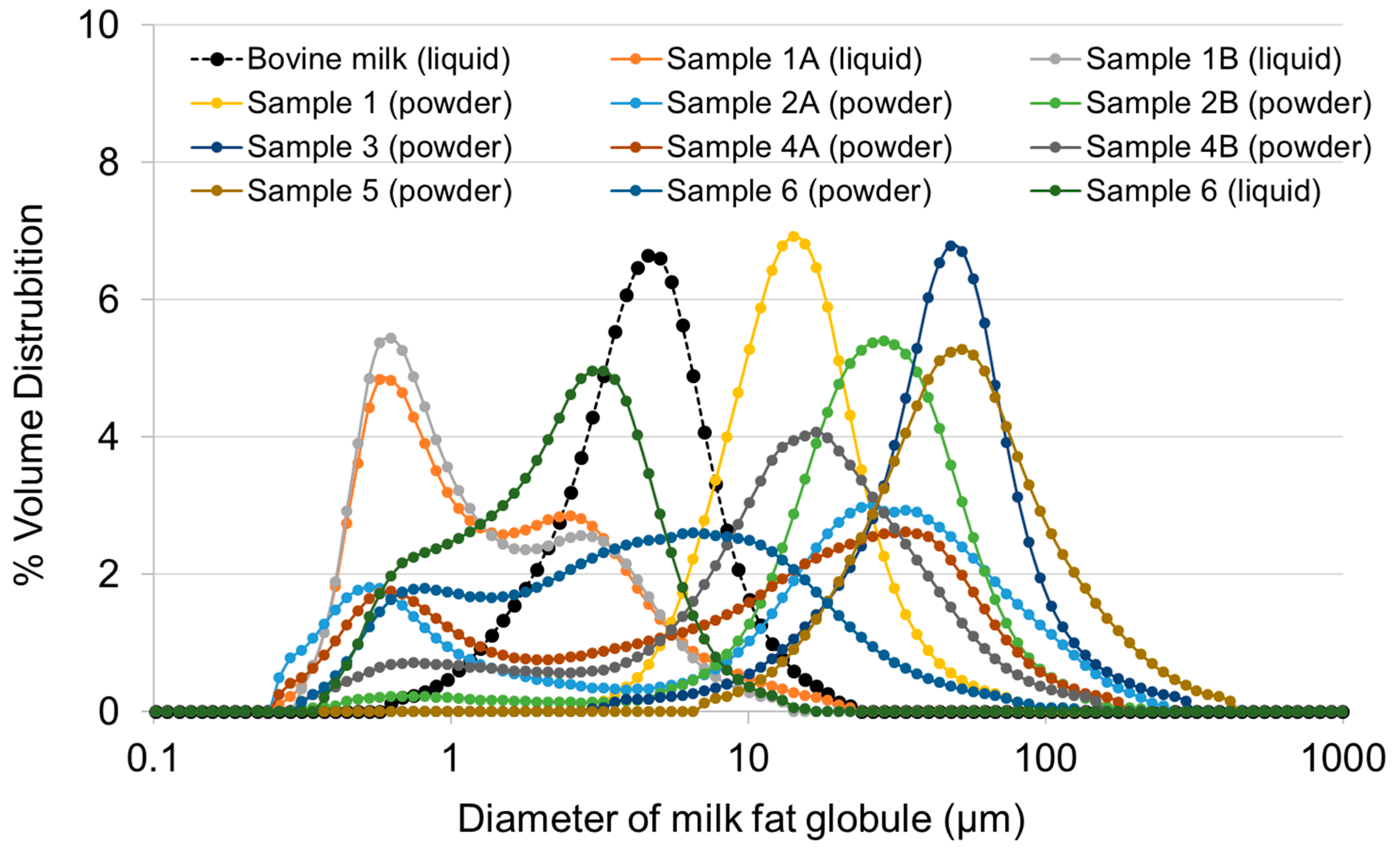
2.10. Measuring Fat Globule Size Distribution in WPPC and a Comparison to Bovine Milk
2.11. Folch Extraction for the Total Lipids
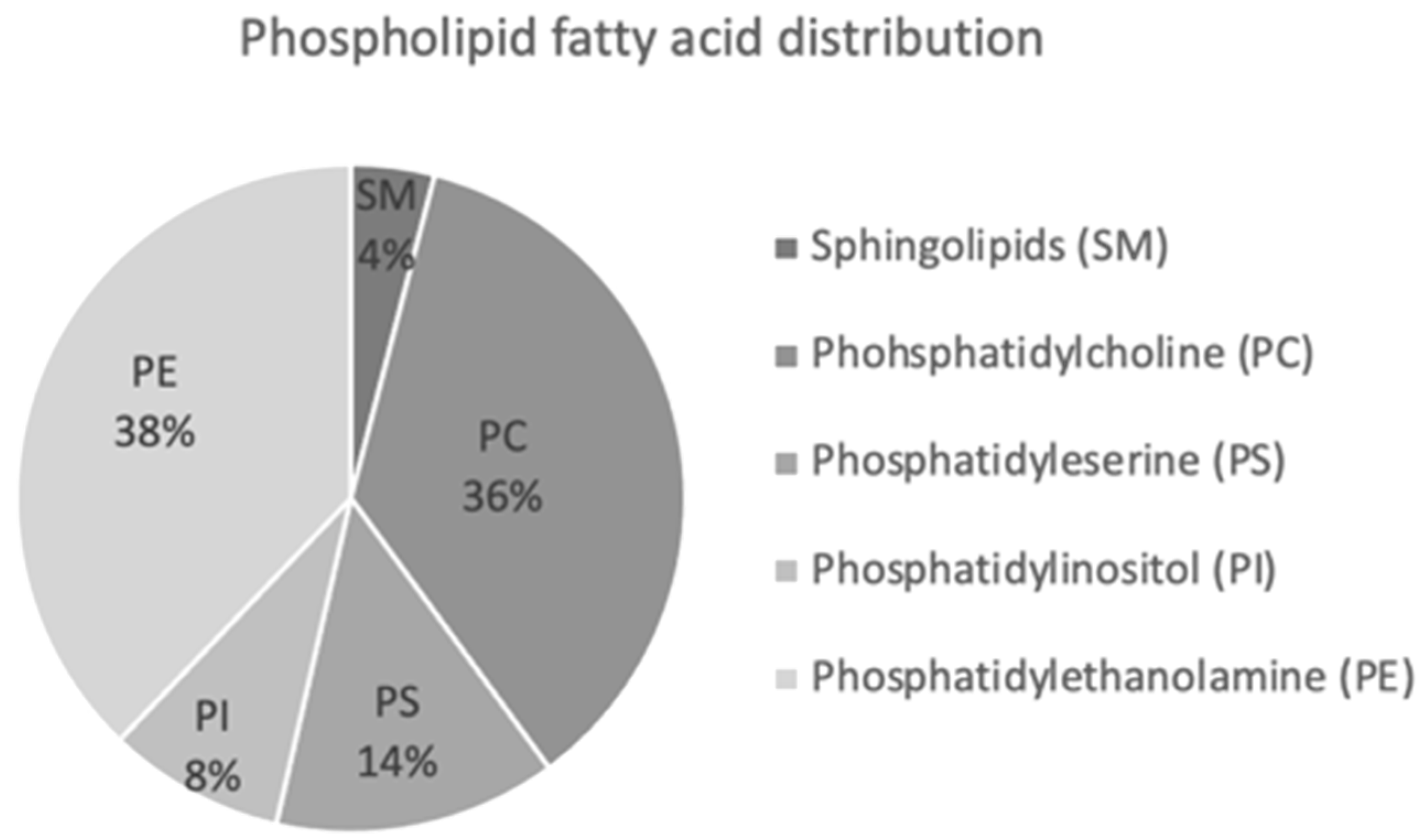
2.12. Thin-Layer Chromatogram (TLC) Separation of the Major Phospholipid Classes
2.13. Derivatization of Total Lipids and TLC-Separated Lipid Fractions
2.14. GC Analysis of Fatty Acid Methyl Ester (FAME)
2.15. Extraction of Free Oxylipins from the WPPC Samples
2.16. Liberation of the Free Oxylipins from Total Oxylipins from the Folch Extract
2.17. UPLC-MS/MS Analysis of the Free and Total Oxylipins
3. Results and Discussion
3.1. Compositional Analysis of Whey Protein Phospholipid Concentrate (WPPC) and a Comparison to Bovine Milk
3.2. Identification of Proteins in WPPC by SDS-PAGE
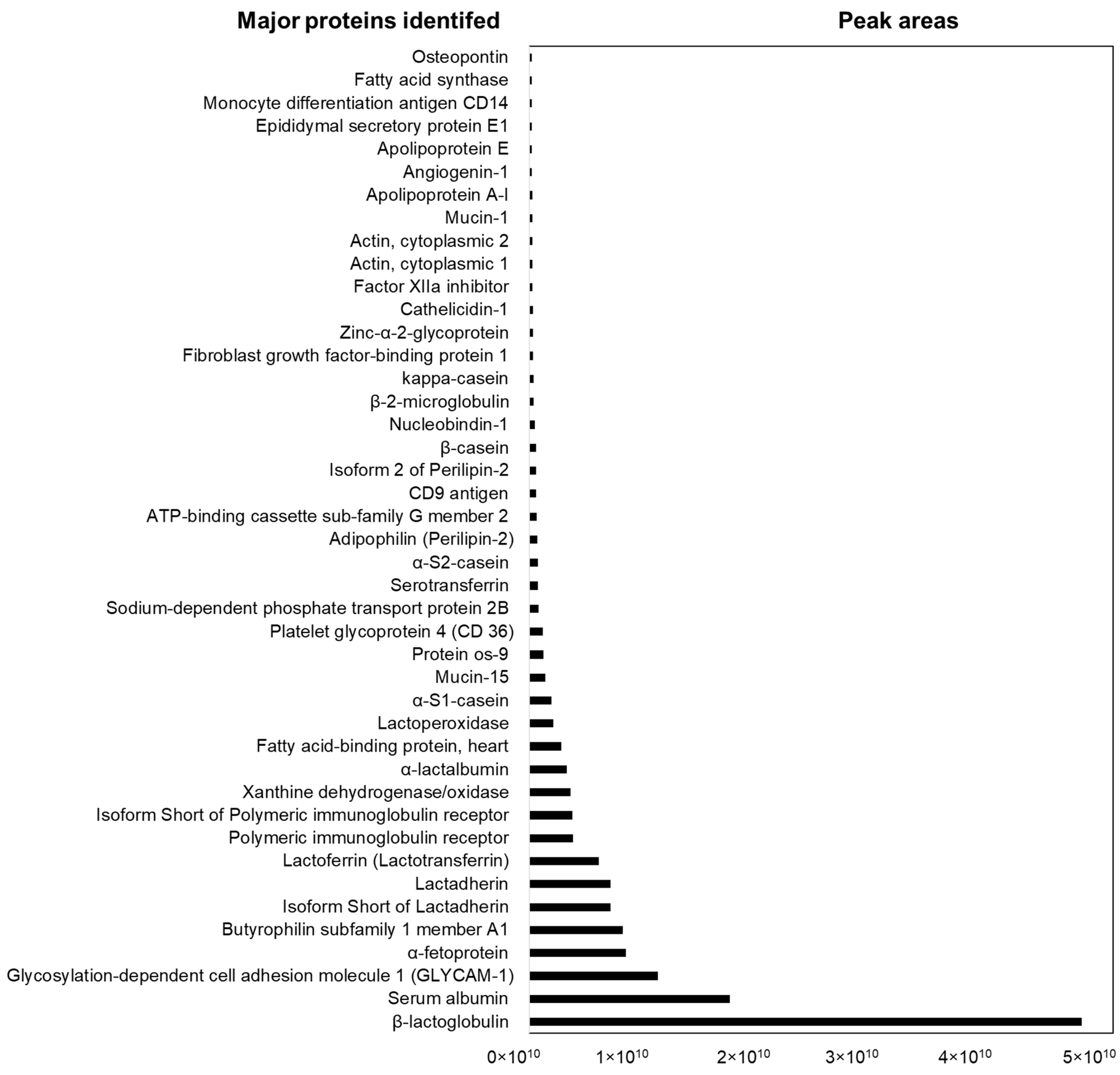
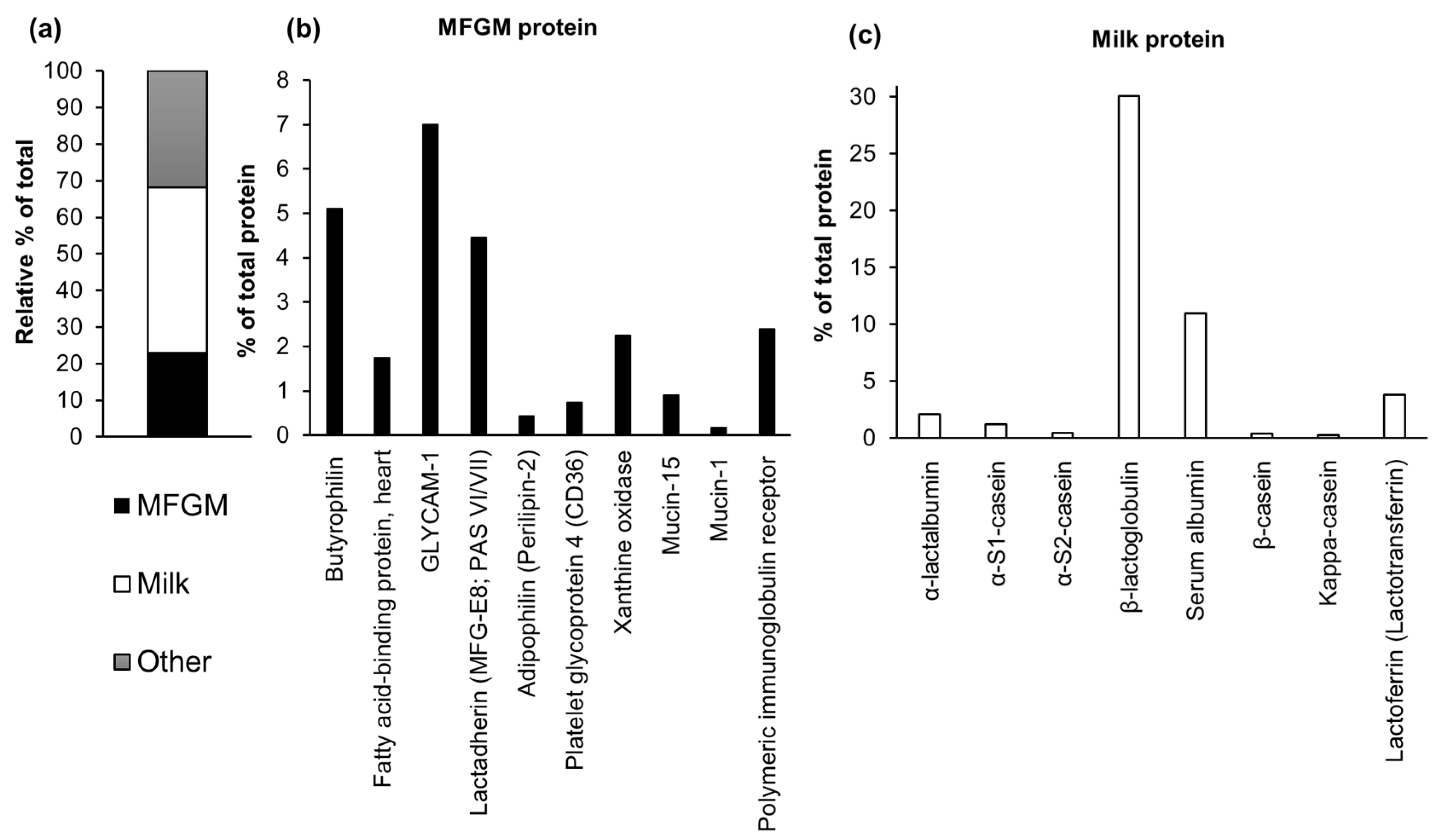
3.3. Proteomic Analysis
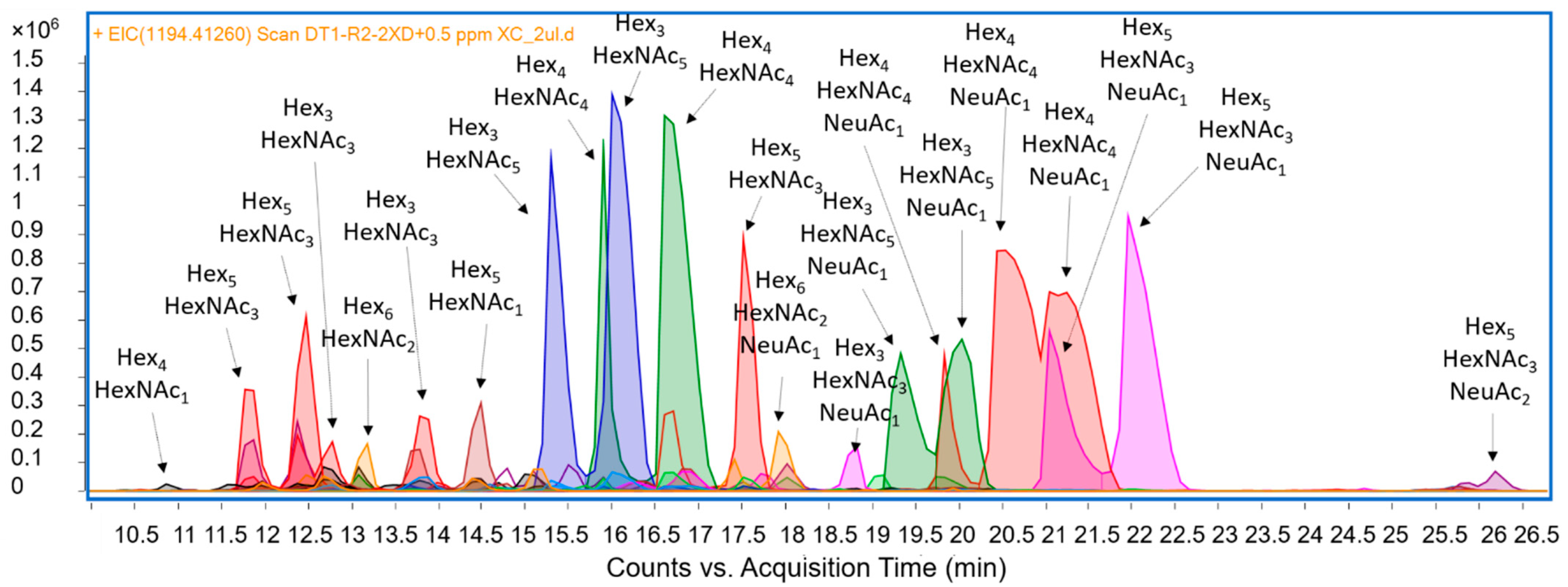
3.4. N-Linked Glycans (N-Glycans)
3.5. Sialic Acid
3.6. Determination of Xanthine Oxidase Activity and Kinetic Parameters in WPPC and a Comparison to Bovine Milk
3.7. Measuring the Fat Globule Size in WPPC
- (i)
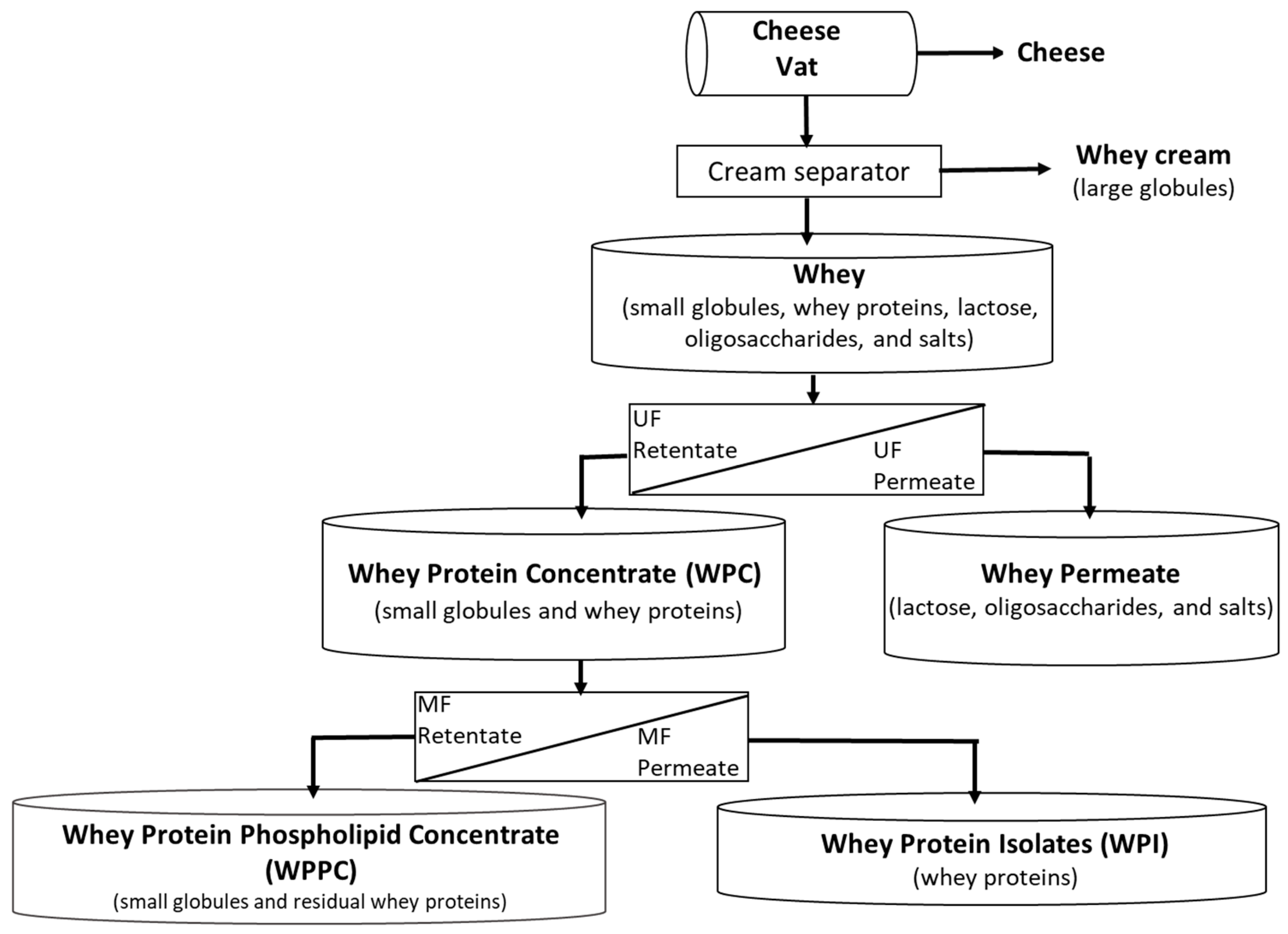
- A centrifugal cream separator, which fractionates MFG by density, and removes the larger, less-dense MFG from the whey. The resulting whey contains 0.4 to 0.5% residual fat (small MFG);
- (ii)
- Ultrafiltration, 10 KDa cut off, separates whey proteins and small MFG from smaller molecules, such as lactose, oligosaccharides, and salts;
- (iii)
- Microfiltration with a 0.1 um pore size was used to produce highly purified whey protein WPI (permeate) by the dairy industry. As a result, there is a significant accumulation of small MFG and residual whey proteins in the microfiltration retentate.
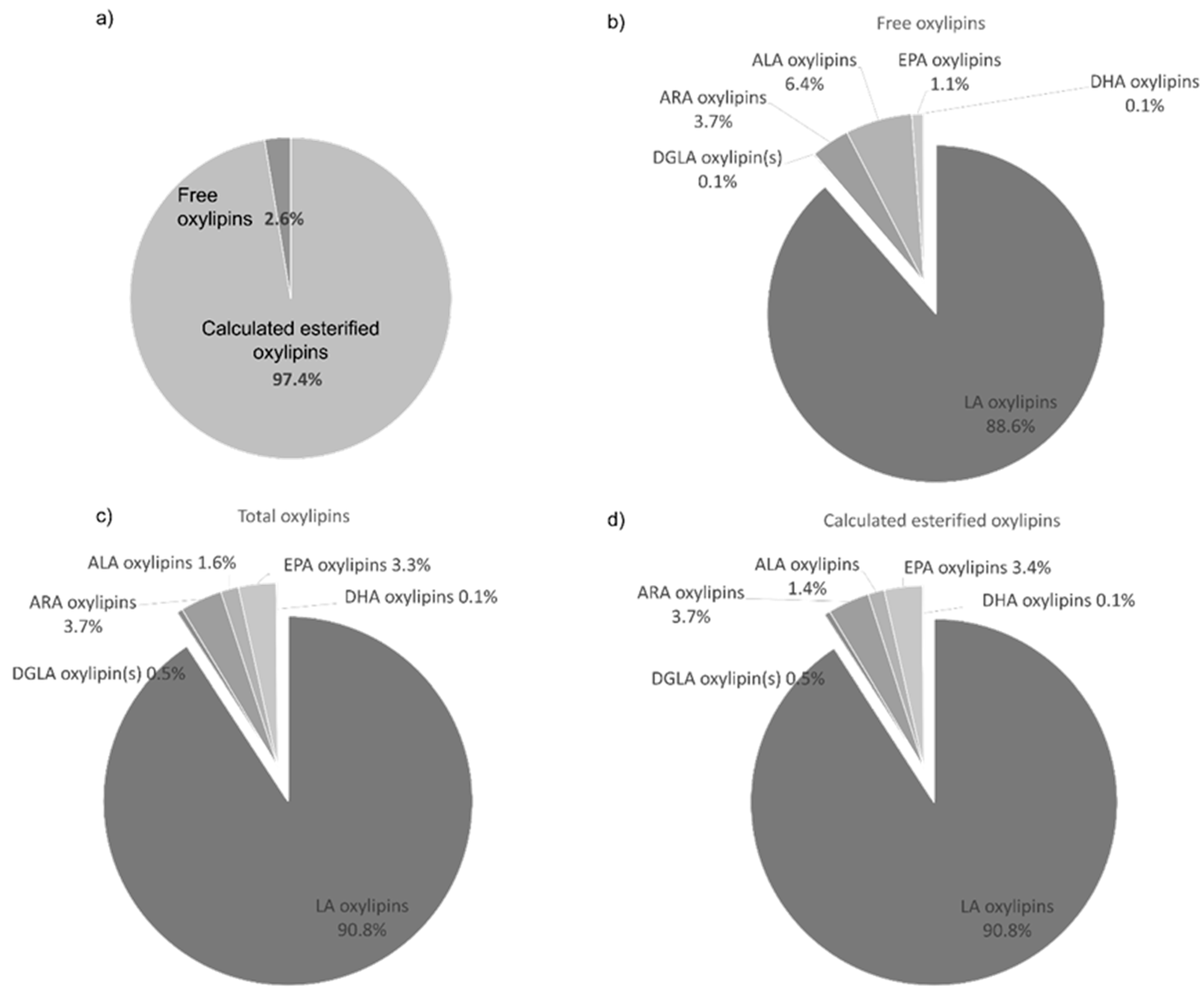
3.8. Lipidomic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hinde, K.; German, J.B. Food in an Evolutionary Context: Insights from Mother’s Milk. J. Sci. Food Agric. 2012, 92, 2219–2223. [Google Scholar] [CrossRef] [PubMed]
- Barile, D.; Tao, N.; Lebrilla, C.B.; Coisson, J.-D.; Arlorio, M.; German, J.B. Permeate from Cheese Whey Ultrafiltration Is a Source of Milk Oligosaccharides. Int. Dairy J. 2009, 19, 524–530. [Google Scholar] [CrossRef]
- Charbonneau, M.R.; O’Donnell, D.; Blanton, L.V.; Totten, S.M.; Davis, J.C.C.; Barratt, M.J.; Cheng, J.; Guruge, J.; Talcott, M.; Bain, J.R.; et al. Sialylated Milk Oligosaccharides Promote Microbiota-Dependent Growth in Models of Infant Undernutrition. Cell 2016, 164, 859–871. [Google Scholar] [CrossRef] [PubMed]
- Pennisi, E. Gut Microbes May Help Malnourished Children. Science 2019, 365, 109. [Google Scholar] [CrossRef] [PubMed]
- Dewettinck, K.; Rombaut, R.; Thienpont, N.; Le, T.T.; Messens, K.; Van Camp, J. Nutritional and Technological Aspects of Milk Fat Globule Membrane Material. Int. Dairy J. 2008, 18, 436–457. [Google Scholar] [CrossRef]
- Lee, H.; Padhi, E.; Hasegawa, Y.; Larke, J.; Parenti, M.; Wang, A.; Hernell, O.; Lönnerdal, B.; Slupsky, C. Compositional Dynamics of the Milk Fat Globule and Its Role in Infant Development. Front. Pediatr. 2018, 6, 313. [Google Scholar] [CrossRef]
- Fontecha, J.; Brink, L.; Wu, S.; Pouliot, Y.; Visioli, F.; Jiménez-Flores, R. Sources, Production, and Clinical Treatments of Milk Fat Globule Membrane for Infant Nutrition and Well-Being. Nutrients 2020, 12, 1607. [Google Scholar] [CrossRef]
- Michalski, M.-C.; Briard, V.; Michel, F. Optical Parameters of Milk Fat Globules for Laser Light Scattering Measurements. Le Lait 2001, 81, 787–796. [Google Scholar] [CrossRef]
- Wiking, L.; Stagsted, J.; Björck, L.; Nielsen, J.H. Milk Fat Globule Size Is Affected by Fat Production in Dairy Cows. Int. Dairy J. 2004, 14, 909–913. [Google Scholar] [CrossRef]
- Heid, H.W.; Keenan, T.W. Intracellular Origin and Secretion of Milk Fat Globules. Eur. J. Cell Biol. 2005, 84, 245–258. [Google Scholar] [CrossRef]
- Argov, N.; Lemay, D.G.; German, J.B. Milk Fat Globule Structure and Function: Nanoscience Comes to Milk Production. Trends Food Sci. Technol. 2008, 19, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Fong, B.Y.; Norris, C.S.; MacGibbon, A.K.H. Protein and Lipid Composition of Bovine Milk-Fat-Globule Membrane. Int. Dairy J. 2007, 17, 275–288. [Google Scholar] [CrossRef]
- Riccio, P. The Proteins of the Milk Fat Globule Membrane in the Balance. Trends Food Sci. Technol. 2004, 15, 458–461. [Google Scholar] [CrossRef]
- Brink, L.R.; Lönnerdal, B. The Role of Milk Fat Globule Membranes in Behavior and Cognitive Function Using a Suckling Rat Pup Supplementation Model. J. Nutr. Biochem. 2018, 58, 131–137. [Google Scholar] [CrossRef]
- Favre, L.; Bosco, N.; Roggero, I.S.; Corthésy, B.; Benyacoub, J. Combination of Milk Fat Globule Membranes and Probiotics to Potentiate Neonatal Immune Maturation and Early Life Protection. Pediatr. Res. 2011, 70, 439. [Google Scholar] [CrossRef]
- Zhang, L.; Chichlowski, M.; Gross, G.; Holle, M.J.; lbarra-Sánchez, L.A.; Wang, S.; Miller, M.J. Milk Fat Globule Membrane Protects Lactobacillus Rhamnosus GG from Bile Stress by Regulating Exopolysaccharide Production and Biofilm Formation. J. Agric. Food Chem. 2020, 68, 6646–6655. [Google Scholar] [CrossRef]
- Mather, I.H. A Review and Proposed Nomenclature for Major Proteins of the Milk-Fat Globule Membrane. J. Dairy Sci. 2000, 83, 203–247. [Google Scholar] [CrossRef]
- Al-Shehri, S.S.; Knox, C.L.; Liley, H.G.; Cowley, D.M.; Wright, J.R.; Henman, M.G.; Hewavitharana, A.K.; Charles, B.G.; Shaw, P.N.; Sweeney, E.L.; et al. Breastmilk-Saliva Interactions Boost Innate Immunity by Regulating the Oral Microbiome in Early Infancy. PLoS ONE 2015, 10, e0135047. [Google Scholar] [CrossRef]
- Ozturk, G.; Shah, I.M.; Mills, D.A.; German, J.B.; de Moura Bell, J.M.L.N. The Antimicrobial Activity of Bovine Milk Xanthine Oxidase. Int. Dairy J. 2020, 102, 104581. [Google Scholar] [CrossRef]
- Navab, M.; Hama, S.; Hough, G.; Fogelman, A.M. Oral Synthetic Phospholipid (DMPC) Raises High-Density Lipoprotein Cholesterol Levels, Improves High-Density Lipoprotein Function, and Markedly Reduces Atherosclerosis in Apolipoprotein E-Null Mice. Circulation 2003, 108, 1735–1739. [Google Scholar] [CrossRef]
- Vors, C.; Joumard-Cubizolles, L.; Lecomte, M.; Combe, E.; Ouchchane, L.; Drai, J.; Raynal, K.; Joffre, F.; Meiller, L.; Barz, M.L.; et al. Milk Polar Lipids Reduce Lipid Cardiovascular Risk Factors in Overweight Postmenopausal Women: Towards a Gut Sphingomyelin-Cholesterol Interplay. Gut 2020, 69, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Kamili, A.; Wat, E.; Chung, R.W.; Tandy, S.; Weir, J.M.; Meikle, P.J.; Cohn, J.S. Hepatic Accumulation of Intestinal Cholesterol Is Decreased and Fecal Cholesterol Excretion Is Increased in Mice Fed a High-Fat Diet Supplemented with Milk Phospholipids. Nutr. Metab. 2010, 7, 90. [Google Scholar] [CrossRef] [PubMed]
- Wat, E.; Tandy, S.; Kapera, E.; Kamili, A.; Chung, R.W.S.; Brown, A.; Rowney, M.; Cohn, J.S. Dietary Phospholipid-Rich Dairy Milk Extract Reduces Hepatomegaly, Hepatic Steatosis and Hyperlipidemia in Mice Fed a High-Fat Diet. Atherosclerosis 2009, 205, 144–150. [Google Scholar] [CrossRef]
- Wang, X.; Kong, X.; Qin, Y.; Zhu, X.; Qu, D.; Han, J. Milk Phospholipid Supplementation Mediates Colonization Resistance of Mice against Salmonella Infection in Association with Modification of Gut Microbiota. Food Funct. 2020, 11, 6078–6090. [Google Scholar] [CrossRef] [PubMed]
- Snow, D.R.; Ward, R.E.; Olsen, A.; Jimenez-Flores, R.; Hintze, K.J. Membrane-Rich Milk Fat Diet Provides Protection against Gastrointestinal Leakiness in Mice Treated with Lipopolysaccharide. J. Dairy Sci. 2011, 94, 2201–2212. [Google Scholar] [CrossRef] [PubMed]
- Lordan, R.; Zabetakis, I. Invited Review: The Anti-Inflammatory Properties of Dairy Lipids. J. Dairy Sci. 2017, 100, 4197–4212. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Li, B.; Gao, S.; Duan, R.-D. Dietary Sphingomyelin Inhibits Colonic Tumorigenesis with an Up-Regulation of Alkaline Sphingomyelinase Expression in ICR Mice. Anticancer Res. 2008, 28, 3631–3635. [Google Scholar]
- Pepeu, G.; Pepeu, I.M.; Amaducci, L. A Review of Phosphatidylserine Pharmacological and Clinical Effects. Is Phosphatidylserine a Drug for the Ageing Brain? Pharmacol. Res. 1996, 33, 73–80. [Google Scholar] [CrossRef]
- Cenacchi, T.; Bertoldin, T.; Farina, C.; Fiori, M.G.; Crepaldi, G. Cognitive Decline in the Elderly: A Double-Blind, Placebo-Controlled Multicenter Study on Efficacy of Phosphatidylserine Administration. Aging Milan Italy 1993, 5, 123–133. [Google Scholar] [CrossRef]
- Tomé-Carneiro, J.; Carmen Crespo, M.; Burgos-Ramos, E.; Tomas-Zapico, C.; García-Serrano, A.; Castro-Gómez, P.; Venero, C.; Pereda-Pérez, I.; Baliyan, S.; Valencia, A.; et al. Buttermilk and Krill Oil Phospholipids Improve Hippocampal Insulin Resistance and Synaptic Signaling in Aged Rats. Mol. Neurobiol. 2018, 55, 7285–7296. [Google Scholar] [CrossRef]
- Brink, L.R.; Gueniot, J.P.; Lönnerdal, B. Effects of Milk Fat Globule Membrane and Its Various Components on Neurologic Development in a Postnatal Growth Restriction Rat Model. J. Nutr. Biochem. 2019, 69, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Hosozawa, M.; Kudo, N.; Yoshikawa, N.; Hisata, K.; Shoji, H.; Shinohara, K.; Shimizu, T. The Pilot Study: Sphingomyelin-Fortified Milk Has a Positive Association with the Neurobehavioural Development of Very Low Birth Weight Infants during Infancy, Randomized Control Trial. Brain Dev. 2013, 35, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Timby, N.; Domellöf, E.; Hernell, O.; Lönnerdal, B.; Domellöf, M. Neurodevelopment, Nutrition, and Growth until 12 Mo of Age in Infants Fed a Low-Energy, Low-Protein Formula Supplemented with Bovine Milk Fat Globule Membranes: A Randomized Controlled Trial. Am. J. Clin. Nutr. 2014, 99, 860–868. [Google Scholar] [CrossRef] [PubMed]
- Higgs, G.A.; Salmon, J.A.; Spayne, J.A. The Inflammatory Effects of Hydroperoxy and Hydroxy Acid Products of Arachidonate Lipoxygenase in Rabbit Skin. Br. J. Pharmacol. 1981, 74, 429–433. [Google Scholar] [CrossRef]
- Tian, Y.; Gou, J.; Zhang, H.; Lu, J.; Jin, Z.; Jia, S.; Bai, L. The Anti-Inflammatory Effects of 15-HETE on Osteoarthritis during Treadmill Exercise. Life Sci. 2021, 273, 119260. [Google Scholar] [CrossRef] [PubMed]
- Rosqvist, F.; Smedman, A.; Lindmark-Månsson, H.; Paulsson, M.; Petrus, P.; Straniero, S.; Rudling, M.; Dahlman, I.; Risérus, U. Potential Role of Milk Fat Globule Membrane in Modulating Plasma Lipoproteins, Gene Expression, and Cholesterol Metabolism in Humans: A Randomized Study. Am. J. Clin. Nutr. 2015, 102, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Pesta, G.; Meyer-Pittroff, R.; Russ, W. Utilization of Whey. In Utilization of By-Products and Treatment of Waste in the Food Industry; Oreopoulou, V., Russ, W., Eds.; Springer: New York, NY, USA, 2007; pp. 193–207. ISBN 978-0-387-33511-7. [Google Scholar]
- Saxena, A.; Tripathi, B.P.; Kumar, M.; Shahi, V.K. Membrane-Based Techniques for the Separation and Purification of Proteins: An Overview. Adv. Colloid Interface Sci. 2009, 145, 1–22. [Google Scholar] [CrossRef]
- Bylund, G. Whey Processing. In Dairy Processing Handbook; Tetra Pak Processing Systems AB: Lund, Sweden, 2015. [Google Scholar]
- Burrington, K.J.; Schoenfuss, T.; Sonia, P. Technical Report: Coproducts of Milk and Whey Processing. Think USA Dairy by the U.S. Dairy Export Council. Available online: https://www.thinkusadairy.org/resources-and-insights/resources-and-insights/application-and-technical-materials/technical-report-coproducts-of-milk-and-whey-processing (accessed on 7 February 2022).
- Mollea, C.; Marmo, L.; Bosco, F. Valorisation of Cheese Whey, a By-Product from the Dairy Industry; IntechOpen: London, UK, 2013; ISBN 978-953-51-0911-2. [Google Scholar]
- Horwitz, W.; AOAC International. Official Methods of Analysis of AOAC International; AOAC International: Rockville, MA, USA, 2003. [Google Scholar]
- Gundry, R.L.; White, M.Y.; Murray, C.I.; Kane, L.A.; Fu, Q.; Stanley, B.A.; Van Eyk, J.E. Preparation of Proteins and Peptides for Mass Spectrometry Analysis in a Bottom-up Proteomics Workflow. In Current Protocols in Molecular Biology; Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., Struhl, K., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2009; ISBN 978-0-471-14272-0. [Google Scholar]
- Dallas, D.C.; Guerrero, A.; Khaldi, N.; Castillo, P.A.; Martin, W.F.; Smilowitz, J.T.; Bevins, C.L.; Barile, D.; German, J.B.; Lebrilla, C.B. Extensive in Vivo Human Milk Peptidomics Reveals Specific Proteolysis Yielding Protective Antimicrobial Peptides. J. Proteome Res. 2013, 12, 2295–2304. [Google Scholar] [CrossRef]
- Karav, S.; Le Parc, A.; Leite Nobrega de Moura Bell, J.M.; Frese, S.A.; Kirmiz, N.; Block, D.E.; Barile, D.; Mills, D.A. Oligosaccharides Released from Milk Glycoproteins Are Selective Growth Substrates for Infant-Associated Bifidobacteria. Appl. Environ. Microbiol. 2016, 82, 3622–3630. [Google Scholar] [CrossRef]
- Hurum, D.C.; Rohrer, J.S. Determination of Sialic Acids in Infant Formula by Chromatographic Methods: A Comparison of High-Performance Anion-Exchange Chromatography with Pulsed Amperometric Detection and Ultra-High-Performance Liquid Chromatography Methods. J. Dairy Sci. 2012, 95, 1152–1161. [Google Scholar] [CrossRef]
- Cerbulis, J.; Farrell, H.M. Xanthine Oxidase Activity in Dairy Products. J. Dairy Sci. 1977, 60, 170–176. [Google Scholar] [CrossRef]
- Avis, P.G.; Bergel, F.; Bray, R.C.; Shooter, K.V. A Crystalline Material with Xanthine Oxidase Activity. Nature 1954, 173, 1230–1231. [Google Scholar] [CrossRef] [PubMed]
- Michaelis, L.; Menten, M.L.; Johnson, K.A.; Goody, R.S. The Original Michaelis Constant: Translation of the 1913 Michaelis-Menten Paper. Biochemistry 2011, 50, 8264–8269. [Google Scholar] [CrossRef] [PubMed]
- Alshehab, M.; Reis, M.G.; Day, L.; Nitin, N. Milk Fat Globules, a Novel Carrier for Delivery of Exogenous Cholecalciferol. Food Res. Int. Ott. Ont 2019, 126, 108579. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, B.F.; Dias, F.F.G.; Vieira, T.M.F.d.; Bell, J.M.L.N.d.; Taha, A.Y. Method Optimization of Oxylipin Hydrolysis in Nonprocessed Bovine Milk Indicates That the Majority of Oxylipins Are Esterified. J. Food Sci. 2021, 86, 1791–1801. [Google Scholar] [CrossRef] [PubMed]
- Taha, A.Y.; Cheon, Y.; Ma, K.; Rapoport, S.I.; Rao, J.S. Altered Fatty Acid Concentrations in Prefrontal Cortex of Schizophrenic Patients. J. Psychiatr. Res. 2013, 47, 636–643. [Google Scholar] [CrossRef] [PubMed]
- Ichihara, K.; Fukubayashi, Y. Preparation of Fatty Acid Methyl Esters for Gas-Liquid Chromatography. J. Lipid Res. 2010, 51, 635–640. [Google Scholar] [CrossRef]
- Shen, Q.; Zhang, Z.; Emami, S.; Chen, J.; Leite Nobrega de Moura Bell, J.M.; Taha, A.Y. Triacylglycerols Are Preferentially Oxidized over Free Fatty Acids in Heated Soybean Oil. NPJ Sci. Food 2021, 5, 7. [Google Scholar] [CrossRef]
- ADPI Product Standards. Available online: https://www.adpi.org/DairyProducts/ADPIProductStandards/tabid/398/Default.aspx (accessed on 3 January 2022).
- Levin, M.A.; Burrington, K.J.; Hartel, R.W. Composition and Functionality of Whey Protein Phospholipid Concentrate and Delactosed Permeate. J. Dairy Sci. 2016, 99, 6937–6947. [Google Scholar] [CrossRef]
- Swaminathan, A.V.; Molitor, M.S.; Burrington, K.J.; Otter, D.; Lucey, J.A. A Study of Various Chemical Pretreatments to Fractionate Lipids from Whey Protein Phospholipid Concentrate. J. Dairy Sci. 2021, 104, 12249–12262. [Google Scholar] [CrossRef]
- Siqueiros-Cendón, T.; Arévalo-Gallegos, S.; Iglesias-Figueroa, B.F.; García-Montoya, I.A.; Salazar-Martínez, J.; Rascón-Cruz, Q. Immunomodulatory Effects of Lactoferrin. Acta Pharmacol. Sin. 2014, 35, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Arena, S.; Renzone, G.; Novi, G.; Scaloni, A. Redox Proteomics of Fat Globules Unveils Broad Protein Lactosylation and Compositional Changes in Milk Samples Subjected to Various Technological Procedures. J. Proteom. 2011, 74, 2453–2475. [Google Scholar] [CrossRef] [PubMed]
- Vorbach, C. The Housekeeping Gene Xanthine Oxidoreductase Is Necessary for Milk Fat Droplet Enveloping and Secretion: Gene Sharing in the Lactating Mammary Gland. Genes Dev. 2002, 16, 3223–3235. [Google Scholar] [CrossRef] [PubMed]
- Ogg, S.L.; Weldon, A.K.; Dobbie, L.; Smith, A.J.H.; Mather, I.H. Expression of Butyrophilin (Btn1a1) in Lactating Mammary Gland Is Essential for the Regulated Secretion of Milk-Lipid Droplets. Proc. Natl. Acad. Sci. USA 2004, 101, 10084–10089. [Google Scholar] [CrossRef] [PubMed]
- Monks, J.; Dzieciatkowska, M.; Bales, E.S.; Orlicky, D.J.; Wright, R.M.; McManaman, J.L. Xanthine Oxidoreductase Mediates Membrane Docking of Milk-Fat Droplets but Is Not Essential for Apocrine Lipid Secretion: Xanthine Oxidoreductase and Milk Fat Secretion. J. Physiol. 2016, 594, 5899–5921. [Google Scholar] [CrossRef]
- Hancock, J.T.; Salisbury, V.; Ovejero-Boglione, M.C.; Cherry, R.; Hoare, C.; Eisenthal, R.; Harrison, R. Antimicrobial Properties of Milk: Dependence on Presence of Xanthine Oxidase and Nitrite. Antimicrob. Agents Chemother. 2002, 46, 3308–3310. [Google Scholar] [CrossRef][Green Version]
- Newburg, D.S.; Peterson, J.A.; Ruiz-Palacios, G.M.; Matson, D.O.; Morrow, A.L.; Shults, J.; Guerrero, M.d.L.; Chaturvedi, P.; Newburg, S.O.; Scallan, C.D.; et al. Role of Human-Milk Lactadherin in Protectoin against Symptomatic Rotavirus Infection. Lancet 1998, 351, 1160–1164. [Google Scholar] [CrossRef]
- Raymond, A.; Ensslin, M.A.; Shur, B.D. SED1/MFG-E8: A Bi-Motif Protein That Orchestrates Diverse Cellular Interactions. J. Cell. Biochem. 2009, 106, 957–966. [Google Scholar] [CrossRef]
- Brandtzaeg, P.; Prydz, H. Direct Evidence for an Integrated Function of J Chain and Secretory Component in Epithelial Transport of Immunoglobulins. Nature 1984, 311, 71–73. [Google Scholar] [CrossRef]
- Kaetzel, C.S. The Polymeric Immunoglobulin Receptor: Bridging Innate and Adaptive Immune Responses at Mucosal Surfaces. Immunol. Rev. 2005, 206, 83–99. [Google Scholar] [CrossRef]
- Pallesen, L.T.; Pedersen, L.R.L.; Petersen, T.E.; Rasmussen, J.T. Characterization of Carbohydrate Structures of Bovine MUC15 and Distribution of the Mucin in Bovine Milk. J. Dairy Sci. 2007, 90, 3143–3152. [Google Scholar] [CrossRef] [PubMed]
- Sando, L.; Pearson, R.; Gray, C.; Parker, P.; Hawken, R.; Thomson, P.C.; Meadows, J.R.S.; Kongsuwan, K.; Smith, S.; Tellam, R.L. Bovine Muc1 Is a Highly Polymorphic Gene Encoding an Extensively Glycosylated Mucin That Binds Bacteria. J. Dairy Sci. 2009, 92, 5276–5291. [Google Scholar] [CrossRef] [PubMed]
- Peeters, R.A.; Veerkamp, J.H.; Demel, R.A. Are Fatty Acid-Binding Proteins Involved in Fatty Acid Transfer? Biochim. Biophys. Acta BBA Lipids Lipid Metab. 1989, 1002, 8–13. [Google Scholar] [CrossRef]
- Bhat, M.Y.; Dar, T.A.; Singh, L.R. Casein Proteins: Structural and Functional Aspects. In Milk Proteins—From Structure to Biological Properties and Health Aspects; Gigli, I., Ed.; IntechOpen: London, UK, 2016; ISBN 978-953-51-2536-5. [Google Scholar]
- Brink, L.R.; Herren, A.W.; McMillen, S.; Fraser, K.; Agnew, M.; Roy, N.; Lönnerdal, B. Omics Analysis Reveals Variations among Commercial Sources of Bovine Milk Fat Globule Membrane. J. Dairy Sci. 2020, 103, 3002–3016. [Google Scholar] [CrossRef]
- Liao, Y.; Alvarado, R.; Phinney, B.; Lönnerdal, B. Proteomic Characterization of Human Milk Fat Globule Membrane Proteins during a 12 Month Lactation Period. J. Proteome Res. 2011, 10, 3530–3541. [Google Scholar] [CrossRef]
- Tsuji, S.; Hirata, Y.; Mukai, F.; Ohtagaki, S. Comparison of Lactoferrin Content in Colostrum Between Different Cattle Breeds. J. Dairy Sci. 1990, 73, 125–128. [Google Scholar] [CrossRef]
- Cheng, J.B.; Wang, J.Q.; Bu, D.P.; Liu, G.L.; Zhang, C.G.; Wei, H.Y.; Zhou, L.Y.; Wang, J.Z. Factors Affecting the Lactoferrin Concentration in Bovine Milk. J. Dairy Sci. 2008, 91, 970–976. [Google Scholar] [CrossRef]
- Varki, A.; Cummings, R.D.; Esko, J.D.; Freeze, H.H.; Stanley, P.; Bertozzi, C.R.; Hart, G.W.; Etzler, M.E. (Eds.) Essentials of Glycobiology, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009; ISBN 978-0-87969-770-9. [Google Scholar]
- Varki, A.; Lowe, J.B. Biological Roles of Glycans. In Essentials of Glycobiology; Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Bertozzi, C.R., Hart, G.W., Etzler, M.E., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009; ISBN 978-0-87969-770-9. [Google Scholar]
- Wu, X.; Jackson, R.T.; Khan, S.A.; Ahuja, J.; Pehrsson, P.R. Human Milk Nutrient Composition in the United States: Current Knowledge, Challenges, and Research Needs. Curr. Dev. Nutr. 2018, 2, nzy025. [Google Scholar] [CrossRef]
- Björck, L.; Claesson, O. Xanthine Oxidase as a Source of Hydrogen Peroxide for the Lactoperoxidase System in Milk. J. Dairy Sci. 1979, 62, 1211–1215. [Google Scholar] [CrossRef]
- Stevens, C.R.; Millar, T.M.; Clinch, J.G.; Kanczler, J.M.; Bodamyali, T.; Blake, D.R. Antibacterial Properties of Xanthine Oxidase in Human Milk. Lancet Lond. Engl. 2000, 356, 829–830. [Google Scholar] [CrossRef]
- Millar, T.M.; Kanczler, J.M.; Bodamyali, T.; Blake, D.R.; Stevens, C.R. Xanthine Oxidase Is a Peroxynitrite Synthase: Newly Identified Roles for a Very Old Enzyme. Redox Rep. 2002, 7, 65–70. [Google Scholar] [CrossRef]
- Harrison, R. Milk Xanthine Oxidase: Properties and Physiological Roles. Int. Dairy J. 2006, 16, 546–554. [Google Scholar] [CrossRef]
- Yao, Y.; Zhao, G.; Yan, Y.; Chen, C.; Sun, C.; Zou, X.; Jin, Q.; Wang, X. Effects of Freeze Drying and Spray Drying on the Microstructure and Composition of Milk Fat Globules. RSC Adv. 2016, 6, 2520–2529. [Google Scholar] [CrossRef]
- Lu, J.; Argov-Argaman, N.; Anggrek, J.; Boeren, S.; van Hooijdonk, T.; Vervoort, J.; Hettinga, K.A. The Protein and Lipid Composition of the Membrane of Milk Fat Globules Depends on Their Size. J. Dairy Sci. 2016, 99, 4726–4738. [Google Scholar] [CrossRef] [PubMed]
- Michalski, M.C.; Leconte, N.; Briard-Bion, V.; Fauquant, J.; Maubois, J.L.; Goudédranche, H. Microfiltration of Raw Whole Milk to Select Fractions with Different Fat Globule Size Distributions: Process Optimization and Analysis. J. Dairy Sci. 2006, 89, 3778–3790. [Google Scholar] [CrossRef]
- Dhungana, P.; Truong, T.; Palmer, M.; Bansal, N.; Bhandari, B. Size-Based Fractionation of Native Milk Fat Globules by Two-Stage Centrifugal Separation. Innov. Food Sci. Emerg. Technol. 2017, 41, 235–243. [Google Scholar] [CrossRef]
- Gan, J.; Zhang, Z.; Kurudimov, K.; German, J.B.; Taha, A.Y. Distribution of Free and Esterified Oxylipins in Cream, Cell, and Skim Fractions of Human Milk. Lipids 2020, 55, 661–670. [Google Scholar] [CrossRef]
| Sample Description | Total Solids (%) | Protein (%) | Fat (%) |
|---|---|---|---|
| Sample 1A (liquid) | 20.47 | 65.46 * | 27.25 * |
| Sample 1B (liquid) | 21.56 | 62.15 * | 28.10 * |
| Sample 1 (powder) | - | 62.7 | 24.4 |
| Sample 2A (powder) | - | >72 | - |
| Sample 2B (powder) | - | >73 | - |
| Sample 3 (powder) | - | 75.09 | 18.11 |
| Sample 4A (powder) | - | 76.19 | 13.08 |
| Sample 4B (powder) | - | 69.39 | 16.45 |
| Sample 5 (powder) | - | 67.61 | - |
| Sample 6A (powder) | - | 52.63 | 13.08 |
| Sample 6B (liquid) | 13.40 | 63.43 | 20.20 |
| Bovine milk (liquid) | 13 | 24.92 * | 26.92 * |
| This Study | Brink et al. (2020) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Whey Based | Formula | Cream Based | |||||||
| WPPC | MFGM-10 | PF | SF | PL-20 | BPC-50 | BSP2 | PLC1 | SM2 | |
| MFGM proteins | 22.81 | 10.71 | 5.06 | 2.85 | 24 | 25.73 | 23.33 | 23.93 | 26.39 |
| Butyrophilin | 5.11 | 1.65 | 1.14 | 0.1 | 4.99 | 5.7 | 5.42 | 5.44 | 5.2 |
| Fatty acid-binding protein | 1.75 | 2.35 | 1.35 | 1.03 | 2.60 | 4.26 | 4 | 4.54 | 3.18 |
| GLYCAM-1 | 7.00 | 3.94 | 2.17 | 1.32 | 7.16 | 3.88 | 3.64 | 4.52 | 7.74 |
| Lactadherin | 4.45 | 1.36 | 0.1 | 0.1 | 5.42 | 4.93 | 4.21 | 4.24 | 4.26 |
| Adipophilin (Perilipin-2) | 0.44 | 0.1 | 0.1 | 0.1 | 2.3 | 3.63 | 2.94 | 3.11 | 3.13 |
| Platelet glycoprotein 4 (CD36) | 0.74 | 1.21 | 0.1 | 0.1 | 0.1 | 1.05 | 1.15 | 0.1 | 1.09 |
| Xanthine oxidase | 2.25 | 0.1 | 0.1 | 0.1 | 1.43 | 2.28 | 1.97 | 1.98 | 1.79 |
| Mucin-15 | 0.90 | - | - | - | - | - | - | - | - |
| Mucin-1 | 0.18 | - | - | - | - | - | - | - | - |
| Milk proteins | 45.42 | 55.64 | 61.64 | 68.5 | 56.5 | 50.58 | 51.39 | 52.4 | 52.34 |
| α-lactalbumin | 2.07 | 0.1 | 1.4 | 2.65 | 1.05 | 1.12 | 1.2 | 0.1 | 1.4 |
| α-S1-casein | 1.22 | 0.1 | 8.20 | 12.23 | 9.80 | 9.45 | 9.90 | 10.45 | 9.07 |
| α-S2-casein | 0.48 | 0.1 | 8.24 | 11.92 | 11.60 | 10.64 | 11.24 | 14.5 | 10.22 |
| β-lactoglobulin | 30.11 | 46.29 | 13.62 | 14.14 | 15.30 | 10.07 | 13.64 | 10.69 | 10.91 |
| Serum albumin | 10.95 | 7.10 | 4.58 | 4.17 | 1.97 | 1.70 | 2.23 | 1.51 | 2.03 |
| β-casein | 0.36 | 0.1 | 15.05 | 24.27 | 16.58 | 17.5 | 12.98 | 13.79 | 18.51 |
| Kappa-casein | 0.24 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 1.26 | 0.1 |
| Lactoferrin (Lactotransferrin) | 3.80 | 1.75 | 10.45 | 1.81 | 0.1 | 0.1 | 1.1 | 0.1 | 0.1 |
| Other proteins | 31.32 | 33.65 | 33.3 | 28.65 | 19.5 | 76.31 | 25.28 | 23.67 | 21.27 |
| Lipid Fraction | Total Fatty Acids | SM | PC | PS | PI | PE |
|---|---|---|---|---|---|---|
| Sum of Fatty acids | 123.91 ± 6.91 | 0.97 ± 0.13 | 8.65 ± 0.70 | 3.30 ± 0.18 | 2.07 ± 0.32 | 9.25 ± 0.23 |
| C8:0 | 0.212 ± 0.029 | ND | ND | ND | ND | 0.003 ± 0.006 |
| C10:0 | 1.413 ± 0.138 | ND | ND | ND | ND | ND |
| C11:0 | 0.065 ± 0.004 | ND | ND | ND | ND | ND |
| C12:0 | 3.286 ± 0.177 | ND | ND | ND | ND | ND |
| C13:0 | 0.460 ± 0.062 | ND | ND | ND | ND | 0.095 ± 0.165 |
| C14:0 | 13.341 ± 0.781 | ND | 0.802 ± 0.099 | ND | ND | 0.074 ± 0.065 |
| C14:1 | 1.010 ± 0.055 | ND | ND | ND | 0.042 ± 0.072 | ND |
| C15:0 | 1.641 ± 0.086 | ND | 0.108 ± 0.095 | ND | ND | ND |
| C16:0 | 44.722 ± 2.473 | 0.182 ± 0.159 | 3.702 ± 0.331 | 0.144 ± 0.125 | 0.211 ± 0.030 | 1.242 ± 0.028 |
| C16:1 | 1.897 ± 0.108 | 0.349 ± 0.034 | 0.291 ± 0.091 | 0.357 ± 0.085 | 0.360 ± 0.087 | 0.340 ± 0.032 |
| C17:0 | 0.820 ± 0.041 | ND | ND | ND | ND | ND |
| C18:0 | 14.493 ± 0.778 | ND | 0.798 ± 0.069 | 1.326 ± 0.097 | 0.748 ± 0.092 | 1.168 ± 0.030 |
| C18:1cis+trans | 30.723 ± 1.655 | ND | 2.155 ± 0.144 | 1.039 ± 0.081 | 0.551 ± 0.054 | 4.259 ± 0.159 |
| C18:2 n6 | 6.213 ± 0.335 | ND | 0.796 ± 0.055 | 0.433 ± 0.043 | 0.155 ± 0.018 | 1.565 ± 0.076 |
| C18:3 n6 | 0.049 ± 0.003 | ND | ND | ND | ND | 0.033 ± 0.058 |
| C18:3 n3 | 0.674 ± 0.036 | ND | ND | ND | ND | 0.034 ± 0.059 |
| C20:0 | 0.263 ± 0.029 | ND | ND | ND | ND | ND |
| C20:1 n9 | 0.222 ± 0.096 | ND | ND | ND | ND | ND |
| C20:2 n6 | 0.057 ± 0.004 | ND | ND | ND | ND | ND |
| C20:3 n6 | 0.498 ± 0.027 | ND | ND | ND | ND | 0.076 ± 0.066 |
| C20:4 n6 | 0.559 ± 0.029 | ND | ND | ND | ND | 0.155 ± 0.007 |
| C20:3 n3 | 0.011 ± 0.003 | ND | ND | ND | ND | 0.010 ± 0.018 |
| C20:5 n3 | 0.119 ± 0.005 | ND | ND | ND | ND | ND |
| C22:0 | 0.343 ± 0.042 | 0.233 ± 0.017 | ND | ND | ND | ND |
| C22:1 n9 | 0.023 ± 0.004 | ND | ND | ND | ND | ND |
| C22:2 n6 | 0.187 ± 0.007 | ND | ND | ND | ND | ND |
| C22:5 n6 | 0.051 ± 0.021 | ND | ND | ND | ND | 0.059 ± 0.103 |
| C22:5 n3 | 0.276 ± 0.017 | ND | ND | ND | ND | 0.013 ± 0.023 |
| C22:6n3 | 0.255 ± 0.036 | 0.209 ± 0.017 | ND | ND | ND | ND |
| C24:1 | 0.029 ± 0.002 | ND | ND | ND | ND | ND |
| cholesterol | 6.768 ± 0.101 | ND | ND | ND | ND | ND |
| Precursors | Abbreviations | Compound Names | Total Oxylipins (n = 5) | Free Oxylipins (n = 5) | p Value |
|---|---|---|---|---|---|
| Ave ± Std | Ave ± Std | (t-Test) | |||
| LA | 9,12,13-TriHOME | 9,12,13-trihydroxyoctadecamonoenoic acid | 57.305 ± 5.531 | 0.120 ± 0.009 | 1.30 × 10−8 |
| 9,10,13-TriHOME | 9,10,13-trihydroxyoctadecamonoenoic acid | 19.147 ± 2.123 | 0.034 ± 0.002 | 3.87 × 10−8 | |
| 12,13-DiHOME | 12,13-dihydroxyoctadecamonoenoic acid | 2.220 ± 0.205 | 0.391 ± 0.046 | 5.06 × 10−8 | |
| 9,10-DiHOME | 9,10-dihydroxyoctadecamonoenoic acid | 3.566 ± 0.331 | 0.498 ± 0.059 | 3.49 × 10−8 | |
| 13-HODE | 13-hydroxyoctadecadienoic acid | 85.360 ± 5.593 | 3.246 ± 0.369 | 8.22 × 10−10 | |
| 9-HODE | 9-hydroxyoctadecadienoic acid | 21.644 ± 2.207 | 1.522 ± 0.181 | 3.59 × 10−8 | |
| 13-oxo-ODE | 13-oxo-octadecadienoic acid | 1.246 ± 0.209 | 0.045 ± 0.005 | 1.26 × 10−6 | |
| 9-oxo-ODE | 9-oxo-octadecadienoic acid | 1.360 ± 0.201 | 0.067 ± 0.008 | 5.39 × 10−7 | |
| 12(13)-EpOME | 12(13)-epoxyoctadecamonoenoic acid | 23.227 ± 5.087 | 0.087 ± 0.050 | 7.47 × 10−6 | |
| 9(10)-EpOME | 9(10)-epoxyoctadecamonoenoic acid | 31.945 ± 7.670 | 0.262 ± 0.150 | 1.53 × 10−5 | |
| DGLA | 15(S)-HETrE | 15(S)-hydroxyeicosatrienoic acid | 1.436 ± 0.107 | 0.010 ± 0.001 | 1.75 × 10−9 |
| ARA | TXB2 | Tromboxane B2 | 0.021 ± 0.004 | 0.001 ± 0.00 | 2.70 × 10−6 |
| 6-trans-LTB4 | 6-trans-leukotriene B4 | 0.029 ± 0.001 | 0.001 ± 0.00 | 5.74 × 10−11 | |
| 14,15-DiHETrE | 14,15-dihydroxyeicosatrienoic acid | 0.028 ± 0.004 | 0.002 ± 0.00 | 2.15 × 10−7 | |
| 11,12-DiHETrE | 11,12-dihydroxyeicosatrienoic acid | 0.028 ± 0.004 | 0.002 ± 0.00 | 7.24 × 10−7 | |
| 8,9-DiHETrE | 8,9-dihydroxyeicosatrienoic acid | 0.007 ± 0.004 | 0.001 ± 0.001 | 4.36 × 10−3 | |
| 20-HETE | 20-hydroxyeicosatetraenoic acid | 0.590 ± 0.117 | 0.009 ± 0.003 | 3.82 × 10−6 | |
| 5,6-DiHETrE | 5,6-dihydroxyeicosatrienoic acid | 0.402 ± 0.038 | 0.001 ± 0.00 | 1.20 × 10−8 | |
| 15-HETE | 15-hydroxyeicosatetraenoic acid | 1.050 ± 0.13 | 0.062 ± 0.006 | 1.45 × 10−7 | |
| 11-HETE | 11-hydroxyeicosatetraenoic acid | 0.282 ± 0.044 | 0.030 ± 0.003 | 1.34 × 10−6 | |
| 15-oxo-ETE | 15-oxo-eicosatetraenoic acid | 0.007 ± 0.002 | 0.006 ± 0.001 | 1.58 × 10−1 | |
| 8-HETE | 8-hydroxyeicosatetraenoic acid | 0.085 ± 0.005 | 0.029 ± 0.003 | 3.05 × 10−8 | |
| 12-HETE | 12-hydroxyeicosatetraenoic acid | 0.116 ± 0.011 | 0.015 ± 0.002 | 3.96 × 10−8 | |
| 9-HETE | 9-hydroxyeicosatetraenoic acid | 0.392 ± 0.043 | 0.005 ± 0.002 | 4.19 × 10−8 | |
| 12-oxo-ETE | 12-oxo-eicosatetraenoic acid | 0.004 ± 0.001 | 0.001 ± 0.00 | 1.21 × 10−3 | |
| 5-HETE | 5-hydroxyeicosatetraenoic acid | 1.614 ± 0.147 | 0.064 ± 0.007 | 1.12 × 10−8 | |
| 14(15)-EpETrE | 14(15)-epoxyeicosatrienoic acid | 1.406 ± 0.248 | 0.001 ± 0.001 | 1.40 × 10−6 | |
| 5-oxo-ETE | 5-oxo-eicosatetraenoic acid | 0.048 ± 0.011 | 0.014 ± 0.005 | 2.84 × 10−4 | |
| 11(12)-EpETrE | 11(12)-epoxyeicosatrienoic acid | 1.236 ± 0.280 | 0.002 ± 0.001 | 9.56 × 10−6 | |
| 8(9)-EpETrE | 8(9)-epoxyeicosatrienoic acid | 1.740 ± 0.131 | 0.013 ± 0.009 | 1.99 × 10−9 | |
| 5(6)-EpETrE | 5(6)-epoxyeicosatrienoic acid | 0.909 ± 0.177 | 0.002 ± 0.001 | 3.01 × 10−6 | |
| ALA | 9-HOTrE | 9-hydroxyoctadecatrienoic acid | 1.877 ± 0.214 | 0.151 ± 0.014 | 9.50 × 10−8 |
| 13-HOTrE | 13-hydroxyoctadecatrienoic acid | 2.381 ± 0.222 | 0.302 ± 0.024 | 2.97 × 10−8 | |
| EPA | 8,15-DiHETE | 8,15-dihydroxyeicosatetraenoic acid | 0.159 ± 0.028 | 0.003 ± 0.001 | 1.77 × 10−6 |
| 5,15-DiHETE | 5,15-dihydroxyeicosatetraenoic acid | 0.032 ± 0.002 | 0.001 ± 0.000 | 2.72 × 10−9 | |
| 5,6-DiHETE | 5,6-dihydroxyeicosatetraenoic acid | 4.228 ± 0.684 | 0.039 ± 0.06 | 8.00 × 10−7 | |
| 17,18-DiHETE | 17,18-dihydroxyeicosatetraenoic acid | 0.079 ± 0.030 | 0.007 ± 0.001 | 7.53 × 10−4 | |
| 14,15-DiHETE | 14,15-dihydroxyeicosatetraenoic acid | 0.018 ± 0.007 | 0.004 ± 0.002 | 2.59 × 10−3 | |
| 11,12-DiHETE | 11,12-dihydroxy-5Z,8Z,14Z,17Z-eicosatetraenoic acid | 0.007 ± 0.001 | 0.001 ± 0.000 | 6.34 × 10−6 | |
| 15-HEPE | 15-hydroxyeicosapentaenoic acid | 0.096 ± 0.013 | 0.001 ± 0.000 | 1.71 × 10−7 | |
| 8-HEPE | 8-hydroxyeicosapentaenoic acid | 0.055 ± 0.009 | 0.006 ± 0.002 | 3.46 × 10−6 | |
| 12-HEPE | 12-hydroxyeicosapentaenoic acid | 0.066 ± 0.007 | 0.003 ± 0.001 | 3.27 × 10−8 | |
| 5-HEPE | 5-hydroxyeicosapentaenoic acid | 1.454 ± 0.070 | 0.012 ± 0.001 | 5.64 × 10−11 | |
| 17(18)-EpETE | 17(18)-epoxyeicosateteaenoic acid | 2.368 ± 0.868 | 0.000 ± 0.00 | 2.91 × 10−4 | |
| 14(15)-EpETE | 14(15)-epoxyeicosatetraenoic acid | 0.127 ± 0.045 | 0.001 ± 0.001 | 2.28 × 10−4 | |
| 11(12)-EpETE | 11(12)-epoxyeicosateteaenoic acid | 0.102 ± 0.019 | 0.000 ± 0.000 | 2.45 × 10−6 | |
| 8(9)-EpETE | 8(9)-epoxyeicosateteaenoic acid | 0.207 ± 0.059 | 0.000 ± 0.000 | 5.03 × 10−5 | |
| DHA | 19(20)-DiHDPA | (±)19,20-dihydroxy−4Z,7Z,10Z,13Z,16Z-docosapentaenoic acid | 0.001 ± 0.001 | 0.000 ± 0.000 | 9.44 × 10−2 |
| 16(17)-DiHDPA | (±)16,17-dihydroxy-4Z,7Z,10Z,13Z,19Z-docosapentaenoic acid | 0.002 ± 0.001 | 0.000 ± 0.000 | 1.88 × 10−4 | |
| 17-HDoHE | 17-hydroxydocosahexaenoic acid | 0.154 ± 0.079 | 0.000 ± 0.000 | 2.34 × 10−3 | |
| 19(20)-EpDPE | 19(20)-epoxydocosapentaenoic acid | 0.082 ± 0.031 | 0.002 ± 0.002 | 4.85 × 10−4 | |
| 16(17)-EpDPE | 16(17)-epoxydocosapentaenoic acid | 0.015 ± 0.010 | 0.000 ± 0.000 | 1.38 × 10−3 | |
| 13(14)-EpDPE | 13(14)-epoxydocosapentaenoic acid | 0.025 ± 0.007 | 0.001 ± 0.001 | 4.27 × 10−5 | |
| 10(11)-EpDPE | 10(11)-epoxydocosapentaenoic acid | 0.045 ± 0.012 | 0.001 ± 0.001 | 3.86 × 10−5 | |
| 7(8)-EpDPE | 7(8)-epoxydocosapentaenoic acid | 0.047 ± 0.027 | 0.000 ± 0.000 | 4.83 × 10−3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ozturk, G.; Liang, N.; Bhattacharya, M.; Robinson, R.C.; Shankar, S.; Huang, Y.-P.; Paviani, B.; Taha, A.Y.; Barile, D. Glycoproteomic and Lipidomic Characterization of Industrially Produced Whey Protein Phospholipid Concentrate with Emphasis on Antimicrobial Xanthine Oxidase, Oxylipins and Small Milk Fat Globules. Dairy 2022, 3, 277-302. https://doi.org/10.3390/dairy3020022
Ozturk G, Liang N, Bhattacharya M, Robinson RC, Shankar S, Huang Y-P, Paviani B, Taha AY, Barile D. Glycoproteomic and Lipidomic Characterization of Industrially Produced Whey Protein Phospholipid Concentrate with Emphasis on Antimicrobial Xanthine Oxidase, Oxylipins and Small Milk Fat Globules. Dairy. 2022; 3(2):277-302. https://doi.org/10.3390/dairy3020022
Chicago/Turabian StyleOzturk, Gulustan, Nuanyi Liang, Mrittika Bhattacharya, Randall C. Robinson, Shalini Shankar, Yu-Ping Huang, Bruna Paviani, Ameer Y. Taha, and Daniela Barile. 2022. "Glycoproteomic and Lipidomic Characterization of Industrially Produced Whey Protein Phospholipid Concentrate with Emphasis on Antimicrobial Xanthine Oxidase, Oxylipins and Small Milk Fat Globules" Dairy 3, no. 2: 277-302. https://doi.org/10.3390/dairy3020022
APA StyleOzturk, G., Liang, N., Bhattacharya, M., Robinson, R. C., Shankar, S., Huang, Y.-P., Paviani, B., Taha, A. Y., & Barile, D. (2022). Glycoproteomic and Lipidomic Characterization of Industrially Produced Whey Protein Phospholipid Concentrate with Emphasis on Antimicrobial Xanthine Oxidase, Oxylipins and Small Milk Fat Globules. Dairy, 3(2), 277-302. https://doi.org/10.3390/dairy3020022






