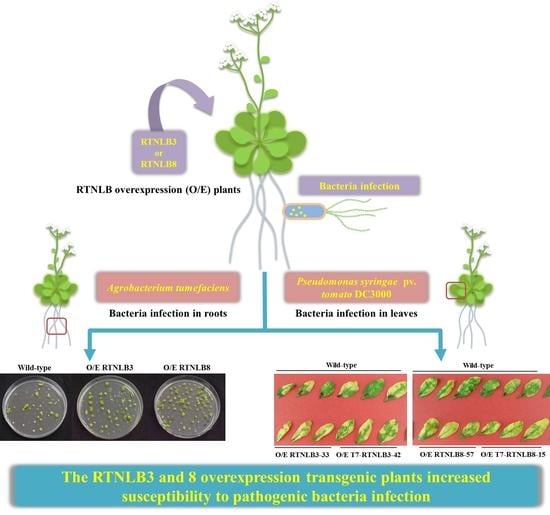
Arabidopsis RETICULON-LIKE3 (RTNLB3) and RTNLB8 Participate in Agrobacterium-Mediated Plant Transformation
Abstract
:1. Introduction
2. Results
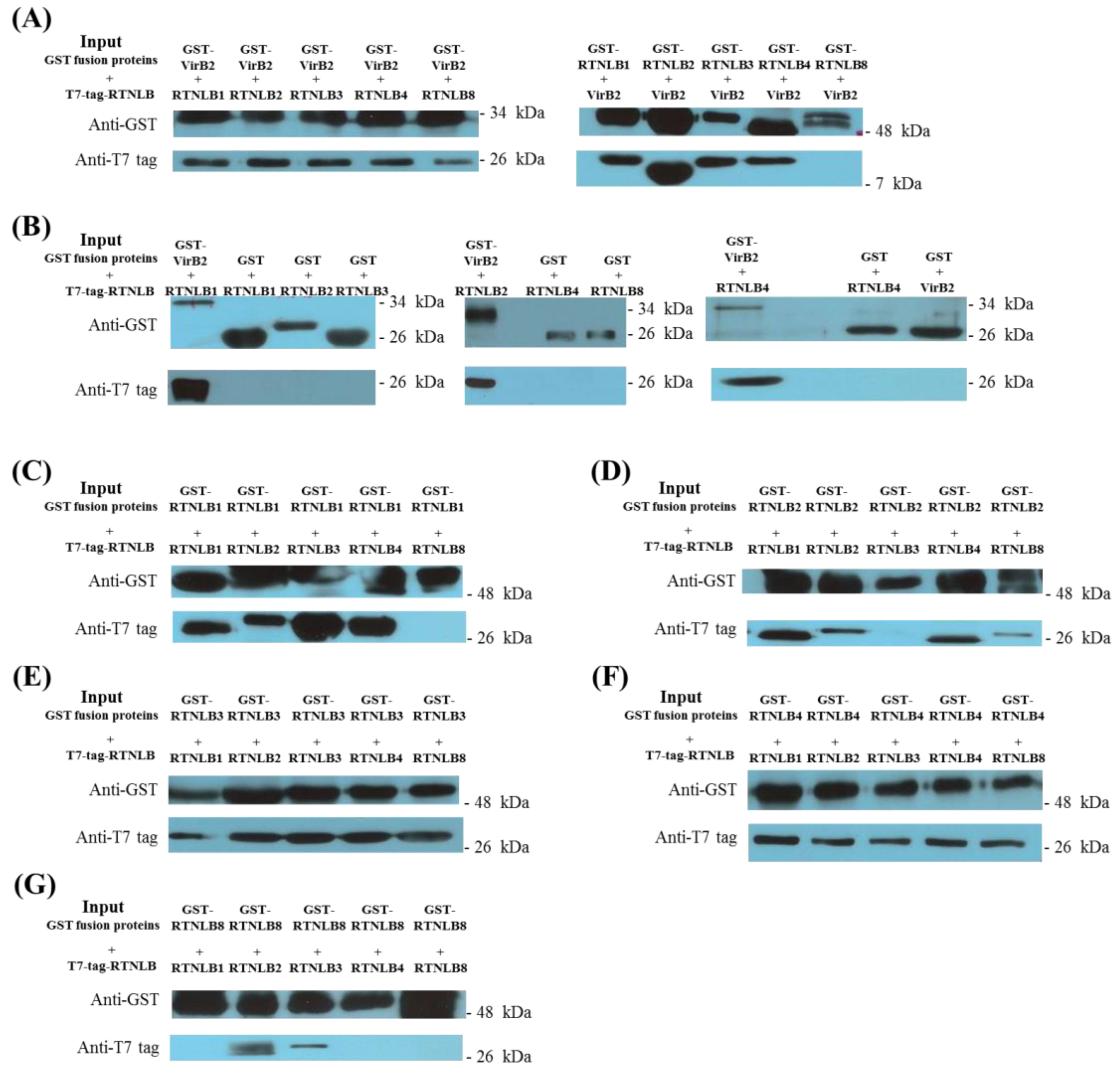
2.1. Interactions Among RTNLB3 and 8 and Vir Proteins in Yeast and In Vitro
2.2. Arabidopsis Rtnlb3 and Rtnlb8 Mutants Showed Reduced Levels of A. tumefaciens-Mediated Transformation Efficiency
2.3. RTNLB3 and 8 Overexpression Increased Plant Susceptibility to A. tumefaciens-Mediated Transformation
2.4. RTNLB3 and 8 Overexpression Plants were More Susceptible to P. syringae Infection
2.5. RTNLB1-4 and 8 Gene Levels Differed in Various Plant Tissues
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture
4.2. Yeast Two-Hybrid Assays
4.3. Glutathione-S-Transferase (GST) Protein Affinity Purification Assays
4.4. DNA Isolation from Arabidopsis Plants and Genomic DNA PCR Analysis
4.5. RNA Isolation from Arabidopsis Plants and RT-PCR Analysis
4.6. Protein Extraction from Arabidopsis Plants and Protein Gel Blot Analysis
4.7. Generation of RTNLB3 and 8 Overexpression Arabidopsis Transgenic Plants
4.8. Agrobacterium Tumefaciens-Mediated Stable, Transient Root and Seedling Transformation Assays of Rtnlb Mutant Plants and Arabidopsis RTNLB Overexpression Plants
4.9. Pseudomonas Syringae Infection Assays of Arabidopsis RTNLB Overexpression Plants
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gelvin, S.B. Plant proteins involved in Agrobacterium-mediated genetic transformation. Annu. Rev. Phytopathol. 2010, 48, 45–68. [Google Scholar] [CrossRef] [PubMed]
- Gelvin, S.B. Traversing the cell: Agrobacterium T-DNA’s journey to the host genome. Front. Plant Sci. 2012, 3, 52. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, B.; Citovsky, V. The roles of bacterial and host plant factors in Agrobacterium-mediated genetic transformation. Int. J. Biochem. Cell Biol. 2013, 57, 467–481. [Google Scholar] [CrossRef] [PubMed]
- Pitzschke, A. Infection and plant defense-transformation success hangs by a thread. Front. Plant Sci. 2013, 4, 519. [Google Scholar] [CrossRef]
- Gohlke, J.; Deeken, R. Plant responses to Agrobacterium tumefaciens and crown gall development. Front. Plant Sci. 2014, 5, 155. [Google Scholar] [CrossRef] [PubMed]
- Brencic, A.; Winans, S.C. Detection of and response to signals involved in host–microbe interactions by plant-associated bacteria. Microbiol. Mol. Biol. Rev. 2005, 69, 155–194. [Google Scholar] [CrossRef] [PubMed]
- McCullen, C.A.; Binns, A.N. Agrobacterium tumefaciens and plant cell interactions and activities required for interkingdom macromolecular transfer. Annu. Rev. Cell Dev. Biol. 2006, 22, 101–127. [Google Scholar] [CrossRef] [PubMed]
- Zechner, E.L.; Lang, S.; Schildbach, J.F. Assembly and mechanisms of bacterial type IV secretion machines. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 1073–1087. [Google Scholar] [CrossRef] [PubMed]
- Bhatty, M.; Laverde Gomez, J.A.; Christie, P.J. The expanding bacterial type IV secretion lexicon. Res. Microbiol. 2013, 164, 620–639. [Google Scholar] [CrossRef] [PubMed]
- Chandran, V. Type IV secretion machinery: Molecular architecture and function. Biochem. Soc. Trans. 2013, 41, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Christie, P.J.; Whitaker, N.; Gonzalez-Rivera, C. Mechanism and structure of the bacterial type IV secretion systems. Biochim. Biophys. Acta 2014, 1843, 1578–1591. [Google Scholar] [CrossRef] [PubMed]
- Wallden, K.; Rivera-Calzada, A.; Waksman, G. Type IV secretion systems: Versatility and diversity in function. Cell Microbiol. 2010, 12, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Waksman, G.; Fronzes, R. Molecular architecture of bacterial type IV secretion systems. Trends Biochem. Sci. 2010, 35, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Waksman, G.; Orlova, E.V. Structural organisation of the type IV secretion systems. Curr. Opin. Microbiol. 2014, 17, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Lai, E.M.; Kado, C.I. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J. Bacteriol. 1998, 180, 2711–2717. [Google Scholar] [PubMed]
- Lai, E.M.; Chesnokova, O.; Banta, L.M.; Kado, C.I. Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J. Bacteriol. 2000, 182, 3705–3716. [Google Scholar] [CrossRef] [PubMed]
- Lai, E.M.; Eisenbrandt, R.; Kalkum, M.; Lanka, E.; Kado, C.I. Biogenesis of T pili in Agrobacterium tumefaciens requires precise VirB2 propilin cleavage and cyclization. J. Bacteriol. 2002, 184, 327–330. [Google Scholar] [CrossRef] [PubMed]
- Eisenbrandt, R.; Kalkum, M.; Lai, E.M.; Lurz, R.; Kado, C.I.; Lanka, E. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J. Biol. Chem. 1999, 274, 22548–22555. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Eisenlohr, H.; Domke, N.; Angerer, C.; Wanner, G.; Zambryski, P.C.; Baron, C. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol. 1999, 181, 7485–7492. [Google Scholar] [PubMed]
- Aly, K.A.; Baron, C. The VirB5 protein localizes to the T-pilus tips in Agrobacterium tumefaciens. Microbiology 2007, 153, 3766–3775. [Google Scholar] [CrossRef] [PubMed]
- Djamei, A.; Pitzschke, A.; Nakagami, H.; Rajh, I.; Hirt, H. Trojan horse strategy in Agrobacterium transformation: Abusing MAPK defense signaling. Science 2007, 318, 453–456. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Lee, L.Y.; Gelvin, S.B. Is VIP1 important for Agrobacterium-mediated transformation? Plant J. 2014, 79, 848–860. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.H.; Gelvin, S.B. Plant proteins that interact with VirB2, the Agrobacterium pilin protein, mediate plant transformation. Plant Cell 2004, 16, 3148–3167. [Google Scholar] [CrossRef] [PubMed]
- Roebroek, A.J.; van de Velde, H.J.; Van Bokhoven, A.; Broers, J.L.; Ramaekers, F.C.; Van de Ven, W.J. Cloning and expression of alternative transcripts of a novel neuroendocrine-specific gene and identification of its 135-kDa translational product. J. Biol. Chem. 1993, 268, 13439–13447. [Google Scholar] [PubMed]
- Van de Velde, H.J.; Senden, N.H.; Roskams, T.A.; Broers, J.L.; Ramaekers, F.C.; Roebroek, A.J.; Van de Ven, W.J. NSP-encoded reticulons are neuroendocrine markers of a novel category in human lung cancer diagnosis. Cancer Res. 1994, 54, 4769–4776. [Google Scholar] [PubMed]
- Senden, N.H.; Timmer, E.D.; Boers, J.E.; van de Velde, H.J.; Roebroek, A.J.; Van de Ven, W.J.; Broers, J.L.; Ramaekers, F.C. Neuroendocrine-specific protein C (NSP-C): Subcellular localization and differential expression in relation to NSP-A. Eur. J. Cell Biol. 1996, 69, 197–213. [Google Scholar] [PubMed]
- Oertle, T.; Schwab, M.E. Nogo and its paRTNers. Trends Cell Biol. 2003, 13, 187–194. [Google Scholar] [CrossRef]
- Voeltz, G.K.; Prinz, W.A.; Shibata, Y.; Rist, J.M.; Rapoport, T.A. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 2006, 124, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Shibata, Y.; Voss, C.; Shemesh, T.; Li, Z.; Coughlin, M.; Kozlov, M.M.; Rapoport, T.A.; Prinz, W.A. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science 2008, 319, 1247–1250. [Google Scholar] [CrossRef] [PubMed]
- Nziengui, H.; Schoefs, B. Functions of reticulons in plants: What we can learn from animals and yeasts. Cell Mol. Life Sci. 2009, 66, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Tolley, N.; Sparkes, I.A.; Aunter, P.R.; Craddock, C.P.; Nuttall, J.; Roberts, L.M.; Hawes, C.; Pedrazzini, E.; Frigerio, L. Overexpression of a plant reticulon remodels the lumen of the cortical endoplasmic reticulum but does not perturb protein transport. Traffic 2008, 9, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Tolley, N.; Sparkes, I.; Craddock, C.P.; Eastmond, P.J.; Runions, J.; Hawes, C.; Frigerio, L. Transmembrane domain length is responsible for the ability of a plant reticulon to shape endoplasmic reticulum tubules in vivo. Plant J. 2010, 64, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Sparkes, I.; Tolley, N.; Aller, I.; Svozil, J.; Osterrieder, A.; Botchway, S.; Mueller, C.; Frigerio, L.; Hawes, C. Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell 2010, 22, 1333–1343. [Google Scholar] [CrossRef] [PubMed]
- Oertle, T.; Klinger, M.; Stuermer, C.A.; Schwab, M.E. A reticular rhapsody: Phylogenic evolution and nomenclature of the RTN/Nogo gene family. FASEB J. 2003, 17, 1238–1247. [Google Scholar] [CrossRef] [PubMed]
- Nziengui, H.; Bouhidel, K.; Pillon, D.; Der, C.; Marty, F.; Schoefs, B. Reticulon-like proteins in Arabidopsis thaliana: Structure organization and ER localization. FEBS Lett. 2007, 581, 3356–3362. [Google Scholar] [CrossRef] [PubMed]
- Marmagne, A.; Rouet, M.A.; Ferro, M.; Rolland, N.; Alcon, C.; Joyard, J.; Garin, J.; Barbier-Brygoo, H.; Ephritikhine, G. Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol. Cell Proteomics 2004, 3, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Knox, K.; Wang, P.; Kriechbaumer, V.; Tilsner, J.; Frigerio, L.; Sparkes, I.; Hawes, C.; Oparka, K.J. Putting the squeeze on plasmodesmata: A role for reticulons in primary plasmodesmata formation. Plant Physiol. 2015, 168, 1563–1572. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.H.; Lee, S.C.; Wang, C.W. Viral protein targeting to the cortical endoplasmic reticulum is required for cell-cell spreading in plants. J. Cell Biol. 2011, 193, 521–535. [Google Scholar] [CrossRef] [PubMed]
- Tilsner, J.; Linnik, O.; Louveaux, M.; Roberts, I.M.; Chapman, S.N.; Oparka, K.J. Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J. Cell Biol. 2013, 201, 981–995. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Bowen, C.H.; Popescu, G.V.; Kang, H.G.; Kato, N.; Ma, S.; Dinesh-Kumar, S.; Snyder, M.; Popescu, S.C. Arabidopsis RTNLB1 and RTNLB2 Reticulon-like proteins regulate intracellular trafficking and activity of the FLS2 immune receptor. Plant Cell 2011, 23, 3374–3391. [Google Scholar] [CrossRef] [PubMed]
- Mysore, K.S.; Bassuner, B.; Deng, X.B.; Darbinian, N.S.; Motchoulski, A.; Ream, W.; Gelvin, S.B. Role of the Agrobacterium tumefaciens VirD2 protein in T-DNA transfer and integration. Mol. Plant Microbe Interact. 1998, 11, 668–683. [Google Scholar] [CrossRef] [PubMed]
- Mysore, K.S.; Kumar, C.T.; Gelvin, S.B. Arabidopsis ecotypes and mutants that are recalcitrant to Agrobacterium root transformation are susceptible to germ-line transformation. Plant J. 2000, 21, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.Y.; Liu, K.H.; Wang, Y.C.; Wu, J.F.; Chiu, W.L.; Chen, C.Y.; Wu, S.H.; Sheen, J.; Lai, E.M. AGROBEST: An efficient Agrobacterium-mediated transient expression method for versatile gene function analyses in Arabidopsis seedlings. Plant Methods 2014, 10, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An ’Electronic fluorescent pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Stefano, G.; Brandizzi, F.; Zheng, H. Arabidopsis RHD3 mediates the generation of the tubular ER network and is required for Golgi distribution and motility in plant cells. J. Cell Sci. 2011, 124, 2241–2252. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Sparkes, I.; Gattolin, S.; Dzimitrowicz, N.; Roberts, L.M.; Hawes, C.; Frigerio, L. An Arabidopsis reticulon and the atlastin homologue RHD3-like2 act together in shaping the tubular endoplasmic reticulum. New Phytol. 2013, 197, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Kriechbaumer, V.; Botchway, S.W.; Slade, S.E.; Knox, K.; Frigerio, L.; Oparka, K.; Hawes, C. Reticulomics: Protein-protein interaction studies with two plasmodesmata-localized reticulon family proteins identify binding partners enriched at plasmodesmata, endoplasmic reticulum, and the plasma membrane. Plant Physiol. 2015, 169, 1933–1945. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.D.; Lazarowitz, S.G. Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc. Natl. Acad. Sci. USA 2010, 107, 2491–2496. [Google Scholar] [CrossRef] [PubMed]
- Feraru, E.; Feraru, M.I.; Asaoka, R.; Paciorek, T.; De Rycke, R.; Tanaka, H.; Nakano, A.; Friml, J. BEX5/RabA1b regulates trans-Golgi network-to-plasma membrane protein trafficking in Arabidopsis. Plant Cell 2012, 24, 3074–3086. [Google Scholar] [CrossRef] [PubMed]
- Asaoka, R.; Uemura, T.; Ito, J.; Fujimoto, M.; Ito, E.; Ueda, T.; Nakano, A. Arabidopsis RABA1 GTPases are involved in transport between the trans-Golgi network and the plasma membrane, and are required for salinity stress tolerance. Plant J. 2013, 73, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.W.; Tamaki, T.; Ebine, K.; Uemura, T.; Ueda, T.; Nakano, A. RABA members act in distinct steps of subcellular trafficking of the FLAGELLIN SENSING2 receptor. Plant Cell 2013, 25, 1174–1187. [Google Scholar] [CrossRef] [PubMed]
- Chinchilla, D.; Bauer, Z.; Regenass, M.; Boller, T.; Felix, G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 2006, 18, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C.; Kunze, G.; Chinchilla, D.; Caniard, A.; Jones, J.D.; Boller, T.; Felix, G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 2006, 125, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signaling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Beck, M.; Heard, W.; Mbengue, M.; Robatzek, S. The INs and OUTs of pattern recognition receptors at the cell surface. Plant Biol. 2012, 15, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Ditt, R.F.; Nester, E.; Comai, L. The plant cell defense and Agrobacterium tumefaciens. FEMS Microbiol. Lett. 2005, 247, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Ditt, R.F.; Nester, E.W.; Comai, L. Plant gene expression response to Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 2001, 98, 10954–10959. [Google Scholar] [CrossRef] [PubMed]
- Veena; Doerge, R.W.; Gelvin, S.B. Transfer of T-DNA and Vir proteins to plant cells by Agrobacterium tumefaciens induces expression of host genes involved in mediating transformation and suppresses host defense gene expression. Plant J. 2003, 35, 219–236. [Google Scholar] [PubMed]
- Pitzschke, A.; Djamei, A.; Teige, M.; Hirt, H. VIP1 response elements mediate mitogen-activated protein kinase 3-induced stress gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 18414–18419. [Google Scholar] [CrossRef] [PubMed]
- Pitzschke, A.; Hirt, H. New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J. 2010, 29, 1021–1032. [Google Scholar] [CrossRef] [PubMed]
- Citovsky, V.; Kapelnikov, A.; Oliel, S.; Zakai, N.; Rojas, M.R.; Gilbertson, R.L.; Tzfira, T.; Loyter, A. Protein interactions involved in nuclear import of the Agrobacterium VirE2 protein in vivo and in vitro. J. Biol. Chem. 2004, 279, 29528–29533. [Google Scholar] [CrossRef] [PubMed]
- Tzfira, T.; Vaidya, M.; Citovsky, V. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 2001, 20, 3596–3607. [Google Scholar] [CrossRef] [PubMed]
- Ben Khaled, S.; Postma, J.; Robatzek, S. A moving view: Subcellular trafficking processes in pattern recognition receptor-triggered plant immunity. Annu. Rev. Phytopathol. 2015, 53, 379–402. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Zavaliev, R.; Dong, X. Membrane trafficking in plant immunity. Mol. Plant. 2017, 10, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
- Gorlich, D.; Kostka, S.; Kraft, R.; Dingwall, C.; Laskey, R.A.; Hartmann, E.; Prehn, S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr. Biol. 1995, 5, 383–392. [Google Scholar] [CrossRef]
- Bednenko, J.; Cingolani, G.; Gerace, L. Importin beta contains a COOH-terminal nucleoporin binding region important for nuclear transport. J. Cell Biol. 2003, 162, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S.; Lee, L.Y.; Oltmanns, H.; Cao, H.; Cuperus, J.; Gelvin, S.B. AtImpa-4, an Arabidopsis importin α isoform, is preferentially involved in Agrobacterium-mediated plant transformation. Plant Cell 2008, 20, 2661–2680. [Google Scholar] [CrossRef] [PubMed]
- Mysore, K.S.; Nam, J.; Gelvin, S.B. An Arabidopsis histone H2A mutant is deficient in Agrobacterium T-DNA integration. Proc. Natl. Acad. Sci. USA 2000, 97, 948–953. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Mysore, K.S.; Gelvin, S.B. Expression of the Arabidopsis histone H2A-1 gene correlates with susceptibility to Agrobacterium transformation. Plant J. 2002, 32, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Tenea, G.N.; Spantzel, J.; Lee, L.Y.; Zhu, Y.; Lin, K.; Johnson, S.J.; Gelvin, S.B. Overexpression of several Arabidopsis histone genes increases Agrobacterium-mediated transformation and transgene expression in plants. Plant Cell 2009, 21, 3350–3367. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Sardesai, N.; Fujinuma, T.; Chan, C.W.; Gelvin, S.B. Constitutive expression exposes functional redundancy between the Arabidopsis histone H2A gene HTA1 and other H2A gene family members. Plant Cell 2006, 18, 1575–1589. [Google Scholar] [CrossRef] [PubMed]
- Sauer, N.; Ludwig, A.; Knoblauch, A.; Rothe, P.; Gahrtz, M.; Klebl, F. AtSUC8 and AtSUC9 encode functional sucrose transporters, but the closely related AtSUC6 and AtSUC7 genes encode aberrant proteins in different Arabidopsis ecotypes. Plant J. 2004, 40, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Lemoine, R. Sucrose transporters in plants: Update on function and structure. Biochimica et Biophysica Acta 2000, 1465, 246–262. [Google Scholar] [CrossRef]
- Williams, L.E.; Lemoine, R.; Sauer, N. Sugar transporters in higher plants—A diversity of roles and complex regulation. Trends Plant Sci. 2000, 5, 283–290. [Google Scholar] [CrossRef]
- Feuerstein, A.; Niedermeier, M.; Bauer, K.; Engelmann, S.; Hoth, S.; Stadler, R.; Sauer, N. Expression of the AtSUC1 gene in the female gametophyte, and ecotype-specific expression differences in male reproductive organs. Plant Biol. 2010, 12, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001; ISBN 978–1-936113-42-2. [Google Scholar]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.G.; Smith, J.A.; Struhl, K. Current Protocols in Molecular Biology; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2003; ISBN 978–0-471-50338-5. [Google Scholar]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA minipreperation: Version 2. Plant Mol. Biol. Rep. 1983, 1, 19–22. [Google Scholar] [CrossRef]
- Koncz, C.; Schell, J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 1986, 204, 383–396. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Nam, J.; Humara, J.M.; Mysore, K.S.; Lee, L.Y.; Cao, H.; Valentine, L.; Li, J.; Kaiser, A.D.; Kopecky, A.L.; et al. Identification of Arabidopsis rat mutants. Plant Physiol. 2003, 132, 494–505. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.H.; Wang, M.H.; Lee, Y.L.; Tsai, Y.L.; Li, Y.H.; Yang, F.J.; Liao, Y.C.; Lin, S.K.; Lai, E.M. Agrobacterium-produced and exogenous cytokinin-modulated Agrobacterium-mediated plant transformation. Mol. Plant Pathol. 2010, 11, 677–690. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.L.; Preston, G.; Collmer, A.; Chang, C.J.; Huang, H.C. Characterization of the hrpC and hrpRS operons of Pseudomonas syringae pathovars syringae, tomato, and glycinea and analysis of the ability of hrpF, hrpG, hrcC, hrpT, and hrpV mutants to elicit the hypersensitive response and disease in plants. J. Bacteriol. 1998, 180, 4523–4531. [Google Scholar] [PubMed]
- Katagiri, F.; Thilmony, R.; He, S.Y. The Arabidopsis thaliana-Pseudomonas syringae interaction. Arabidopsis Book 2002, 1, e0039. [Google Scholar] [CrossRef] [PubMed]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, F.-C.; Fu, B.-J.; Liu, Y.-T.; Chang, Y.-R.; Chi, S.-F.; Chien, P.-R.; Huang, S.-C.; Hwang, H.-H. Arabidopsis RETICULON-LIKE3 (RTNLB3) and RTNLB8 Participate in Agrobacterium-Mediated Plant Transformation. Int. J. Mol. Sci. 2018, 19, 638. https://doi.org/10.3390/ijms19020638
Huang F-C, Fu B-J, Liu Y-T, Chang Y-R, Chi S-F, Chien P-R, Huang S-C, Hwang H-H. Arabidopsis RETICULON-LIKE3 (RTNLB3) and RTNLB8 Participate in Agrobacterium-Mediated Plant Transformation. International Journal of Molecular Sciences. 2018; 19(2):638. https://doi.org/10.3390/ijms19020638
Chicago/Turabian StyleHuang, Fan-Chen, Bi-Ju Fu, Yin-Tzu Liu, Yao-Ren Chang, Shin-Fei Chi, Pei-Ru Chien, Si-Chi Huang, and Hau-Hsuan Hwang. 2018. "Arabidopsis RETICULON-LIKE3 (RTNLB3) and RTNLB8 Participate in Agrobacterium-Mediated Plant Transformation" International Journal of Molecular Sciences 19, no. 2: 638. https://doi.org/10.3390/ijms19020638
APA StyleHuang, F.-C., Fu, B.-J., Liu, Y.-T., Chang, Y.-R., Chi, S.-F., Chien, P.-R., Huang, S.-C., & Hwang, H.-H. (2018). Arabidopsis RETICULON-LIKE3 (RTNLB3) and RTNLB8 Participate in Agrobacterium-Mediated Plant Transformation. International Journal of Molecular Sciences, 19(2), 638. https://doi.org/10.3390/ijms19020638