Screening of Azo-Dye-Degrading Bacteria from Textile Industry Wastewater-Activated Sludge
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Bacterial Isolation
2.3. Microbiome Identification
2.4. Prospection of Potential Azo-Dye-Degrading Bacteria in Solid and Liquid Mediums
2.5. Evaluation of Carbon Sources on the Kinetic Degradation of Azo Dye
2.6. Phytotoxicity Assay
2.7. Detection of Azo Dye
2.8. Enzymatic Azo Dye Degradation
3. Results and Discussion
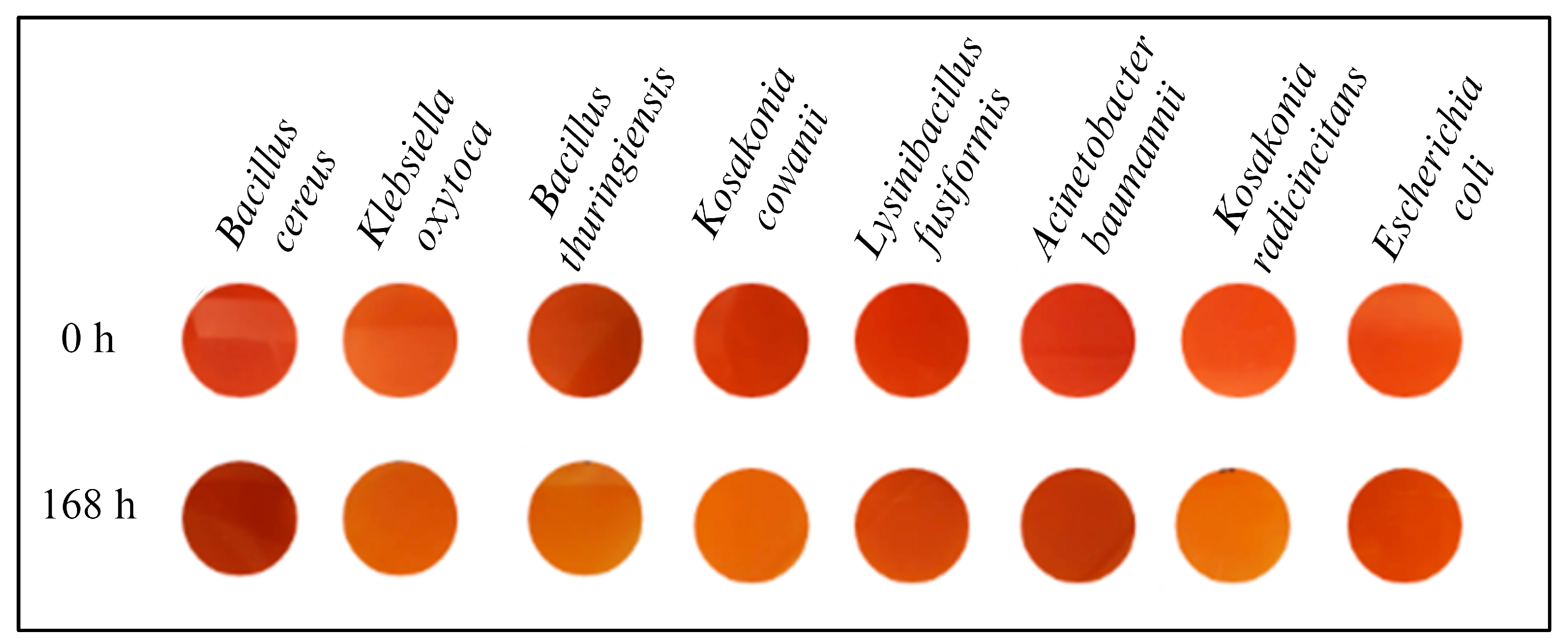
3.1. Screening of Dye Decolourization in Solid and Liquid Mediums
3.2. Effect of Carbon Sources and Dye-Decolorizing Kinetics by B. thuringiensis or K. radicincitans
3.3. Enzymatic Azo Dye Degradation
3.4. Dye-Decolorizing Kinetics by B. thuringiensis and K. radicincitans as a Consortium
3.5. Biodegradation Analysis
3.6. Phytotoxicity Assay
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Scanning Electron Microscope (SEM)

References
- Grand View Research Textile Market Size, Share & Trends Analysis Report by Raw Material (Wool, Chemical, Silk), by Product (Natural Fibers, Polyester), by Application (Household, Technical), by Region, and Segment Forecasts, 2021–2028; Grand View Research: San Francisco, CA, USA, 2021.
- de Vasconcelos, G.M.D.; Mulinari, J.; Souza, S.M.d.A.G.U.d.; de Souza, A.A.U.; de Oliveira, D.; de Andrade, C.J. Biodegradation of Azo Dye-Containing Wastewater by Activated Sludge: A Critical Review. World J. Microbiol. Biotechnol. 2021, 37, 101. [Google Scholar] [CrossRef] [PubMed]
- Lade, H.S.; Waghmode, T.R.; Kadam, A.A.; Govindwar, S.P. Enhanced Biodegradation and Detoxification of Disperse Azo Dye Rubine GFL and Textile Industry Effluent by Defined Fungal-Bacterial Consortium. Int. Biodeterior. Biodegrad. 2012, 72, 94–107. [Google Scholar] [CrossRef]
- Bilińska, L.; Gmurek, M.; Ledakowicz, S. Comparison between Industrial and Simulated Textile Wastewater Treatment by AOPs—Biodegradability, Toxicity and Cost Assessment. Chem. Eng. J. 2016, 306, 550–559. [Google Scholar] [CrossRef]
- Paździor, K.; Bilińska, L.; Ledakowicz, S.; Pa, K.; Bili, L.; Paździor, K.; Bilińska, L.; Ledakowicz, S. A Review of the Existing and Emerging Technologies in the Combination of AOPs and Biological Processes in Industrial Textile Wastewater Treatment. Chem. Eng. J. 2019, 376. [Google Scholar] [CrossRef]
- Paździor, K.; Wrębiak, J.; Klepacz-Smółka, A.; Gmurek, M.; Bilińska, L.; Kos, L.; Sójka-Ledakowicz, J.; Ledakowicz, S. Influence of Ozonation and Biodegradation on Toxicity of Industrial Textile Wastewater. J. Environ. Manag. 2017, 195, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Xu, J.; Cao, X.; Cheng, Y.; Zhu, T. Characterization of Functional Microbial Communities Involved in Diazo Dyes Decolorization and Mineralization Stages. Int. Biodeterior. Biodegrad. 2018, 132, 166–177. [Google Scholar] [CrossRef]
- Rodrigues, F.K.; Salau, N.P.G.; Dotto, G.L. New Insights about Reactive Red 141 Adsorption onto Multi–Walled Carbon Nanotubes Using Statistical Physics Coupled with Van Der Waals Equation. Sep. Purif. Technol. 2019, 224, 290–294. [Google Scholar] [CrossRef]
- Manekar, P.; Patkar, G.; Aswale, P.; Mahure, M.; Nandy, T. Detoxifying of High Strength Textile Effluent through Chemical and Bio-Oxidation Processes. Bioresour. Technol. 2014, 157, 44–51. [Google Scholar] [CrossRef]
- Zhang, B.; Xu, X.; Zhu, L. Activated Sludge Bacterial Communities of Typical Wastewater Treatment Plants: Distinct Genera Identification and Metabolic Potential Differential Analysis. AMB Express 2018, 8, 184. [Google Scholar] [CrossRef]
- Li, W.; Mu, B.; Yang, Y. Feasibility of Industrial-Scale Treatment of Dye Wastewater via Bio-Adsorption Technology. Bioresour. Technol. 2019, 277, 157–170. [Google Scholar] [CrossRef]
- Meerbergen, K.; Willems, K.A.; Dewil, R.; Van Impe, J.; Appels, L.; Lievens, B. Isolation and Screening of Bacterial Isolates from Wastewater Treatment Plants to Decolorize Azo Dyes. J. Biosci. Bioeng. 2018, 125, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Ashfaq, M.Y.; Da’na, D.A.; Al-Ghouti, M.A. Application of MALDI-TOF MS for Identification of Environmental Bacteria: A Review. J. Environ. Manag. 2022, 305, 114359. [Google Scholar] [CrossRef] [PubMed]
- Avanzi, I.R.; Gracioso, L.H.; Baltazar, M.d.P.G.; Karolski, B.; Perpetuo, E.A.; Nascimento, C.A.O.D. Rapid Bacteria Identification from Environmental Mining Samples Using MALDI-TOF MS Analysis. Environ. Sci. Pollut. Res. 2017, 24, 3717–3726. [Google Scholar] [CrossRef] [PubMed]
- Abdel Samad, R.; Al Disi, Z.; Mohammad Ashfaq, M.Y.; Wahib, S.M.; Zouari, N. The Use of Principle Component Analysis and MALDI-TOF MS for the Differentiation of Mineral Forming: Virgibacillus and Bacillus Species Isolated from Sabkhas. RSC Adv. 2020, 10, 14606–14616. [Google Scholar] [CrossRef] [PubMed]
- Marklein, G.; Josten, M.; Klanke, U.; Müller, E.; Horré, R.; Maier, T.; Wenzel, T.; Kostrzewa, M.; Bierbaum, G.; Hoerauf, A.; et al. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for Fast and Reliable Identification of Clinical Yeast Isolates. J. Clin. Microbiol. 2009, 47, 2912–2917. [Google Scholar] [CrossRef]
- Alves, L.A.C.; Souza, R.C.; da Silva, T.M.C.; Watanabe, A.; Dias, M.; Mendes, M.A.; Ciamponi, A.L. Identification of Microorganisms in Biofluids of Individuals with Periodontitis and Chronic Kidney Disease Using Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry. Rapid Commun. Mass. Spectrom. 2016, 30, 1228–1232. [Google Scholar] [CrossRef]
- Han, H.W.; Chang, H.C.; Hunag, A.H.; Chang, T.C. Optimization of the Score Cutoff Value for Routine Identification of Staphylococcus Species by Matrix-Assisted Laser Desorption Ionization-Time-of-Flight Mass Spectrometry. Diagn. Microbiol. Infect. Dis. 2015, 83, 349–354. [Google Scholar] [CrossRef]
- Normand, A.C.; Cassagne, C.; Gautier, M.; Becker, P.; Ranque, S.; Hendrickx, M.; Piarroux, R. Decision Criteria for MALDI-TOF MS-Based Identification of Filamentous Fungi Using Commercial and in-House Reference Databases. BMC Microbiol. 2017, 17, 1–17. [Google Scholar] [CrossRef]
- Sreedharan, V.; Saha, P.; Rao, K.V.B. Dye Degradation Potential of Acinetobacter Baumannii Strain VITVB against Commercial Azo Dyes. Bioremediation J. 2021, 25, 347–368. [Google Scholar] [CrossRef]
- Unnikrishnan, S.; Khan, M.H.; Ramalingam, K. Dye-Tolerant Marine Acinetobacter Baumannii-Mediated Biodegradation of Reactive Red. Water Sci. Eng. 2018, 11, 265–275. [Google Scholar] [CrossRef]
- Ameenudeen, S.; Unnikrishnan, S.; Ramalingam, K. Statistical Optimization for the Efficacious Degradation of Reactive Azo Dyes Using Acinetobacter Baumannii JC359. J. Environ. Manag. 2021, 279, 111512. [Google Scholar] [CrossRef] [PubMed]
- Adebajo, S.; Balogun, S.; Akintokun, A. Decolourization of Vat Dyes by Bacterial Isolates Recovered from Local Textile Mills in Southwest, Nigeria. Microbiol. Res. J. Int. 2017, 18, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Cao, M.; Wang, P.; Wang, S.; Yue, Y.; Yuan, W.; Qiao, W.; Wang, F.; Song, X. Simultaneous Decolorization and Biohydrogen Production from Xylose by Klebsiella Oxytoca GS-4-08 in the Presence of Azo Dyes with Sulfonate and Carboxyl Groups. Appl. Environ. Microbiol. 2017, 83, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Balraj, B.; Hussain, Z.; King, P. Experimental Study on Non Sporulating Escherichia Coli Bacteria in Removing Methylene Blue. Int. J. Pharma Bio Sci. 2016, 7, B629–B637. [Google Scholar]
- Hilda Josephine, S.; Sekar, A.S.S. A Comparative Study of Biodegradation of Textile Azo Dyes by Escherichia Coli and Pseudomonas Putida. Nat. Environ. Pollut. Technol. 2014, 13, 417–420. [Google Scholar]
- Haque, M.M.; Haque, M.A.; Mosharaf, M.K.; Marcus, P.K. Novel Bacterial Biofilm Consortia That Degrade and Detoxify the Carcinogenic Diazo Dye Congo Red. Arch. Microbiol. 2021, 203, 643–654. [Google Scholar] [CrossRef]
- Kiayi, Z.; Lotfabad, T.B.; Heidarinasab, A.; Shahcheraghi, F. Microbial Degradation of Azo Dye Carmoisine in Aqueous Medium Using Saccharomyces Cerevisiae ATCC 9763. J. Hazard. Mater. 2019, 373, 608–619. [Google Scholar] [CrossRef]
- dos Santos, F.E.; Carvalho, M.S.S.; Silveira, G.L.; Correa, F.F.; Cardoso, M.d.G.; Andrade-Vieira, L.F.; Vilela, L.R. Phytotoxicity and Cytogenotoxicity of Hydroalcoholic Extracts from Solanum Muricatum Ait. and Solanum Betaceum Cav. (Solanaceae) in the Plant Model Lactuca Sativa. Environ. Sci. Pollut. Res. 2019, 26, 27558–27568. [Google Scholar] [CrossRef]
- Peduto, T.A.G.; de Jesus, T.A.; Kohatsu, M.Y. Sensibilidade de Diferentes Sementes Em Ensaio de Fitotoxicidade. Rev. Bras. De Ciência Tecnol. E Inovação 2019, 4, 200. [Google Scholar] [CrossRef]
- Barros, F.F.C.; Simiqueli, A.P.R.; de Andrade, C.J.; Pastore, G.M. Production of Enzymes from Agroindustrial Wastes by Biosurfactant-Producing Strains of Bacillus Subtilis. Biotechnol. Res. Int. 2013, 2013, 103960. [Google Scholar] [CrossRef]
- Kandelbauer, A.; Guebitz, G.M. Bioremediation for the Decolorization of Textile Dyes-A Review. Environ. Chem. Green. Chem. Pollut. Ecosyst. 2005, 269–288. [Google Scholar] [CrossRef]
- Haque, M.M.; Haque, M.A.; Mosharaf, M.K.; Rahman, A.; Islam, M.S.; Nahar, K.; Molla, A.H. Enhanced Biofilm-Mediated Degradation of Carcinogenic and Mutagenic Azo Dye by Novel Bacteria Isolated from Tannery Wastewater. J. Environ. Chem. Eng. 2023, 11. [Google Scholar] [CrossRef]
- Haque, M.M.; Hossen, M.N.; Rahman, A.; Roy, J.; Talukder, M.R.; Ahmed, M.; Ahiduzzaman, M.; Haque, M.A. Decolorization, Degradation and Detoxification of Mutagenic Dye Methyl Orange by Novel Biofilm Producing Plant Growth-Promoting Rhizobacteria. Chemosphere 2024, 346. [Google Scholar] [CrossRef]
- Khan, R.; Bhawana, P.; Fulekar, M.H. Microbial Decolorization and Degradation of Synthetic Dyes: A Review. Rev. Environ. Sci. Biotechnol. 2013, 12, 75–97. [Google Scholar] [CrossRef]
- Kapoor, R.T.; Danish, M.; Singh, R.S.; Rafatullah, M.; Abdul, A.K. Exploiting Microbial Biomass in Treating Azo Dyes Contaminated Wastewater: Mechanism of Degradation and Factors Affecting Microbial Efficiency. J. Water Process Eng. 2021, 43, 102255. [Google Scholar] [CrossRef]
- Bal, G.; Thakur, A. Distinct Approaches of Removal of Dyes from Wastewater: A Review. In Proceedings of the Materials Today: Proceedings; Elsevier Ltd.: Amsterdam, The Netherlands, 2021; Volume 50, pp. 1575–1579. [Google Scholar]
- Saratale, G.; Kalme, S.; Bhosale, S.; Govindwar, S. Biodegradation of Kerosene by Aspergillus Ochraceus NCIM-1146. J. Basic. Microbiol. 2007, 47, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Shah, B.; Jain, K.; Jiyani, H.; Mohan, V.; Madamwar, D. Microaerophilic Symmetric Reductive Cleavage of Reactive Azo Dye—Remazole Brilliant Violet 5R by Consortium VIE6: Community Synergism. Appl. Biochem. Biotechnol. 2016, 180, 1029–1042. [Google Scholar] [CrossRef]
- Mullai, P.; Yogeswari, M.K.; Vishali, S.; Tejas Namboodiri, M.M.; Gebrewold, B.D.; Rene, E.R.; Pakshirajan, K. Aerobic Treatment of Effluents From Textile Industry. In Current Developments in Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2017; pp. 3–34. ISBN 9780444636768. [Google Scholar]
- Chandanshive, V.V.; Kadam, S.K.; Khandare, R.V.; Kurade, M.B.; Jeon, B.H.; Jadhav, J.P.; Govindwar, S.P. In Situ Phytoremediation of Dyes from Textile Wastewater Using Garden Ornamental Plants, Effect on Soil Quality and Plant Growth. Chemosphere 2018, 210, 968–976. [Google Scholar] [CrossRef]
- Chen, Y.; Feng, L.; Li, H.; Wang, Y.; Chen, G.; Zhang, Q. Biodegradation and Detoxification of Direct Black G Textile Dye by a Newly Isolated Thermophilic Microflora. Bioresour. Technol. 2018, 250, 650–657. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, H.; Xue, G.; Liu, Y.; Chen, S.; Jia, C. A Critical Review of the Aniline Transformation Fate in Azo Dye Wastewater Treatment. J. Clean. Prod. 2021, 321. [Google Scholar] [CrossRef]
- Karim, M.E.; Dhar, K.; Hossain, M.T. Decolorization of Textile Reactive Dyes by Bacterial Monoculture and Consortium Screened from Textile Dyeing Effluent. J. Genet. Eng. Biotechnol. 2018, 16, 375–380. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, S.; Khalid, A.; Arshad, M.; Mahmood, T.; Crowley, D.E. Detoxification of Azo Dyes by Bacterial Oxidoreductase Enzymes. Crit. Rev. Biotechnol. 2016, 36, 639–651. [Google Scholar] [CrossRef] [PubMed]
- Sari, I.P.; Simarani, K. Decolorization of Selected Azo Dye by Lysinibacillus Fusiformis W1B6: Biodegradation Optimization, Isotherm, and Kinetic Study Biosorption Mechanism. Adsorpt. Sci. Technol. 2019, 37, 492–508. [Google Scholar] [CrossRef]
- Pandey, A.; Singh, P.; Iyengar, L. Bacterial Decolorization and Degradation of Azo Dyes. Int. Biodeterior. Biodegrad. 2007, 59, 73–84. [Google Scholar] [CrossRef]
- Misal, S.A.; Gawai, K.R. Azoreductase: A Key Player of Xenobiotic Metabolism. Bioresour. Bioprocess. 2018, 5. [Google Scholar] [CrossRef]
- Khlifi, R.; Belbahri, L.; Woodward, S.; Ellouz, M.; Dhouib, A.; Sayadi, S.; Mechichi, T. Decolourization and Detoxification of Textile Industry Wastewater by the Laccase-Mediator System. J. Hazard. Mater. 2010, 175, 802–808. [Google Scholar] [CrossRef]
- Wong, Y.; Yu, J. Laccase-Catalyzed Decolorization of Synthetic Dyes. Water Res. 1999, 33, 3512–3520. [Google Scholar] [CrossRef]
- Chivukula, M.; Renganathan, V. Phenolic Azo Dye Oxidation by Laccase from Pyricularia Oryzae. Appl. Environ. Microbiol. 1995, 61, 4374–4377. [Google Scholar] [CrossRef]
- Neifar, M.; Sghaier, I.; Guembri, M.; Chouchane, H.; Mosbah, A.; Ouzari, H.I.; Jaouani, A.; Cherif, A. Recent Advances in Textile Wastewater Treatment Using Microbial Consortia. J. Text. Eng. Fash. Technol. 2019, 5, 134–146. [Google Scholar] [CrossRef]
- Shindhal, T.; Rakholiya, P.; Varjani, S.; Pandey, A.; Ngo, H.H.; Guo, W.; Ng, H.Y.; Taherzadeh, M.J. A Critical Review on Advances in the Practices and Perspectives for the Treatment of Dye Industry Wastewater. Bioengineered 2021, 12, 70–87. [Google Scholar] [CrossRef]
- Samuchiwal, S.; Gola, D.; Malik, A. Decolourization of Textile Effluent Using Native Microbial Consortium Enriched from Textile Industry Effluent. J. Hazard. Mater. 2021, 402, 123835. [Google Scholar] [CrossRef] [PubMed]
- Thakur, J.K.; Paul, S.; Dureja, P.; Annapurna, K.; Padaria, J.C.; Gopal, M. Degradation of Sulphonated Azo Dye Red HE7B by Bacillus Sp. and Elucidation of Degradative Pathways. Curr. Microbiol. 2014, 69, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Eslami, H.; Shariatifar, A.; Rafiee, E.; Shiranian, M.; Salehi, F.; Hosseini, S.S.; Eslami, G.; Ghanbari, R.; Ebrahimi, A.A. Decolorization and Biodegradation of Reactive Red 198 Azo Dye by a New Enterococcus Faecalis–Klebsiella Variicola Bacterial Consortium Isolated from Textile Wastewater Sludge. World J. Microbiol. Biotechnol. 2019, 35. [Google Scholar] [CrossRef] [PubMed]
- Al-Tohamy, R.; Ali, S.S.; Xie, R.; Schagerl, M.; Khalil, M.A.; Sun, J. Decolorization of Reactive Azo Dye Using Novel Halotolerant Yeast Consortium HYC and Proposed Degradation Pathway. Ecotoxicol. Environ. Saf. 2023, 263. [Google Scholar] [CrossRef] [PubMed]
- Khandare, S.D.; Teotia, N.; Kumar, M.; Diyora, P.; Chaudhary, D.R. Biodegradation and Decolorization of Trypan Blue Azo Dye by Marine Bacteria Vibrio Sp. JM-17. Biocatal. Agric. Biotechnol. 2023, 51. [Google Scholar] [CrossRef]
- Tizazu, S.; Tesfaye, G.; Wang, A.; Guadie, A.; Andualem, B. Microbial Diversity, Transformation and Toxicity of Azo Dye Biodegradation Using Thermo-Alkaliphilic Microbial Consortia. Heliyon 2023, 9. [Google Scholar] [CrossRef] [PubMed]
- Pearce, C.I.; Lloyd, J.R.; Guthrie, J.T. The Removal of Colour from Textile Wastewater Using Whole Bacterial Cells: A Review. Dye. Pigment. 2003, 58, 179–196. [Google Scholar] [CrossRef]
- Rodrigues, C.S.D.; Madeira, L.M.; Boaventura, R.A.R. Synthetic Textile Dyeing Wastewater Treatment by Integration of Advanced Oxidation and Biological Processes—Performance Analysis with Costs Reduction. J. Environ. Chem. Eng. 2014, 2, 1027–1039. [Google Scholar] [CrossRef]
- Siddiqui, S.I.; Fatima, B.; Tara, N.; Rathi, G.; Chaudhry, S.A. Recent Advances in Remediation of Synthetic Dyes from Wastewaters Using Sustainable and Low-Cost Adsorbents; Elsevier Ltd.: Amsterdam, The Netherlands, 2018; ISBN 9780081024911. [Google Scholar]
- Foster, J.E.; Adamovsky, G.; Gucker, S.N.; Blankson, I.M. A Comparative Study of the Time-Resolved Decomposition of Methylene Blue Dye under the Action of a Nanosecond Repetitively Pulsed Dbd Plasma Jet Using Liquid Chromatography and Spectrophotometry. IEEE Trans. Plasma Sci. 2013, 41, 503–512. [Google Scholar] [CrossRef]
- Ali, H. Biodegradation of Synthetic Dyes—A Review. Water Air Soil. Pollut. 2010, 213, 251–273. [Google Scholar] [CrossRef]
- Sompark, C.; Singkhonrat, J.; Sakkayawong, N. Biotransformation of Reactive Red 141 by Paenibacillus Terrigena KKW2-005 and Examination of Product Toxicity. J. Microbiol. Biotechnol. 2021, 31, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Kalme, S.; Ghodake, G.; Govindwar, S. Red HE7B Degradation Using Desulfonation by Pseudomonas Desmolyticum NCIM 2112. Int Biodeterior Biodegrad. 2007, 60, 327–333. [Google Scholar] [CrossRef]
- Chung, K.T. Mutagenicity and Carcinogenicity of Aromatic Amines Metabolically Produced from Azo Dyes. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2000, 18, 51–74. [Google Scholar] [CrossRef]
- Brown, M.A.; De Vito, S.C. Predicting Azo Dye Toxicity. Crit. Rev. Environ. Sci. Technol. 1993, 23, 249–324. [Google Scholar] [CrossRef]





| Strain | Score | DOAJ * | JSTOR * | Science Direct * | Scopus * | Springer * | Google Scholar * |
|---|---|---|---|---|---|---|---|
| Bacillus cereus | 2.16 | 0 | 1 | 92 | 7 | 335 | 11,600 |
| Klebsiella oxytoca | 1.74 | 0 | 0 | 8 | 1 | 57 | 605 |
| Bacillus thuringiensis | 2.21 | 0 | 1 | 31 | 2 | 112 | 1190 |
| Kosakonia cowanii | 1.96 | 0 | 0 | 1 | 0 | 1 | 11 |
| Lysinibacillus fusiformis | 1.81 | 0 | 0 | 4 | 1 | 17 | 198 |
| Acinetobacter baumannii | 2.16 | 0 | 0 | 10 | 1 | 43 | 676 |
| Kosakonia radicincitans | 1.86 | 0 | 0 | 0 | 0 | 0 | 7 |
| Escherichia coli | 1.75 | 7 | 1 | 318 | 29 | 1028 | 17,200 |
| Bacillus thuringiensis | Kosakonia radicincitans | |
|---|---|---|
| Øcolony (cm) | 0.250 ± 0.056 | 0.549 ± 0.031 |
| Øcolony + halo (cm) | 0.339 ± 0.052 | 0.675 ± 0.051 |
| Øhalo (cm) | 0.091 ± 0.017 | 0.126 ± 0.031 |
| Micro-Organisms | Decolorization Percentage of RR 141 Dye | Removal Percentage of RR 141 Dye | |||
|---|---|---|---|---|---|
| BHI | MSG | MS | BHI | MSG | |
| B. thuringiensis | 29% | 38% | 4% | - | 19% |
| K. radicincitans | 26% | 15% | 10% | 38% | - |
| Consortium | 18% | 40% | - | - | 10% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Vasconcelos, G.M.D.; Della-Flora, I.K.; Kelbert, M.; de Andrade, L.M.; Oliveira, D.d.; Guelli Ulson de Souza, S.M.d.A.; Ulson de Souza, A.A.; Andrade, C.J.d. Screening of Azo-Dye-Degrading Bacteria from Textile Industry Wastewater-Activated Sludge. Eng 2024, 5, 116-132. https://doi.org/10.3390/eng5010008
de Vasconcelos GMD, Della-Flora IK, Kelbert M, de Andrade LM, Oliveira Dd, Guelli Ulson de Souza SMdA, Ulson de Souza AA, Andrade CJd. Screening of Azo-Dye-Degrading Bacteria from Textile Industry Wastewater-Activated Sludge. Eng. 2024; 5(1):116-132. https://doi.org/10.3390/eng5010008
Chicago/Turabian Stylede Vasconcelos, Grazielly Maria Didier, Isabela Karina Della-Flora, Maikon Kelbert, Lidiane Maria de Andrade, Débora de Oliveira, Selene Maria de Arruda Guelli Ulson de Souza, Antônio Augusto Ulson de Souza, and Cristiano José de Andrade. 2024. "Screening of Azo-Dye-Degrading Bacteria from Textile Industry Wastewater-Activated Sludge" Eng 5, no. 1: 116-132. https://doi.org/10.3390/eng5010008
APA Stylede Vasconcelos, G. M. D., Della-Flora, I. K., Kelbert, M., de Andrade, L. M., Oliveira, D. d., Guelli Ulson de Souza, S. M. d. A., Ulson de Souza, A. A., & Andrade, C. J. d. (2024). Screening of Azo-Dye-Degrading Bacteria from Textile Industry Wastewater-Activated Sludge. Eng, 5(1), 116-132. https://doi.org/10.3390/eng5010008









