Effect of Degree of Substitution and Molecular Weight on Transfection Efficacy of Starch-Based siRNA Delivery System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Starch Quaternization
2.2. Quaternized Starch Chemical Analysis
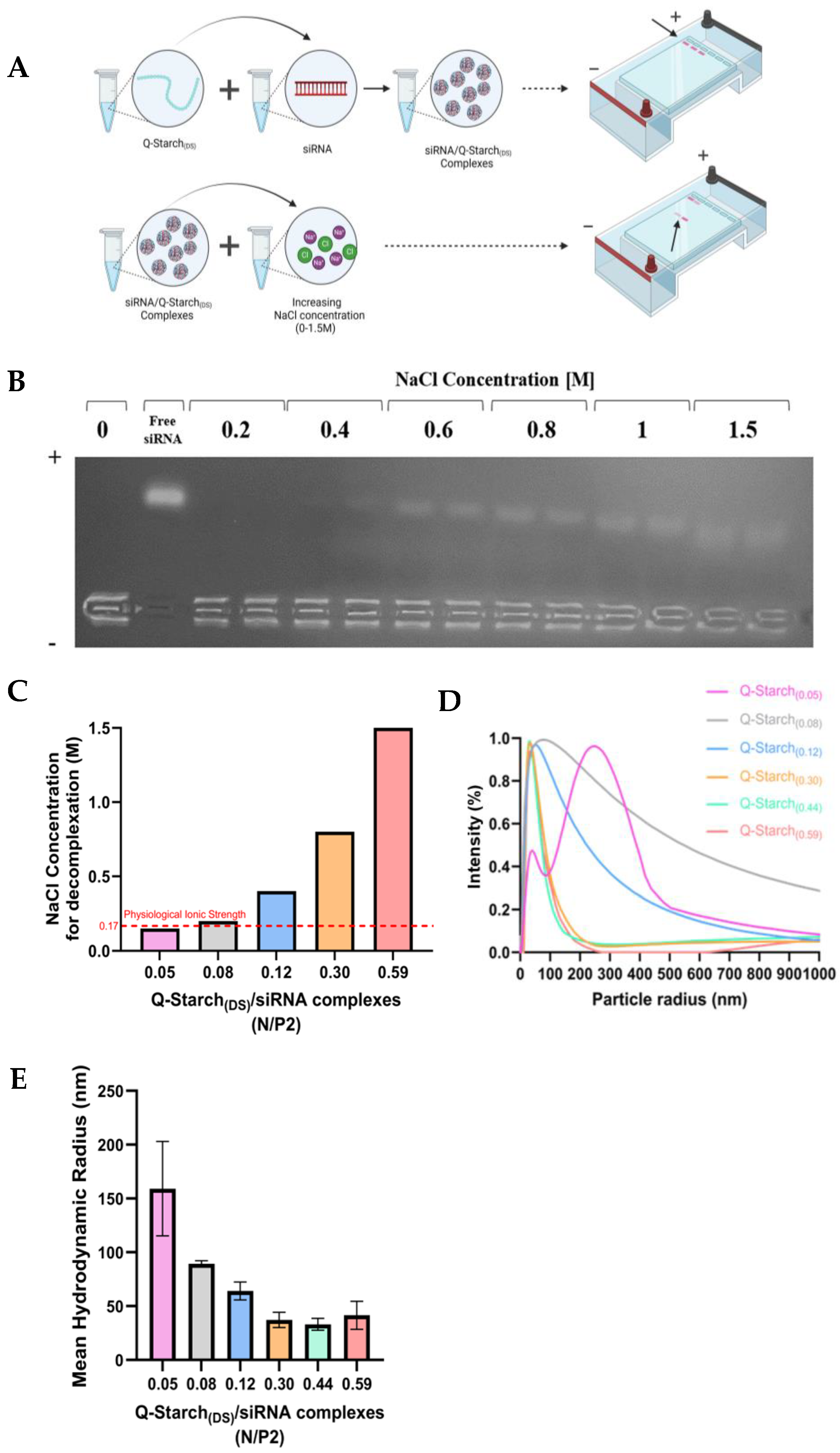
2.3. Q-Starch(DS)/siRNA Complex Preparation
2.4. Q-Starch(DS)/siRNA Complex Characterization
2.4.1. Agarose Gel Electrophoresis
2.4.2. Dynamic Light Scattering (DLS)
2.4.3. Ionic Strength Agarose Gel Electrophoresis
2.4.4. Zeta Potential
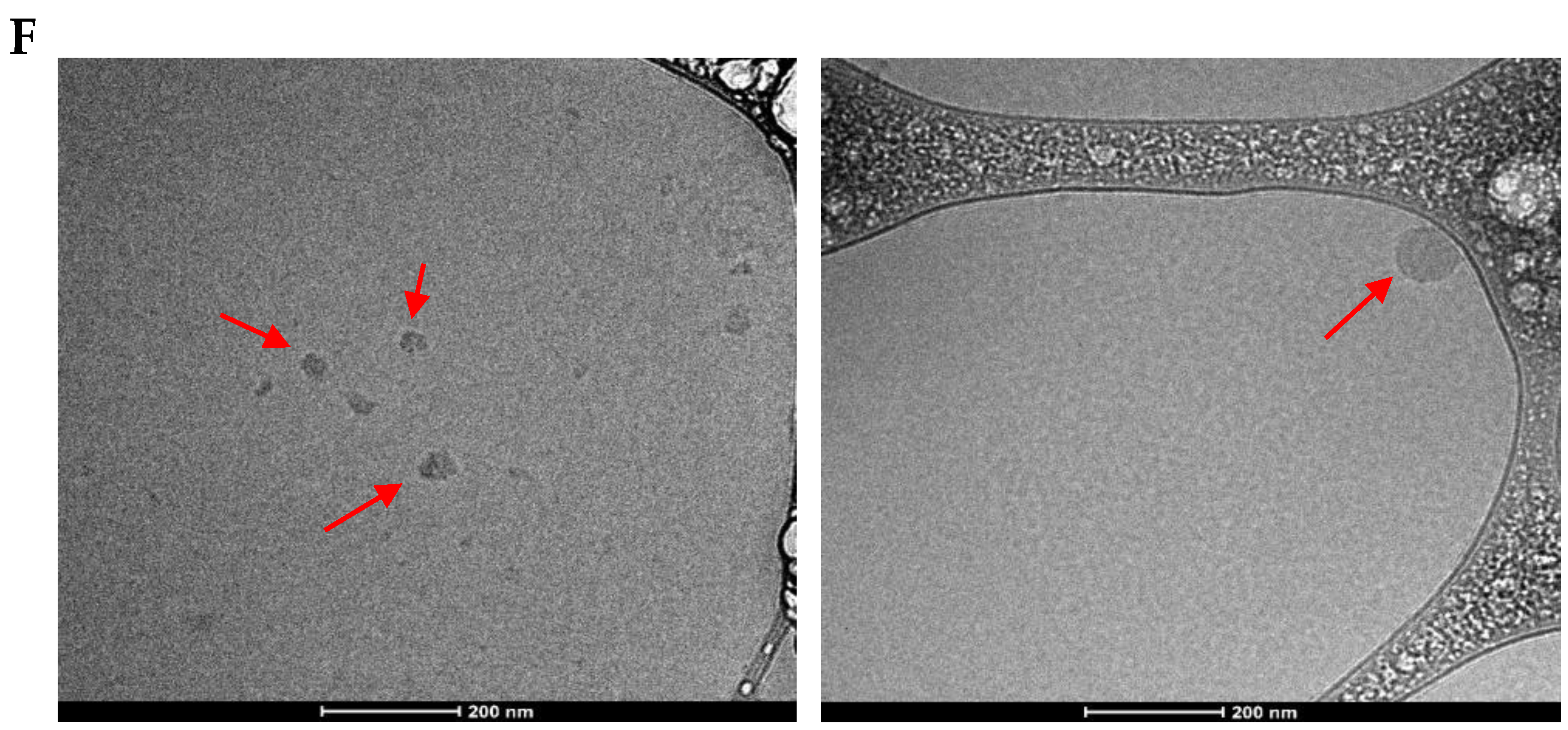
2.4.5. Cryo-Transmission Electron Microscopy (cryoTEM)
2.5. Cell Culture Handling
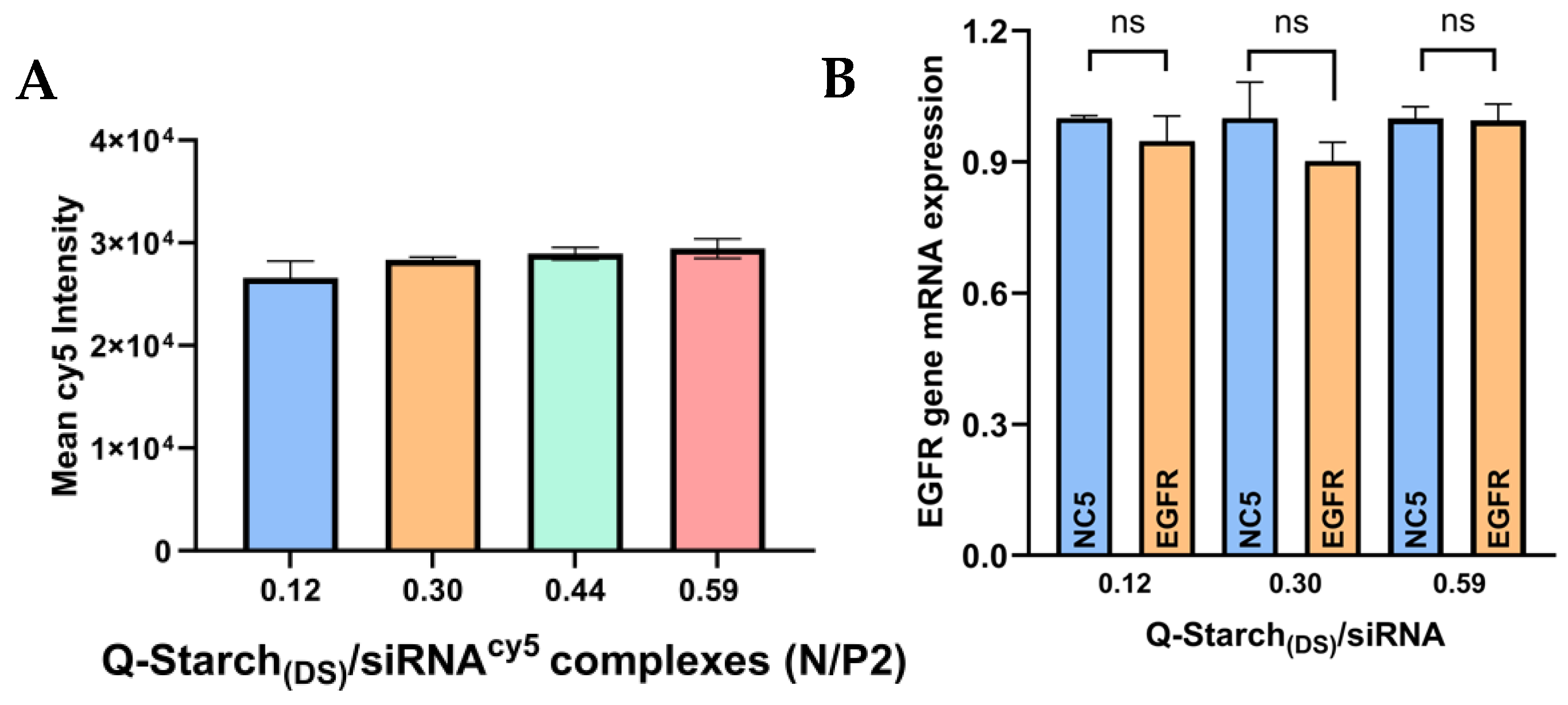
2.6. In Vitro Cellular Uptake
2.7. siRNA: EGFR Gene Silencing
2.8. Iodine Starch Test
2.9. Viscosity Measurements
2.10. Statistical Analysis
3. Results and Discussion
3.1. Q-Starch Synthesis and Chemical Characterization
3.2. Q-Starch(DS)/siRNA Complex Formation and Characterization
3.3. Q-Starch(DS)/siRNA Electrostatic Strengths and Size Distributions
3.4. Q-Starch(DS)/siRNA Cellular Uptake and Gene Silencing Capabilities
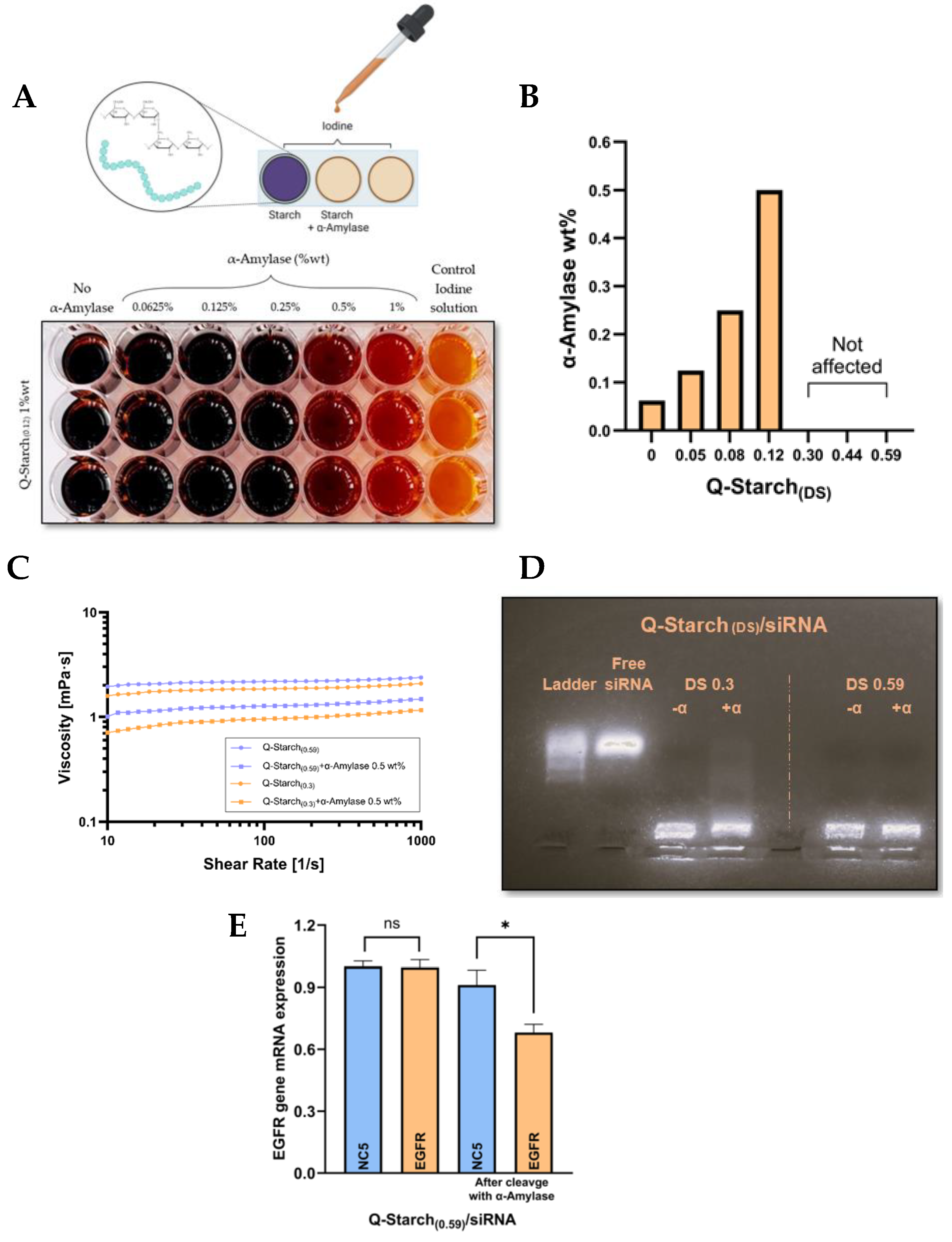
3.5. Enzymatic Cleavage of Q-Starch(DS) by α-Amylase
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Egorova, A.; Petrosyan, M.; Maretina, M.; Bazian, E.; Krylova, I.; Baranov, V.; Kiselev, A. IRGD-Targeted Peptide Nanoparticles for Anti-Angiogenic RNAi-Based Therapy of Endometriosis. Pharmaceutics 2023, 15, 2108. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.; Xiao, H.; Zhang, J.; Liang, X.J.; Huang, Y. RNAi Therapeutic and Its Innovative Biotechnological Evolution. Biotechnol. Adv. 2019, 37, 801–825. [Google Scholar] [CrossRef]
- Xie, X.; Yue, T.; Gu, W.; Cheng, W.; He, L.; Ren, W.; Li, F.; Piao, J.-G. Recent Advances in Mesoporous Silica Nanoparticles Delivering SiRNA for Cancer Treatment. Pharmaceutics 2023, 15, 2483. [Google Scholar] [CrossRef] [PubMed]
- Hülsmann, J.; Lindemann, H.; Wegener, J.; Kühne, M.; Godmann, M.; Koschella, A.; Coldewey, S.M.; Heinze, T.; Heinzel, T. Dually Modified Cellulose as a Non-Viral Vector for the Delivery and Uptake of HDAC3 SiRNA. Pharmaceutics 2023, 15, 2659. [Google Scholar] [CrossRef]
- Scott, L.J. Givosiran: First Approval. Drugs 2020, 80, 335–339. [Google Scholar] [CrossRef]
- Akinc, A.; Maier, M.A.; Manoharan, M.; Fitzgerald, K.; Jayaraman, M.; Barros, S.; Ansell, S.; Du, X.; Hope, M.J.; Madden, T.D.; et al. The Onpattro Story and the Clinical Translation of Nanomedicines Containing Nucleic Acid-Based Drugs. Nat. Nanotechnol. 2019, 14, 1084–1087. [Google Scholar] [CrossRef] [PubMed]
- Yonezawa, S.; Koide, H.; Asai, T. Recent Advances in SiRNA Delivery Mediated by Lipid-Based Nanoparticles. Adv. Drug Deliv. Rev. 2020, 154–155, 64–78. [Google Scholar] [CrossRef]
- Zhang, M.; Huang, Y. SiRNA Modification and Delivery for Drug Development. Trends Mol. Med. 2022, 28, 892–893. [Google Scholar] [CrossRef]
- Layzer, J.M.; Mccaffrey, A.P.; Tanner, A.K.; Huang, Z.; Kay, M.A.; Sullenger, B.A. In Vivo Activity of Nuclease-Resistant SiRNAs. RNA 2004, 10, 766–771. [Google Scholar] [CrossRef]
- Arshad, R.; Fatima, I.; Sargazi, S.; Rahdar, A.; Karamzadeh-Jahromi, M.; Pandey, S.; Díez-Pascual, A.M.; Bilal, M. Novel Perspectives towards Rna-Based Nano-Theranostic Approaches for Cancer Management. Nanomaterials 2021, 11, 3330. [Google Scholar] [CrossRef]
- Zhao, W.; Hou, X.; Vick, O.G.; Dong, Y. RNA Delivery Biomaterials for the Treatment of Genetic and Rare Diseases. Biomaterials 2019, 217, 119291. [Google Scholar] [CrossRef] [PubMed]
- Auriemma, G.; Russo, P.; Del Gaudio, P.; García-González, C.A.; Landín, M.; Aquino, R.P. Technologies and Formulation Design of Polysaccharide-Based Hydrogels for Drug Delivery. Molecules 2020, 25, 3156. [Google Scholar] [CrossRef] [PubMed]
- Barclay, T.G.; Day, C.M.; Petrovsky, N.; Garg, S. Review of Polysaccharide Particle-Based Functional Drug Delivery. Carbohydr. Polym. 2019, 221, 94–112. [Google Scholar] [CrossRef]
- Zhang, M.; Ma, H.; Wang, X.; Yu, B.; Cong, H.; Shen, Y. Polysaccharide-Based Nanocarriers for Efficient Transvascular Drug Delivery. J. Control. Release 2023, 354, 167–187. [Google Scholar] [CrossRef] [PubMed]
- Dattilo, M.; Patitucci, F.; Prete, S.; Parisi, O.I.; Puoci, F. Polysaccharide-Based Hydrogels and Their Application as Drug Delivery Systems in Cancer Treatment: A Review. J. Funct. Biomater. 2023, 14, 55. [Google Scholar] [CrossRef]
- Jurak, M.; Wiącek, A.E.; Ładniak, A.; Przykaza, K.; Szafran, K. What Affects the Biocompatibility of Polymers? Adv. Colloid Interface Sci. 2021, 294, 102451. [Google Scholar] [CrossRef]
- Naveed, M.; Phil, L.; Sohail, M.; Hasnat, M.; Baig, M.M.F.A.; Ihsan, A.U.; Shumzaid, M.; Kakar, M.U.; Mehmood Khan, T.; Akabar, M.D.; et al. Chitosan Oligosaccharide (COS): An Overview. Int. J. Biol. Macromol. 2019, 129, 827–843. [Google Scholar] [CrossRef]
- Li, S.; Xiong, Q.; Lai, X.; Li, X.; Wan, M.; Zhang, J.; Yan, Y.; Cao, M.; Lu, L.; Guan, J.; et al. Molecular Modification of Polysaccharides and Resulting Bioactivities. Compr. Rev. Food Sci. Food Saf. 2016, 15, 237–250. [Google Scholar] [CrossRef]
- Wiącek, A.E.; Dul, K. Effect of Surface Modification on Starch/Phospholipid Wettability. Colloids Surf. A Physicochem. Eng. Asp. 2015, 480, 351–359. [Google Scholar] [CrossRef]
- Prasher, P.; Sharma, M.; Mehta, M.; Satija, S.; Aljabali, A.A.; Tambuwala, M.M.; Anand, K.; Sharma, N.; Dureja, H.; Jha, N.K.; et al. Current-Status and Applications of Polysaccharides in Drug Delivery Systems. Colloid Interface Sci. Commun. 2021, 42, 100418. [Google Scholar] [CrossRef]
- Engelberth, S.A.; Hempel, N.; Bergkvist, M. Chemically Modified Dendritic Starch: A Novel Nanomaterial for SiRNA Delivery. Bioconjug Chem. 2015, 26, 1766–1774. [Google Scholar] [CrossRef] [PubMed]
- Engelberth, S.A.; Hempel, N.; Bergkvist, M. Cationic Dendritic Starch as a Vehicle for Photodynamic Therapy and SiRNA Co-Delivery. J. Photochem. Photobiol. B 2017, 168, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Amar-Lewis, E.; Azagury, A.; Chintakunta, R.; Goldbart, R.; Traitel, T.; Prestwood, J.; Landesman-Milo, D.; Peer, D.; Kost, J. Quaternized Starch-Based Carrier for SiRNA Delivery: From Cellular Uptake to Gene Silencing. J. Control. Release 2014, 185, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Blitsman, Y.; Benafsha, C.; Yarza, N.; Zorea, J.; Goldbart, R.; Traitel, T.; Elkabets, M.; Kost, J. Cargo-Dependent Targeted Cellular Uptake Using Quaternized Starch as a Carrier. Nanomaterials 2023, 13, 1988. [Google Scholar] [CrossRef]
- Sieradzki, R.; Traitel, T.; Goldbart, R.; Geresh, S.; Kost, J. Tailoring Quaternized Starch as a Non-viral Carrier for Gene Delivery Applications. Polym. Adv. Technol. 2014, 25, 552–561. [Google Scholar] [CrossRef]
- Lifshiz Zimon, R.; Lerman, G.; Elharrar, E.; Meningher, T.; Barzilai, A.; Masalha, M.; Chintakunta, R.; Hollander, E.; Goldbart, R.; Traitel, T.; et al. Ultrasound Targeting of Q-Starch/MiR-197 Complexes for Topical Treatment of Psoriasis. J. Control. Release 2018, 284, 103–111. [Google Scholar] [CrossRef]
- Faggio, C.; Morabito, M.; Minicante, S.A.; Lo Piano, G.; Pagano, M.; Genovese, G. Potential Use of Polysaccharides from the Brown Alga Undaria Pinnatifida as Anticoagulants. Braz. Arch. Biol. Technol. 2015, 58, 798–804. [Google Scholar] [CrossRef]
- Zheng, Y.; Bai, L.; Zhou, Y.; Tong, R.; Zeng, M.; Li, X.; Shi, J. Polysaccharides from Chinese Herbal Medicine for Anti-Diabetes Recent Advances. Int. J. Biol. Macromol. 2019, 121, 1240–1253. [Google Scholar] [CrossRef]
- Milo, R.; Phillips, R. Cell Biology by the Numbers; Garland Science: New York, NY, USA, 2015; ISBN 9781317230694. [Google Scholar]
- Melkikh, A.V.; Sutormina, M.I. Model of Active Transport of Ions in Cardiac Cell. J. Theor. Biol. 2008, 252, 247–254. [Google Scholar] [CrossRef]
- Terry, J. The Major Electrolytes: Sodium, Potassium, and Chloride. J. Intraven. Nurs. 1994, 17, 240–247. [Google Scholar]
- Wei, W.-H.; Dong, X.-M.; Liu, C.-G. In Vitro Investigation of Self-Assembled Nanoparticles Based on Hyaluronic Acid-Deoxycholic Acid Conjugates for Controlled Release Doxorubicin: Effect of Degree of Substitution of Deoxycholic Acid. Int. J. Mol. Sci. 2015, 16, 7195–7209. [Google Scholar] [CrossRef] [PubMed]
- Al-Absi, M.Y.; Caprifico, A.E.; Calabrese, G. Chitosan and Its Structural Modifications for SiRNA Delivery. Adv. Pharm. Bull. 2023, 13, 275–282. [Google Scholar] [CrossRef]
- Alameh, M.; Lavertu, M.; Tran-Khanh, N.; Chang, C.-Y.; Lesage, F.; Bail, M.; Darras, V.; Chevrier, A.; Buschmann, M.D. SiRNA Delivery with Chitosan: Influence of Chitosan Molecular Weight, Degree of Deacetylation, and Amine to Phosphate Ratio on in Vitro Silencing Efficiency, Hemocompatibility, Biodistribution, and in Vivo Efficacy. Biomacromolecules 2018, 19, 112–131. [Google Scholar] [CrossRef]
- Cao, Y.; Tan, Y.F.; Wong, Y.S.; Liew, M.W.J.; Venkatraman, S. Recent Advances in Chitosan-Based Carriers for Gene Delivery. Mar. Drugs 2019, 17, 381. [Google Scholar] [CrossRef]
- Techaarpornkul, S.; Wongkupasert, S.; Opanasopit, P.; Apirakaramwong, A.; Nunthanid, J.; Ruktanonchai, U. Chitosan-Mediated SiRNA Delivery In Vitro: Effect of Polymer Molecular Weight, Concentration and Salt Forms. AAPS PharmSciTech 2010, 11, 64–72. [Google Scholar] [CrossRef]
- Ewe, A.; Noske, S.; Karimov, M.; Aigner, A. Polymeric Nanoparticles Based on Tyrosine-Modified, Low Molecular Weight Polyethylenimines for SiRNA Delivery. Pharmaceutics 2019, 11, 600. [Google Scholar] [CrossRef]
- Jin, Y.; Adams, F.; Möller, J.; Isert, L.; Zimmermann, C.M.; Keul, D.; Merkel, O.M. Synthesis and Application of Low Molecular Weight PEI-Based Copolymers for SiRNA Delivery with Smart Polymer Blends. Macromol. Biosci. 2023, 23, 2200409. [Google Scholar] [CrossRef]
- Scopes, R.K. Enzyme Activity and Assays. In Encyclopedia of Life Sciences; Wiley: Hoboken, NJ, USA, 2002. [Google Scholar]
- Kwon, D.; Lee, S.H.; Kim, J.; Yoon, T.H. Dispersion, Fractionation and Characterization of Sub-100nm TiO2 Nanoparticles in Aqueous Media. Toxicol. Environ. Health Sci. 2010, 2, 78–85. [Google Scholar] [CrossRef]
- Kalyankrishna, S.; Grandis, J.R. Epidermal Growth Factor Receptor Biology in Head and Neck Cancer. J. Clin. Oncol. 2006, 24, 2666–2672. [Google Scholar] [CrossRef]
- Fouissac, E.; Milas, M.; Rinaudo, M. Shear-Rate, Concentration, Molecular Weight, and Temperature Viscosity Dependences of Hyaluronate, a Wormlike Polyelectrolyte. Macromolecules 1993, 26, 6945–6951. [Google Scholar] [CrossRef]
- Kwaambwa, H.M.; Goodwin, J.W.; Hughes, R.W.; Reynolds, P.A. Viscosity, Molecular Weight and Concentration Relationships at 298K of Low Molecular Weight Cis-Polyisoprene in a Good Solvent. Colloids Surf. A Physicochem. Eng. Asp. 2007, 294, 14–19. [Google Scholar] [CrossRef]
- Fernandes, J.C.; Qiu, X.; Winnik, F.M.; Benderdour, M.; Zhang, X.; Dai, K.; Shi, Q. Low Molecular Weight Chitosan Conjugated with Folate for SiRNA Delivery in Vitro: Optimization Studies. Int. J. Nanomed. 2012, 7, 5833–5845. [Google Scholar] [CrossRef]
- Liu, X.; Howard, K.A.; Dong, M.; Andersen, M.Ø.; Rahbek, U.L.; Johnsen, M.G.; Hansen, O.C.; Besenbacher, F.; Kjems, J. The Influence of Polymeric Properties on Chitosan/SiRNA Nanoparticle Formulation and Gene Silencing. Biomaterials 2007, 28, 1280–1288. [Google Scholar] [CrossRef] [PubMed]
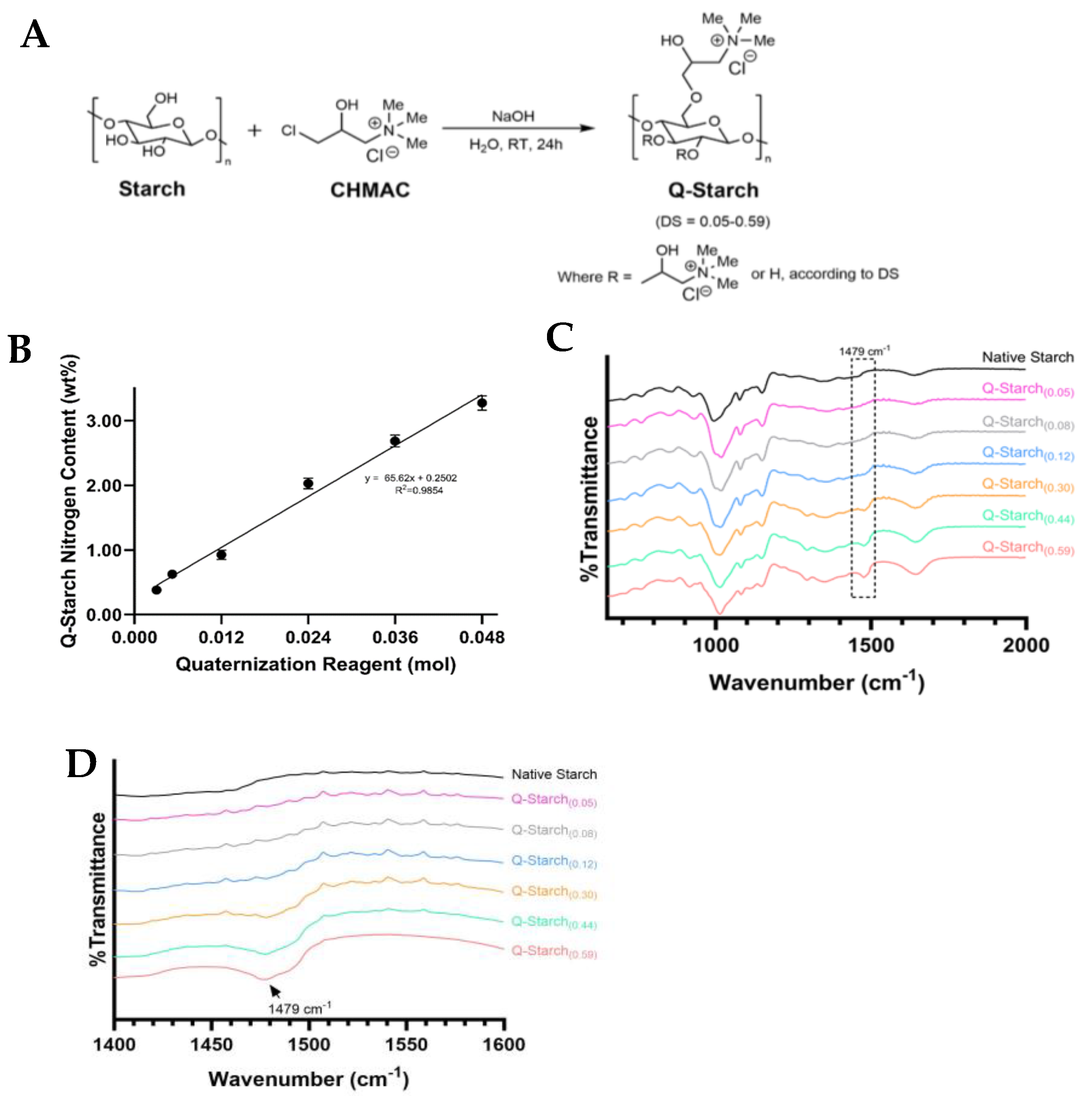

| Q-Starch Average Nitrogen Content (wt. %) | DS * | (DS)−1 |
|---|---|---|
| 0.38 ± 0.00 | 0.05 | 21.71 |
| 0.63 ± 0.00 | 0.08 | 12.87 |
| 0.93 ± 0.07 | 0.12 | 8.39 |
| 2.03 ± 0.08 | 0.30 | 3.32 |
| 2.68 ± 0.09 | 0.44 | 2.29 |
| 3.3 ± 0.1 | 0.59 | 1.70 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Regev, A.; Benafsha, C.; Goldbart, R.; Traitel, T.; Elkabets, M.; Kost, J. Effect of Degree of Substitution and Molecular Weight on Transfection Efficacy of Starch-Based siRNA Delivery System. Polysaccharides 2024, 5, 580-597. https://doi.org/10.3390/polysaccharides5040037
Regev A, Benafsha C, Goldbart R, Traitel T, Elkabets M, Kost J. Effect of Degree of Substitution and Molecular Weight on Transfection Efficacy of Starch-Based siRNA Delivery System. Polysaccharides. 2024; 5(4):580-597. https://doi.org/10.3390/polysaccharides5040037
Chicago/Turabian StyleRegev, Amir, Chen Benafsha, Riki Goldbart, Tamar Traitel, Moshe Elkabets, and Joseph Kost. 2024. "Effect of Degree of Substitution and Molecular Weight on Transfection Efficacy of Starch-Based siRNA Delivery System" Polysaccharides 5, no. 4: 580-597. https://doi.org/10.3390/polysaccharides5040037
APA StyleRegev, A., Benafsha, C., Goldbart, R., Traitel, T., Elkabets, M., & Kost, J. (2024). Effect of Degree of Substitution and Molecular Weight on Transfection Efficacy of Starch-Based siRNA Delivery System. Polysaccharides, 5(4), 580-597. https://doi.org/10.3390/polysaccharides5040037







