Effects of SARS-CoV-2 Spike S1 Subunit on the Interplay Between Hepatitis B and Hepatocellular Carcinoma Related Molecular Processes in Human Liver
Abstract
1. Introduction
2. Materials and Methods
2.1. BioGRID
2.2. STRING
2.3. Protein Enrichment
2.4. Cytoscape and Network Topology Analysis
2.5. Highlighting the Nodes of a STRING Network Involved in the Same Biological Process (GO)
2.6. Enrichment Analysis
2.7. Data Merging Process
3. Results
3.1. Starting Conditions
3.1.1. Interactome-12
3.1.2. Main Features of the Interactome-12
3.1.3. Analysis of KEGG Terms hsa05161-Hepatitis B and hsa05225-Hepatocellular Carcinoma
- Common Pathways to Divergent Outcomes: HBV and HCC may share early molecular triggers, particularly related to inflammation, immune evasion, or cell survival [42]. However, HCC would require additional oncogenic events (mutations, dysregulated signaling) that go beyond the viral impact, resulting in its independent progression;
- Staged Evolution of Disease: It is possible that HBV creates a favorable environment for HCC development [43,44], with S1 inducing early changes that lead to hepatitis but also laying the groundwork for carcinogenesis in susceptible cells. The shared genes might represent pathways involved in liver damage, inflammation, and immune signaling that predispose cells to oncogenic transformation;
- Independent Evolution of Overlapping Pathways: Though HBV and HCC share pathways, they may develop independently once started [45,46]. HBV may follow a chronic inflammatory or immune-evasion route, while HCC could progress through mutations and other cancer-related alterations despite the initial similarity in gene expression patterns.
3.2. Comparisons Between Enrichment Analysis Terms
3.3. Analysis of the Cell Death Present in the Interactomes Examined
3.4. Data Merging
3.5. Genes That Control Cell Death in the Liver
4. Discussion
5. Conclusions: Integrating These Mechanisms
Supplementary Materials
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Colonna, G. Understanding the SARS-CoV-2–Human Liver Interactome Using a Comprehensive Analysis of the Individual Virus–Host Interactions. Livers 2024, 4, 209–239. [Google Scholar] [CrossRef]
- Trypsteen, W.; Van Cleemput, J.; Snippenberg, W.V.; Gerlo, S.; Vandekerckhove, L. On the whereabouts of SARS-CoV-2 in the human body: A systematic review. PLoS Pathog. 2020, 16, e1009037. [Google Scholar] [CrossRef]
- Letarov, A.V.; Babenko, V.V.; Kulikov, E.E. Free SARS-CoV-2 spike protein S1 particles may play a role in the pathogenesis of COVID-19 infection. Biochemistry 2021, 86, 257–261. [Google Scholar] [CrossRef]
- Kopańska, M.; Barnaś, E.; Błajda, J.; Kuduk, B.; Łagowska, A.; Banaś-Ząbczyk, A. Effects of SARS-CoV-2 inflammation on selected organ systems of the human body. Int. J. Mol. Sci. 2022, 23, 4178. [Google Scholar] [CrossRef] [PubMed]
- Iyer, A.S.; Jones, F.K.; Nodoushani, A.; Kelly, M.; Becker, M.; Slater, D.; Mills, R.; Teng, E.; Kamruzzaman, M.; Charles, R.C. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci. Immunol. 2020, 5, eabe0367. [Google Scholar] [CrossRef] [PubMed]
- Frank, M.G.; Ball, J.B.; Hopkins, S.; Kelley, T.; Kuzma, A.J.; Thompson, R.S.; Fleshner, M.; Maier, S.F. SARS-CoV-2 S1 subunit produces a protracted priming of the neuroinflammatory, physiological, and behavioral responses to a remote immune challenge: A role for corticosteroids. Brain Behav. Immun. 2024, 121, 87–103. [Google Scholar] [CrossRef]
- Al-Aly, Z.; Davis, H.; McCorkell, L.; Soares, L.; Wulf-Hanson, S.; Iwasaki, A.; Topol, E.J. Long COVID science, research and policy. Nat. Med. 2024, 30, 2148–2164. [Google Scholar] [CrossRef]
- Hallak, J.; Caldini, E.G.; Teixeira, T.A.; Mendes Correa, M.; Duarte-Neto, A.N.; Zambrano, F.; Taubert, A.; Hermosilla, C.; Drevet, J.; Dolhnikoff, M.; et al. Trasmission electron microscopy reveals the presence of SARS-CoV-2 in human spermatozoa associated with an ETosis-like response. Andrology 2024, 12, 1799–1807. [Google Scholar] [CrossRef]
- Madjunkov, M.; Dviri, M.; Librach, C. A comprehensive review of the impact of COVID-19 on human reproductive biology, assisted reproduction care and pregnancy: A Canadian perspective. J. Ovarian Res. 2020, 13, 140. [Google Scholar] [CrossRef]
- Mansueto, G.; Fusco, G.; Colonna, G. A Tiny Viral Protein, SARS-CoV-2-ORF7b: Functional Molecular Mechanisms. Biomolecules 2024, 14, 541. [Google Scholar] [CrossRef]
- Colonna, G. Interactomic Analyses, and a Reverse Engineering Study Identify Specific Functional Activities of One-to-One Interactions of the S1 Subunit of the SARS-CoV-2 Spike Protein with the Human Proteome. Biomolecules 2024, 14, 1549. [Google Scholar] [CrossRef]
- Cosentino, M.; Marino, F. The spike hypothesis in vaccine-induced adverse effects: Questions and answers. Trends Mol. Med. 2022, 28, 797–799. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Lin, C.; Zhang, J.; Khaing Oo, M.K.; Fan, X. Rapid and quantitative detection of COVID-19 markers in micro-liter sized samples. BioRxiv 2020. [Google Scholar] [CrossRef]
- Bošnjak, B.; Stein, S.C.; Willenzon, S.; Cordes, A.K.; Puppe, W.; Bernhardt, G.; Ravens, I.; Ritter, C.; Schultze-Florey, C.; Godecke, N.; et al. Low serum neutralizing anti-SARS-CoV-2 S antibody levels in mildly affected COVID-19 convalescent patients revealed by two different detection methods. Cell. Mol. Immunol. 2021, 18, 936–944. [Google Scholar] [CrossRef]
- Yonker, L.M.; Swank, Z.; Bartsch, Y.C.; Burns, M.D.; Kane, A.; Boribong, B.P.; Davis, J.P.; Loiselle, M.; Novak, T.; Senussi, Y.; et al. Circulating spike protein detected in post–COVID-19 mRNA vaccine myocarditis. Circulation 2023, 147, 867–876. [Google Scholar] [CrossRef]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2020, 49, D605–D612, Erratum in Nucleic Acids Res. 2021, 49, 10800. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2022, 51, D638–D646. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2018, 18, 623–632. [Google Scholar] [CrossRef]
- Chung, F.; Lu, L.; Dewey, T.G.; Galas, D.J. Duplication Models for Biological Networks. J. Comput. Biol. 2003, 10, 677–687. [Google Scholar] [CrossRef]
- Barabási, A.-L. Network Science, 1st ed.; Cambridge University Press: Cambridge, UK, 2016; ISBN 9781107076266. [Google Scholar]
- Arakawa, K.; Tomita, M. Merging Multiple Omics Datasets In Silico: Statistical Analyses and Data Interpretation. In Systems Metabolic Engineering; Alper, H., Ed.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2013; Volume 985. [Google Scholar] [CrossRef]
- Glazier, D.S. Metabolic scaling in complex living systems. Systems 2014, 2, 451–540. [Google Scholar] [CrossRef]
- De Las Rivas, J.; Fontanillo, C. Protein-protein interactions essentials: Key concepts to building and analyzing interactome networks. PLoS Comput. Biol. 2010, 6, e1000807. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Grassmann, G.; Miotto, M.; Desantis, F.; Di Rienzo, L.; Tartaglia, G.G.; Pastore, A.; Ruocco, G.; Monti, M.; Milanetti, E. Computational Approaches to Predict Protein–Protein Interactions in Crowded Cellular Environments. Chem. Rev. 2024, 124, 3932–3977. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Wallmeroth, N.; Berendzen, K.W.; Grefen, C. Techniques for the Analysis of Protein-Protein Interactions in Vivo. Plant Physiol. 2016, 171, 727–758. [Google Scholar] [CrossRef] [PubMed]
- Lite, T.V.; Grant, R.A.; Nocedal, I.; Littlehale, M.L.; Guo, M.S.; Laub, M.T. Uncovering the basis of protein-protein interaction specificity with a combinatorially complete library. Elife 2020, 9, e60924. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, D.; Lü, L.; Shang, M.S.; Zhang, Y.C.; Zhou, T. Identifying influential nodes in complex networks. Phys. A Stat. Mech. Its Appl. 2012, 391, 1777–1787. [Google Scholar] [CrossRef]
- Barabási, A.L. Network science. Philosophical Transactions of the Royal Society A: Mathematical. Phys. Eng. Sci. 2024, 371, 20120375. [Google Scholar] [CrossRef]
- Guthrie, C.R.; Skâlhegg, B.S.; McKnight, G.S. Two novel brain-specific splice variants of the murine beta gene of cAMP-dependent protein kinase. J. Biol. Chem. 1997, 272, 29560–29565. [Google Scholar] [CrossRef] [PubMed]
- Alqahtani, S.A.; Buti, M. COVID-19 and hepatitis B infection. Antivir. Ther. 2020, 25, 389–397. [Google Scholar] [CrossRef]
- Song, C.I.; Lv, J.; Liu, Y.; Chen, J.G.; Ge, Z.; Zhu, J.; Dai, J.; Du, L.B.; Yu, C.; Guo, Y.; et al. Associations between hepatitis B virus infection and risk of all cancer types. JAMA Netw. Open 2019, 2, e195718. [Google Scholar] [CrossRef]
- Li, Y.; Li, C.; Wang, J.; Zhu, C.; Zhu, L.; Ji, F.; Liu, L.; Xu, T.; Zhang, B.; Xue, L.; et al. A case series of COVID-19 patients with chronic hepatitis B virus infection. J. Med. Virol. 2020, 92, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Li, X.; Wan, T. Effects of hepatitis B virus infection on patients with COVID-19: A meta-analysis. Dig. Dis. Sci. 2023, 68, 1615–1631. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.; Yu, J.; Cheon, M.; Tak, S. Evaluation of the acute hepatitis B surveillance system in the Republic of Korea following the transition to mandatory surveillance. Osong Public Health Res. Perspect. 2024, 15, 353. [Google Scholar] [CrossRef] [PubMed]
- Essam, S.; Hassany, M.; Maged, A.; Mannaa, M.; Sayed, A.; Essam, S.; Magdy, M. Assessment of Hepatocellular Carcinoma Patients Infected with Covid-19 Infection during the Pandemic. Int. J. Chem. Biochem. Sci. (IJCBS) 2024, 25, 1065–1069. [Google Scholar] [CrossRef]
- Nasir, N.; Khanum, I.; Habib, K.; Wagley, A.; Arshad, A.; Majeed, A. Insight into COVID-19 associated liver injury: Mechanisms, evaluation, and clinical implications. Hepatol. Forum 2024, 5, 139. [Google Scholar] [CrossRef]
- Mihai, N.; Olariu, M.C.; Ganea, O.A.; Adamescu, A.I.; Molagic, V.; Aramă, Ș.S.; Tilișcan, C.; Aramă, V. Risk of Hepatitis B Virus Reactivation in COVID-19 Patients Receiving Immunosuppressive Treatment: A Prospective Study. J. Clin. Med. 2024, 13, 6032. [Google Scholar] [CrossRef]
- Chang, H.C.; Su, T.H.; Huang, Y.T.; Hong, C.M.; Sheng, W.H.; Hsueh, P.R.; Kao, J.H. Liver dysfunction and clinical outcomes of unvaccinated COVID-19 patients with and without chronic hepatitis B. J. Microbiol. Immunol. Infect. 2024, 57, 55–63. [Google Scholar] [CrossRef]
- Mushtaq, M.; Colletier, K.; Moghe, A. Hepatitis B Reactivation and Liver Failure Because of COVID-19 Infection. ACG Case Rep. J. 2024, 11, e01397. [Google Scholar] [CrossRef]
- Wu, H.Y.; Su, T.H.; Liu, C.J.; Yang, H.C.; Tsai, J.H.; Wei, M.H.; Chen, C.C.; Tung, C.C.; Kao, J.H.; Chen, P.J. Hepatitis B reactivation: A possible cause of coronavirus disease 2019 vaccine induced hepatitis. J. Formos. Med. Assoc. 2024, 123, 88–97. [Google Scholar] [CrossRef]
- D’souza, S.; Lau, K.C.; Coffin, C.S.; Patel, T.R. Molecular mechanisms of viral hepatitis induced hepatocellular carcinoma. World J. Gastroenterol. 2020, 26, 5759. [Google Scholar] [CrossRef]
- Pollicino, T.; Saitta, C.; Raimondo, G. Hepatocellular carcinoma: The point of view of the hepatitis B virus. Carcinogenesis 2011, 32, 1122–1132. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.B.; Wu, J.F.; Du, Y.; Cao, G.W. Cancer Evolution–Development: Experience of hepatitis B virus–induced hepatocarcinogenesis. Curr. Oncol. 2016, 23, e49–e56. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guerrero, R.B.; Roberts, L.R. The role of hepatitis B virus integrations in the pathogenesis of human hepatocellular carcinoma. J. Hepatol. 2005, 42, 760–777. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y. Hepatitis B virus-associated hepatocellular carcinoma. In Infectious Agents Associated Cancers: Epidemiology and Molecular Biology; Cai, Q., Yuan, Z., Lan, K., Eds.; Advances in Experimental Medicine and Biology; Springer: Singapore, 2017; Volume 1018. [Google Scholar] [CrossRef]
- Ham, S.W.; Jeon, H.Y.; Jin, X.; Kim, E.J.; Kim, J.K.; Shin, Y.J.; Lee, Y.; Kim, S.H.; Lee, S.Y.; Seo, S.; et al. TP53 gain-of-function mutation promotes inflammation in glioblastoma. Cell Death Differ. 2019, 26, 409–425. [Google Scholar] [CrossRef]
- Zhou, P.; Lu, S.; Luo, Y.; Wang, S.; Yang, K.; Zhai, Y.; Sun, G.; Sun, X. Attenuation of TNF-α-induced inflammatory injury in endothelial cells by ginsenoside Rb1 via inhibiting NF-κB, JNK and p38 signaling pathways. Front. Pharmacol. 2017, 8, 464. [Google Scholar] [CrossRef]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef]
- Huebner, K. The Role of the Activating Transcription Factor 2 (ATF2) in Colorectal Carcinogenesis. Doctoral Thesis, Friedrich-Alexander-Universitaet Erlangen-Nuernberg, Erlangen, Germany, 2023. Available online: https://nbn-resolving.org/urn:nbn:de:bvb:29-opus4-167132 (accessed on 15 October 2024).
- Ji, L.; Li, T.; Chen, H.; Yang, Y.; Lu, E.; Liu, J.; Qiao, W.; Chen, H. The crucial regulatory role of type I interferon in inflammatory diseases. Cell Biosci. 2023, 13, 230. [Google Scholar] [CrossRef]
- Moysidou, C.M.; Barberio, C.; Owens, R.M. Advances in engineering human tissue models. Front. Bioeng. Biotechnol. 2021, 8, 620962. [Google Scholar] [CrossRef]
- Tan, Y.J. Hepatitis B virus infection and the risk of hepatocellular carcinoma. World J. Gastroenterol. 2011, 17, 4853–4857. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Arbuthnot, P.; Kew, M. Hepatitis B virus and hepatocellular carcinoma. Int. J. Exp. Pathol. 2001, 82, 77–100. [Google Scholar] [CrossRef]
- Kouroumalis, E.; Tsomidis, I.; Voumvouraki, A. Pathogenesis of Hepatocellular Carcinoma: The Interplay of Apoptosis and Autophagy. Biomedicines 2023, 11, 1166. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- De Re, V.; Rossetto, A.; Rosignoli, A.; Muraro, E.; Racanelli, V.; Tornesello, M.L.; Zompicchiatti, A.; Uzzau, A. Hepatocellular Carcinoma Intrinsic Cell Death Regulates Immune Response and Prognosis. Front. Oncol. 2022, 12, 897703. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Luedde, T.; Kaplowitz, N.; Schwabe, R.F. Cell death and cell death responses in liver disease: Mechanisms and clinical relevance. Gastroenterology 2014, 147, 765–783. [Google Scholar] [CrossRef] [PubMed]
- Imre, G. Cell death signaling in virus infection. Cell. Signal. 2020, 76, 109772. [Google Scholar] [CrossRef]
- Bertheloot, D.; Latz, E.; Franklin, B.S. Necroptosis, pyroptosis and apoptosis: An intricate game of cell death. Cell Mol. Immunol. 2021, 18, 1106–1121. [Google Scholar] [CrossRef]
- Wu, X.; Cao, J.; Wan, X.; Du, S. Programmed cell death in hepatocellular carcinoma: Mechanisms and therapeutic perspectives. Cell Death Discov. 2024, 10, 356. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- García-Pras, E.; Fernández-Iglesias, A.; Gracia-Sancho, J.; Pérez-del-Pulgar, S. Cell death in hepatocellular carcinoma: Pathogenesis and therapeutic opportunities. Cancers 2021, 14, 48. [Google Scholar] [CrossRef]
- Fabregat, I. Dysregulation of apoptosis in hepatocellular carcinoma cells. World J. Gastroenterol. 2009, 15, 513–520. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, G.; Jiang, X.; Torabian, P.; Yang, Z. Investigating autophagy and intricate cellular mechanisms in hepatocellular carcinoma: Emphasis on cell death mechanism crosstalk. Cancer Lett. 2024, 588, 216744. [Google Scholar] [CrossRef]
- Gregory, C.D.; Ford, C.A.; Voss, J.J. Microenvironmental effects of cell death in malignant disease. In Apoptosis in Cancer Pathogenesis and Anti-Cancer Therapy; Gregory, C., Ed.; Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2016; Volume 930. [Google Scholar] [CrossRef]
- Luo, G.; Liu, N. An integrative theory for cancer. Int. J. Mol. Med. 2019, 43, 647–656. [Google Scholar] [CrossRef]
- Osuchowski, M.F.; Winkler, M.S.; Skirecki, T.; Cajander, S.; Shankar-Hari, M.; Lachmann, G.; Monneret, G.; Venet, F.; Bauer, M.; Brunkhorst, F.M.; et al. The COVID-19 puzzle: Deciphering pathophysiology and phenotypes of a new disease entity. Lancet Respir. Med. 2021, 9, 622–642. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Quan, X.B.; Zeng, W.J.; Yang, X.O.; Wang, M.J. Mechanism of hepatocyte apoptosis. J. Cell Death 2016, 9, JCD-S39824. [Google Scholar] [CrossRef] [PubMed]
- Schwabe, R.F.; Luedde, T. Apoptosis and necroptosis in the liver: A matter of life and death. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 738–752. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Luo, Z.; Lin, L.; Sui, X.; Yu, L.; Xu, C.; Zhang, R.; Zhao, Z.; Zhu, Q.; An, B.; et al. Anoikis-Associated Lung Cancer Metastasis: Mechanisms and Therapies. Cancers 2022, 14, 4791. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Adeshakin, F.O.; Adeshakin, A.O.; Afolabi, L.O.; Yan, D.; Zhang, G.; Wan, X. Mechanisms for Modulating Anoikis Resistance in Cancer and the Relevance of Metabolic Reprogramming. Front. Oncol. 2021, 11, 626577. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Paoli, P.; Giannoni, E.; Chiarugi, P. Anoikis molecular pathways and its role in cancer progression. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2013, 1833, 3481–3498. [Google Scholar] [CrossRef]
- Jin, L.; Zuo, X.Y.; Su, W.Y.; Zhao, X.L.; Yuan, M.Q.; Han, L.Z.; Zhao, X.; Chen, Y.D.; Rao, S.Q. Pathway-based analysis tools for complex diseases: A review. Genom. Proteom. Bioinform. 2014, 12, 210–220. [Google Scholar] [CrossRef]
- Brazhnik, P.; De La Fuente, A.; Mendes, P. Gene networks: How to put the function in genomics. TRENDS Biotechnol. 2002, 20, 467–472. [Google Scholar] [CrossRef]
- Hecker, M.; Lambeck, S.; Toepfer, S.; Van Someren, E.; Guthke, R. Gene regulatory network inference: Data integration in dynamic models—A review. Biosystems 2009, 96, 86–103. [Google Scholar] [CrossRef]
- Carpenter, A.E.; Sabatini, D.M. Systematic genome-wide screens of gene function. Nat. Rev. Genet. 2004, 5, 11–22. [Google Scholar] [CrossRef]
- Kelley, R.; Ideker, T. Systematic interpretation of genetic interactions using protein networks. Nat. Biotechnol. 2005, 23, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Kabir, M.H.; Patrick, R.; Ho, J.W.; O’Connor, M.D. Identification of active signaling pathways by integrating gene expression and protein interaction data. BMC Syst. Biol. 2018, 12, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Kuenzi, B.M.; Ideker, T. A census of pathway maps in cancer systems biology. Nat. Rev. Cancer 2020, 20, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, S.; Min, J.; Nigsch, F.; Camargo, M.; Hutz, J.; Cornett, A.; Cleaver, S.; Buckler, A.; Jenkins, J.L. Causal network models for predicting compound targets and driving pathways in cancer. J. Biomol. Screen. 2014, 19, 791–802. [Google Scholar] [CrossRef]
- Buneman, P.; Chapman, A.; Cheney, J. Provenance management in curated databases. In Proceedings of the 2006 ACM SIGMOD International Conference on Management of Data, New York, NY, USA, 27–29 June 2006; pp. 539–550. [Google Scholar] [CrossRef]
- Goudey, B.; Geard, N.; Verspoor, K.; Zobel, J. Propagation, detection and correction of errors using the sequence database network. Brief. Bioinform. 2022, 23, bbac416. [Google Scholar] [CrossRef]
- Azeroual, O. Data wrangling in database systems: Purging of dirty data. Data 2020, 5, 50. [Google Scholar] [CrossRef]
- Li, C.; Liakata, M.; Rebholz-Schuhmann, D. Biological network extraction from scientific literature: State of the art and challenges. Brief. Bioinform. 2014, 15, 856–877. [Google Scholar] [CrossRef]
- Klein, B.; Hoel, E.; Swain, A.; Griebenow, R.; Levin, M. Evolution and emergence: Higher order information structure in protein interactomes across the tree of life. Integr. Biol. 2021, 13, 283–294. [Google Scholar] [CrossRef]
- Wautelet, M. Scaling laws in the macro-, micro-and nanoworlds. Eur. J. Phys. 2001, 22, 601. [Google Scholar] [CrossRef]
- Haken, H.; Haken, H. From the Microscopic to the Macroscopic World. In Information and Self-Organization; Springer Series in Synergetics; Springer: Berlin/Heidelberg, Germany, 1988. [Google Scholar] [CrossRef]
- Bizzarri, M.; Palombo, A.; Cucina, A. Theoretical aspects of systems biology. Prog. Biophys. Mol. Biol. 2013, 112, 33–43. [Google Scholar] [CrossRef]
- Gosak, M.; Markovič, R.; Dolenšek, J.; Rupnik, M.S.; Marhl, M.; Stožer, A.; Perc, M. Network science of biological systems at different scales: A review. Phys. Life Rev. 2018, 24, 118–135. [Google Scholar] [CrossRef] [PubMed]
- Moerner, W.E. A dozen years of single-molecule spectroscopy in physics, chemistry, and biophysics. J. Phys. Chem. B 2002, 106, 910–927. [Google Scholar] [CrossRef]
- Muller, H.J. Variation due to change in the individual gene. Am. Nat. 1922, 56, 32–50. [Google Scholar] [CrossRef]
- El-Hani, C.N. Between the cross and the sword: The crisis of the gene concept. Genet. Mol. Biol. 2007, 30, 297–307. [Google Scholar] [CrossRef]
- Kirschner, M.W. The Meaning of Systems Biology. Cell 2005, 121, 503–504. [Google Scholar] [CrossRef]
- Salehi-Reyhani, A.; Ces, O.; Elani, Y. Artificial cell mimics as simplified models for the study of cell biology. Exp. Biol. Med. 2017, 242, 1309–1317. [Google Scholar] [CrossRef]
- Joyce, A.R.; Palsson, B.Ø. The model organism as a system: Integrating‘omics’ data sets. Nat. Rev. Mol. Cell Biol. 2006, 7, 198–210. [Google Scholar] [CrossRef]
- Hunter, P. The paradox of model organisms: The use of model organisms in research will continue despite their shortcomings. EMBO Rep. 2008, 9, 717–720. [Google Scholar] [CrossRef]
- Bándi, G.; Ramsden, J.J. Emulating biology: The virtual living organism. J. Biol. Phys. Chem. 2011, 11, 97–106, ISSN 1512-0856. [Google Scholar] [CrossRef]
- Hatmal, M.M.M.; Alshaer, W.; Al-Hatamleh, M.A.; Hatmal, M.; Smadi, O.; Taha, M.O.; Oweida, A.J.; Boer, J.; Mohamud, R.; Plebanski, M. Comprehensive structural and molecular comparison of spike proteins of SARS-CoV-2, SARS-CoV and MERS-CoV, and their interactions with ACE2. Cells 2020, 9, 2638. [Google Scholar] [CrossRef]
- Arbuthnot, P.; Capovilla, A.; Kew, M. Putative role of hepatitis B virus X protein in hepatocarcinogenesis: Effects on apoptosis, DNA repair, mitogen-activated protein kinase and JAK/STAT pathways. J. Gastroenterol. Hepatol. 2000, 15, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Moolamalla, S.T.R.; Balasubramanian, R.; Chauhan, R.; Priyakumar, U.D.; Vinod, P.K. Host metabolic reprogramming in response to SARS-CoV-2 infection: A systems biology approach. Microb. Pathog. 2021, 158, 105114. [Google Scholar] [CrossRef] [PubMed]
- Juanola, O.; Martínez-López, S.; Francés, R.; Gómez-Hurtado, I. Non-alcoholic fatty liver disease: Metabolic, genetic, epigenetic and environmental risk factors. Int. J. Environ. Res. Public Health 2021, 18, 5227. [Google Scholar] [CrossRef] [PubMed]
- Hlady, R.A.; Robertson, K.D. Epigenetic memory of environmental exposures as a mediator of liver disease. Hepatology 2024, 80, 451–464. [Google Scholar] [CrossRef]
- Miller, J.L.; Grant, P.A. The role of DNA methylation and histone modifications in transcriptional regulation in humans. In Epigenetics: Development and Disease; Kundu, T., Ed.; Subcellular Biochemistry; Springer: Dordrecht, The Netherlands, 2012; Volume 61. [Google Scholar] [CrossRef]
- Esteller, M.; Herman, J.G. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J. Pathol. J. Pathol. Soc. Great Br. Irel. 2002, 196, 1–7. [Google Scholar] [CrossRef]
- Guerrieri, F.; Belloni, L.; Pediconi, N.; Levrero, M. Molecular mechanisms of HBV-associated hepatocarcinogenesis. In Seminars in Liver Disease; Thieme Medical Publishers: New York, NY, USA, 2013; Volume 33, pp. 147–156. [Google Scholar] [CrossRef]
- Zeisel, M.B.; Guerrieri, F.; Levrero, M. Host epigenetic alterations and hepatitis B virus-associated hepatocellular carcinoma. J. Clin. Med. 2021, 10, 1715. [Google Scholar] [CrossRef]
- Ozyerli-Goknar, E.; Bagci-Onder, T. Epigenetic deregulation of apoptosis in cancers. Cancers 2021, 13, 3210. [Google Scholar] [CrossRef]
- Gao, A.; Zuo, X.; Song, S.; Guo, W.; Tian, L. Epigenetic modification involved in benzene-induced apoptosis through regulating apoptosis-related genes expression. Cell Biol. Int. 2011, 35, 391–396. [Google Scholar] [CrossRef]
- Elpek, G.O. Molecular pathways in viral hepatitis-associated liver carcinogenesis: An update. World J. Clin. Cases 2021, 9, 4890. [Google Scholar] [CrossRef]
- Yang, S.; Pang, L.; Dai, W.; Wu, S.; Ren, T.; Duan, Y.; Zheng, Y.; Bi, S.; Zhang, X.; Kong, J. Role of forkhead box O proteins in hepatocellular carcinoma biology and progression. Front. Oncol. 2021, 11, 667730. [Google Scholar] [CrossRef]
- Gong, Z.; Yu, J.; Yang, S.; Lai, P.B.; Chen, G.G. FOX transcription factor family in hepatocellular carcinoma. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2020, 1874, 188376. [Google Scholar] [CrossRef] [PubMed]
- Sun, E.J.; Wankell, M.; Palamuthusingam, P.; McFarlane, C.; Hebbard, L. Targeting the PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Biomedicines 2021, 9, 1639. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.Y.; Smit, D.J.; Jücker, M. The role of PI3K/AKT/mTOR signaling in hepatocellular carcinoma metabolism. Int. J. Mol. Sci. 2023, 24, 2652. [Google Scholar] [CrossRef] [PubMed]
- Kishor Roy, N.; Bordoloi, D.; Monisha, J.; Padmavathi, G.; Kotoky, J.; Golla, R.; Kunnumakkara, A.B. Specific targeting of Akt kinase isoforms: Taking the precise path for prevention and treatment of cancer. Curr. Drug Targets 2017, 18, 421–435. [Google Scholar] [CrossRef]
- Dong, Y.; Wang, A. Aberrant DNA methylation in hepatocellular carcinoma tumor suppression. Oncol. Lett. 2014, 8, 963–968. [Google Scholar] [CrossRef]
- Rybicka, M.; Verrier, E.R.; Baumert, T.F.; Bielawski, K.P. Polymorphisms within DIO2 and GADD45A genes increase the risk of liver disease progression in chronic hepatitis b carriers. Sci. Rep. 2023, 13, 6124. [Google Scholar] [CrossRef]
- Huebner, K.; Procházka, J.; Monteiro, A.C.; Mahadevan, V.; Schneider-Stock, R. The activating transcription factor 2: An influencer of cancer progression. Mutagenesis 2019, 34, 375–389. [Google Scholar] [CrossRef]
- Rajan, P.K.; Udoh, U.-A.; Sanabria, J.D.; Banerjee, M.; Smith, G.; Schade, M.S.; Sanabria, J.; Sodhi, K.; Pierre, S.; Xie, Z.; et al. The role of histone acetylation-/methylation-mediated apoptotic gene regulation in hepatocellular carcinoma. Int. J. Mol. Sci. 2020, 21, 8894. [Google Scholar] [CrossRef]
- Huang, G.; Chen, J.; Zhou, J.; Xiao, S.; Zeng, W.; Xia, J.; Zeng, X. Epigenetic modification and BRAF gene mutation in thyroid carcinoma. Cancer Cell Int. 2021, 21, 687. [Google Scholar] [CrossRef]
- Kim, H.C.; Choi, K.C.; Choi, H.K.; Kang, H.B.; Kim, M.J.; Lee, Y.H.; Lee, O.H.; Lee, J.; Kim, Y.-J.; Jun, W.; et al. HDAC3 selectively represses CREB3-mediated transcription and migration of metastatic breast cancer cells. Cell. Mol. Life Sci. 2010, 67, 3499–3510. [Google Scholar] [CrossRef]
- Groner, B.; von Manstein, V. Jak Stat signaling and cancer: Opportunities, benefits and side effects of targeted inhibition. Mol. Cell. Endocrinol. 2017, 451, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Masliah-Planchon, J.; Garinet, S.; Pasmant, E. RAS-MAPK pathway epigenetic activation in cancer: miRNAs in action. Oncotarget 2016, 7, 38892. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xie, Z.; Wang, Q.; Hu, S. Coordination of PRKCA/PRKCA-AS1 interplay facilitates DNA methyltransferase 1 recruitment on DNA methylation to affect protein kinase C alpha transcription in mitral valve of rheumatic heart disease. Bioengineered 2021, 12, 5904–5915. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.M.; Nikolic-Paterson, D.J.; Lan, H.Y. TGF-β: The master regulator of fibrosis. Nat. Rev. Nephrol. 2016, 12, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, H.W.; Trautwein, C.; Tacke, F. Functional role of monocytes and macrophages for the inflammatory response in acute liver injury. Front. Physiol. 2012, 3, 56. [Google Scholar] [CrossRef]
- Hildebrand, F.; Pape, H.C.; van Griensven, M.; Meier, S.; Hasenkamp, S.; Krettek, C.; Stuhrmann, M. Genetic predisposition for a compromised immune system after multiple trauma. Shock 2005, 24, 518–522. [Google Scholar] [CrossRef]
- Hazeldine, J.; Lord, J.M. Immunesenescence: A predisposing risk factor for the development of COVID-19? Front. Immunol. 2020, 11, 573662. [Google Scholar] [CrossRef]
- Lee, P.; Chandel, N.S.; Simon, M.C. Cellular adaptation to hypoxia through hypoxia inducible factors and beyond. Nat. Rev. Mol. Cell Biol. 2020, 21, 268–283. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Panda, P.K.; Sinha, N.; Das, D.N.; Bhutia, S.K. Autophagy and apoptosis: Where do they meet? Apoptosis 2014, 19, 555–566. [Google Scholar] [CrossRef]
- Das, S.; Shukla, N.; Singh, S.S.; Kushwaha, S.; Shrivastava, R. Mechanism of interaction between autophagy and apoptosis in cancer. Apoptosis 2021, 26, 512–533. [Google Scholar] [CrossRef]
- Moreira, R.K. Hepatic stellate cells and liver fibrosis. Arch. Pathol. Lab. Med. 2007, 131, 1728–1734. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Alvarez-Dominguez, J.R.; Lodish, H.F. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012, 13, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef] [PubMed]
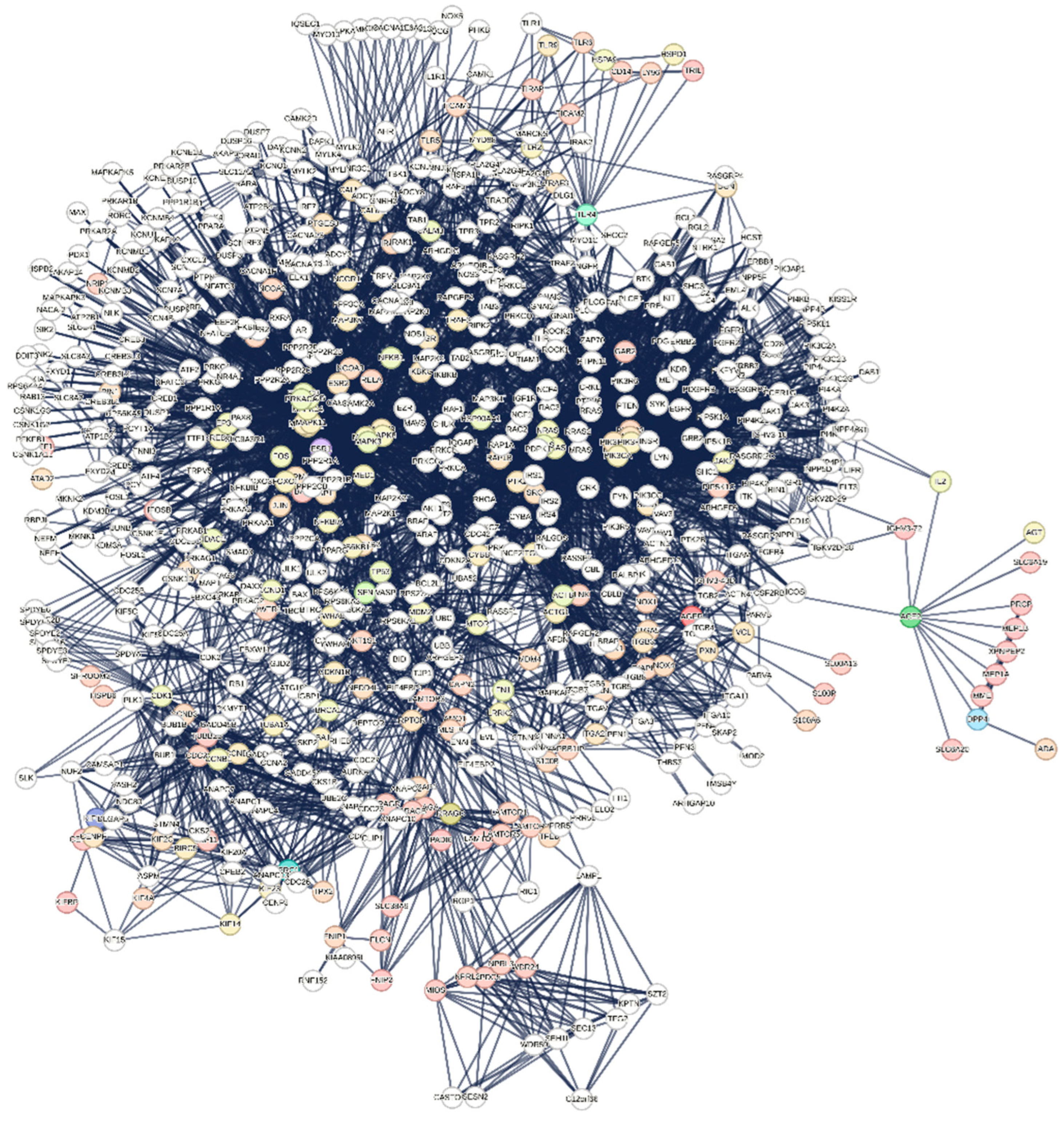
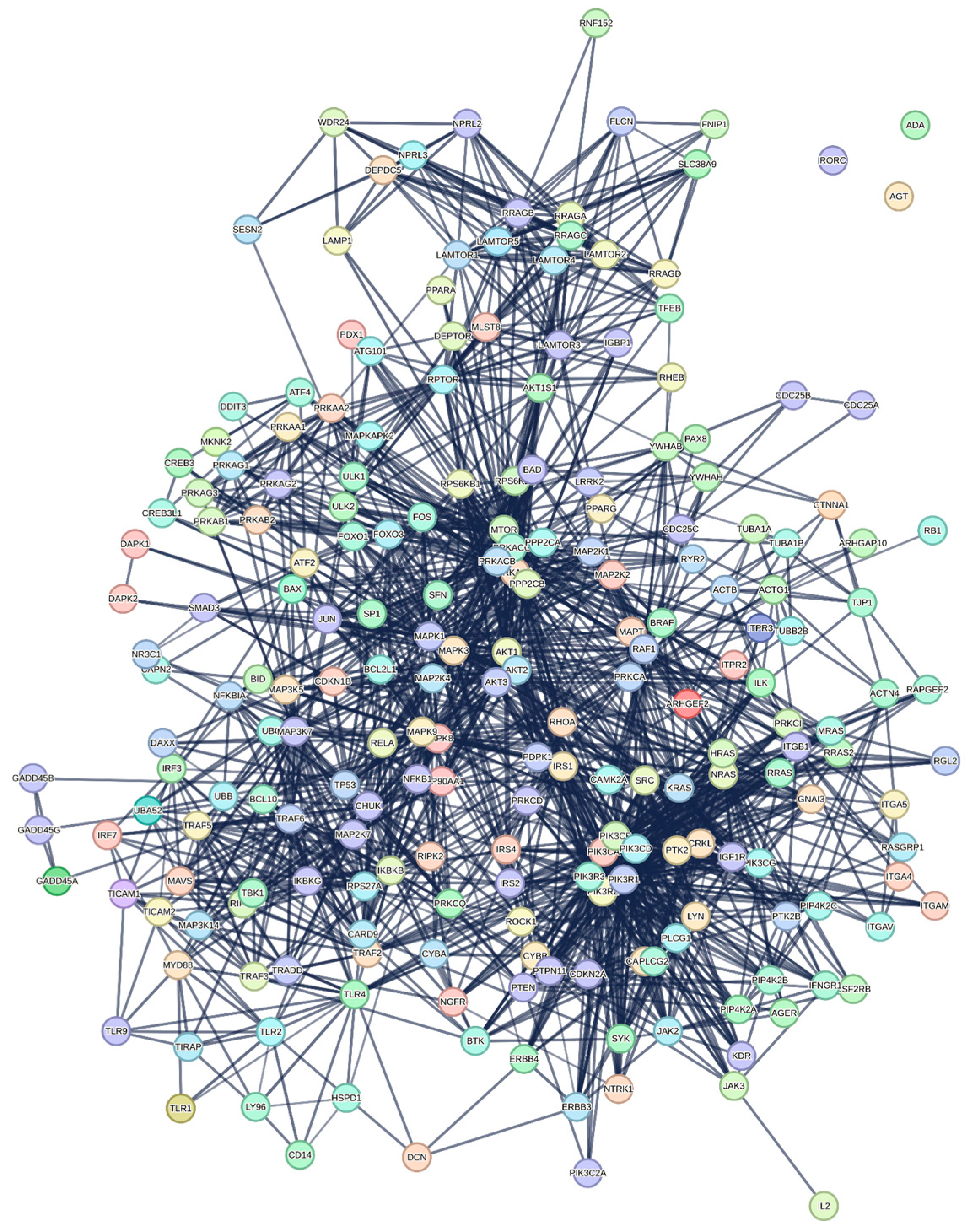
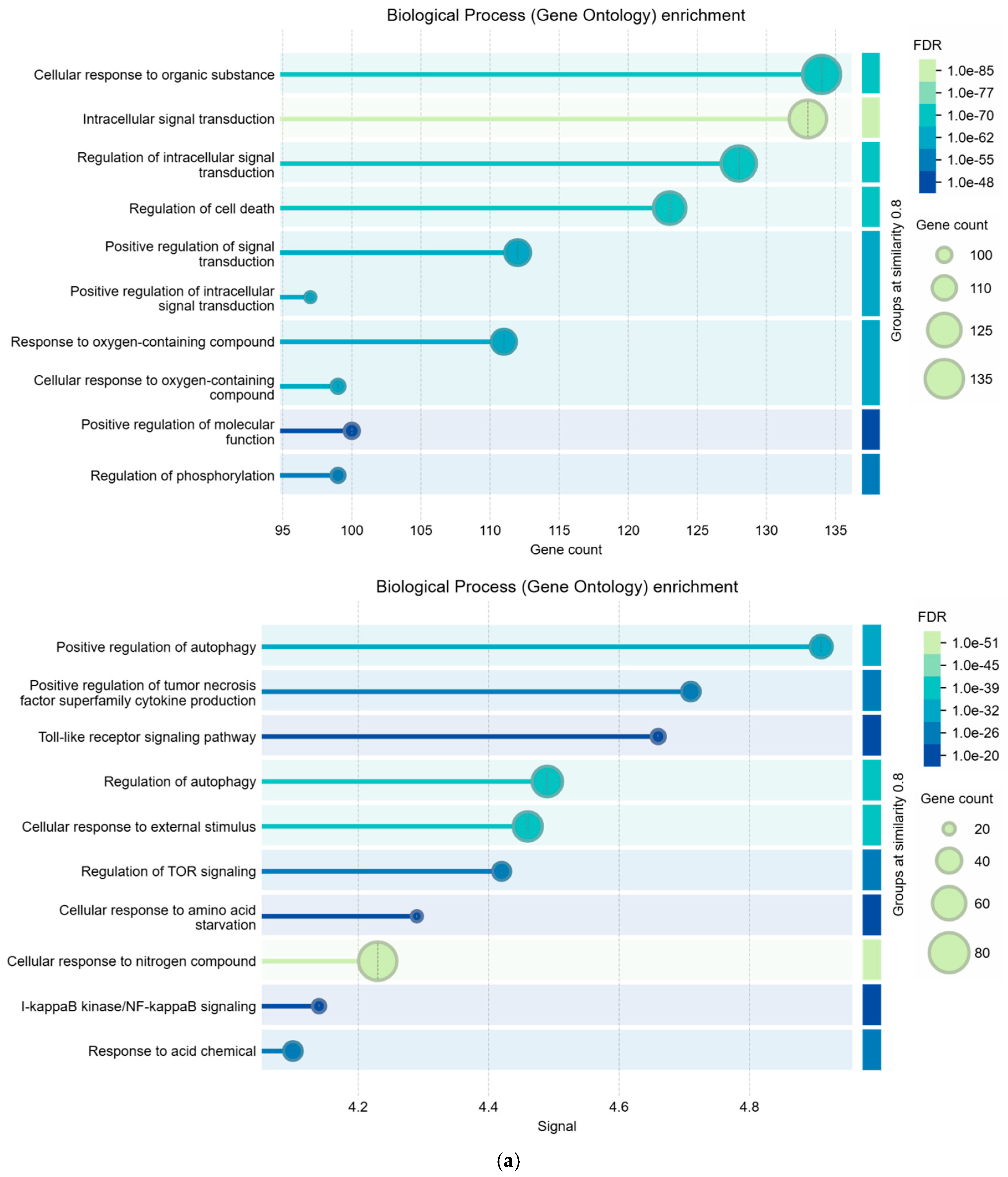
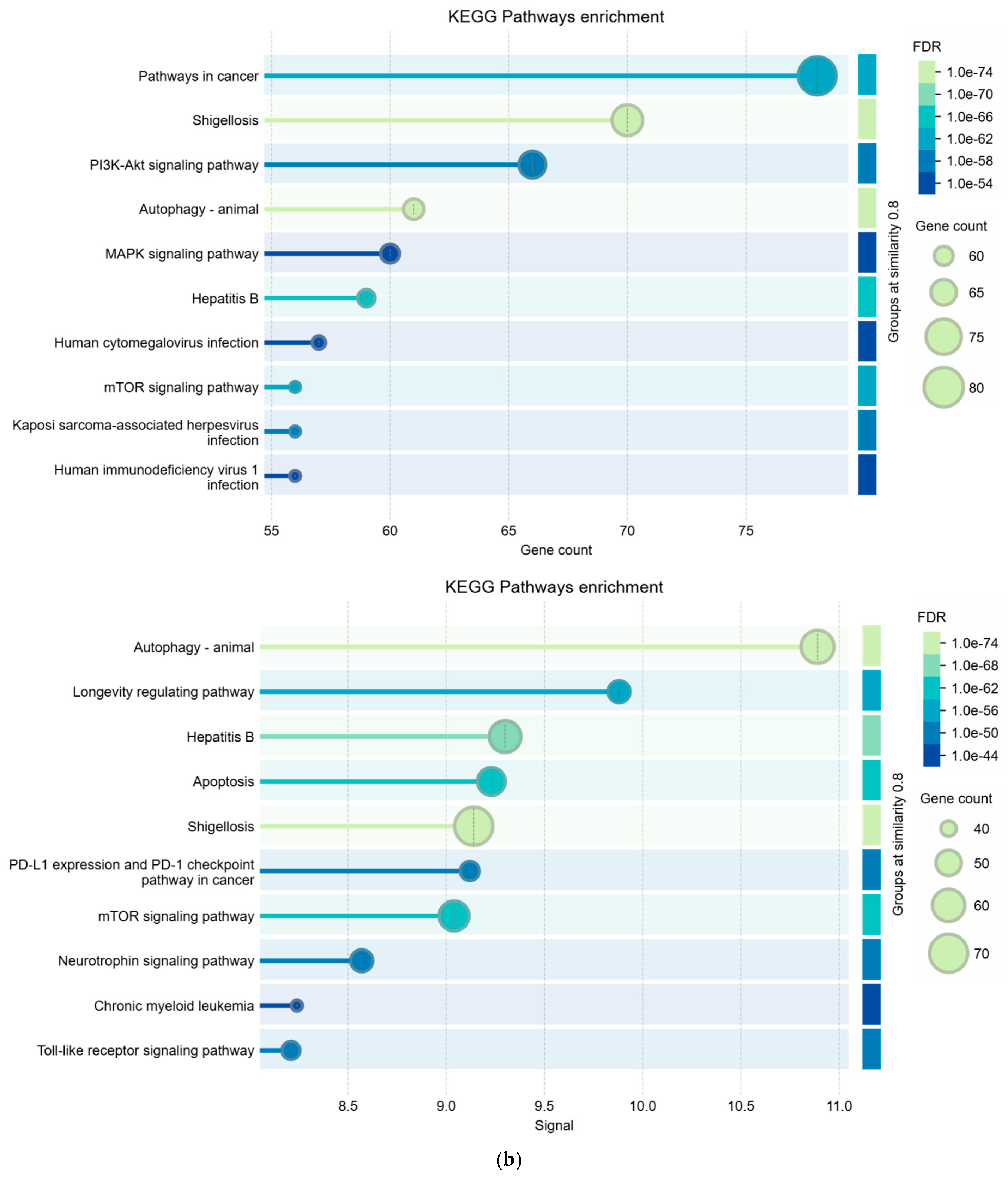

| Biological Process (Gene Ontology) | 2015 GO-terms significantly enriched; |
| Molecular Function (Gene Ontology) | 276 GO-terms significantly enriched; |
| Cellular Component (Gene Ontology) | 217 GO-terms significantly enriched; |
| Reference Publications (PubMed) | 10,000 publications significantly enriched; |
| Local Network Cluster (STRING) | 193 clusters significantly enriched; |
| KEGG Pathways | 199 pathways significantly enriched; |
| Reactome Pathways | 802 pathways significantly enriched; |
| WikiPathways | 388 pathways significantly enriched; |
| Disease-gene Associations (DISEASES) | 137 diseases significantly enriched; |
| Tissue Expression (TISSUES) | 162 tissues significantly enriched; |
| Subcellular Localization (COMPARTMENTS) | 205 compartments significantly enriched; |
| Human Phenotype (Monarch) | 1013 phenotypes significantly enriched; |
| Annotated Keywords (UniProt) | 87 keywords significantly enriched; |
| Protein Domains (Pfam) | 9 domains significantly enriched; |
| Protein Domains and Features (InterPro) | 187 domains significantly enriched; |
| Protein Domains (SMART) | 46 domains significantly enriched; |
| All enriched terms (without PubMed) | 5936 enriched terms in 15 categories. |
| Key Genes Linked to Epigenetic Phenomena in HCC | Key Genes Linked to Epigenetic Phenomena in HBV | ||||
|---|---|---|---|---|---|
| Name | Function | Bibliography | Name | Function | Bibliography |
| TP53 | DNA methylation patterns and histone modifications. | [102,103] | TP53 | also plays a critical role in HBV-associated carcinogenesis. | [104,105] |
| BAX, BCL2 | Apoptosis-related genes which undergo epigenetic regulation of their expression. | [106,107] | BAX, BCL2 | also involved in HBV-related apoptosis regulation influenced by viral-mediated epigenetic modifications. | [108] |
| FOXO1, FOXO3 | Members of the FOXO family are involved in histone modifications and can influence cell proliferation in HCC. | [109,110] | AKT1, AKT2, AKT3 | Epigenetically regulated in response to HBV infection, these genes modulate survival and proliferation pathways. | [111,112] |
| AKT1, AKT2, AKT3 | AKT isoforms are involved in epigenetically regulated signaling pathways, particularly in cancer processes such as HCC. | [111,113] | PTEN | PTEN is often epigenetically silenced via methylation in HBV. | [105] |
| PTEN | Tumor suppressor gene, regulated via promoter methylation in HCC. | [114] | GADD45A, GADD45B, GADD45G | These genes are involved in DNA repair and can influence epigenetic modifications through their role in response to cellular stress. | [115] |
| Other key genes usually linked to epigenetic phenomena. | |||||
| ATF2: This gene is involved in regulating gene expression through chromatin remodeling and can influence cancer progression [116]. | |||||
| BCL2L1: While primarily known for its role in apoptosis, it may also have implications in epigenetic regulation through interactions with chromatin-modifying complexes [117]. | |||||
| BRAF: Known for its role in cell signaling, its mutations are also associated with epigenetic changes in various cancers [118]. | |||||
| CREB3: Involved in transcriptional regulation linked to epigenetic modifications in various cancers [119]. | |||||
| GADD45A, GADD45B, GADD45G: These genes are involved in DNA repair and can influence epigenetic modifications through their role in response to cellular stress [115]. | |||||
| JAK2: While primarily part of the signaling pathway, it can influence gene expression and epigenetic modifications indirectly [120] | |||||
| MAPK1 and MAPK3: These genes are part of signaling pathways that can lead to changes in gene expression and implicated in epigenetic modifications [121]. | |||||
| NRAS: Like KRAS and BRAF, it is involved in signaling pathways that can lead to epigenetic alterations [121]. | |||||
| PRKCA: Plays a role in various signaling pathways and can influence epigenetic changes by modulating gene expression [122]. | |||||
| SMAD3: Involved in TGF-β signaling, which can lead to epigenetic modifications related to fibrosis and cancer progression [123]. | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colonna, G. Effects of SARS-CoV-2 Spike S1 Subunit on the Interplay Between Hepatitis B and Hepatocellular Carcinoma Related Molecular Processes in Human Liver. Livers 2025, 5, 1. https://doi.org/10.3390/livers5010001
Colonna G. Effects of SARS-CoV-2 Spike S1 Subunit on the Interplay Between Hepatitis B and Hepatocellular Carcinoma Related Molecular Processes in Human Liver. Livers. 2025; 5(1):1. https://doi.org/10.3390/livers5010001
Chicago/Turabian StyleColonna, Giovanni. 2025. "Effects of SARS-CoV-2 Spike S1 Subunit on the Interplay Between Hepatitis B and Hepatocellular Carcinoma Related Molecular Processes in Human Liver" Livers 5, no. 1: 1. https://doi.org/10.3390/livers5010001
APA StyleColonna, G. (2025). Effects of SARS-CoV-2 Spike S1 Subunit on the Interplay Between Hepatitis B and Hepatocellular Carcinoma Related Molecular Processes in Human Liver. Livers, 5(1), 1. https://doi.org/10.3390/livers5010001




