Exosome-Derived microRNAs: Bridging the Gap Between Obesity and Type 2 Diabetes in Diagnosis and Treatment
Abstract
1. Introduction—Obesity and Type 2 Diabetes
1.1. Obesity as the Beginning of Diabetes
1.2. Adipose Tissue—A Critical Player in the Disturbance of Immunometabolism
1.3. Type 2 Diabetes
2. Exosomes and miRNAs
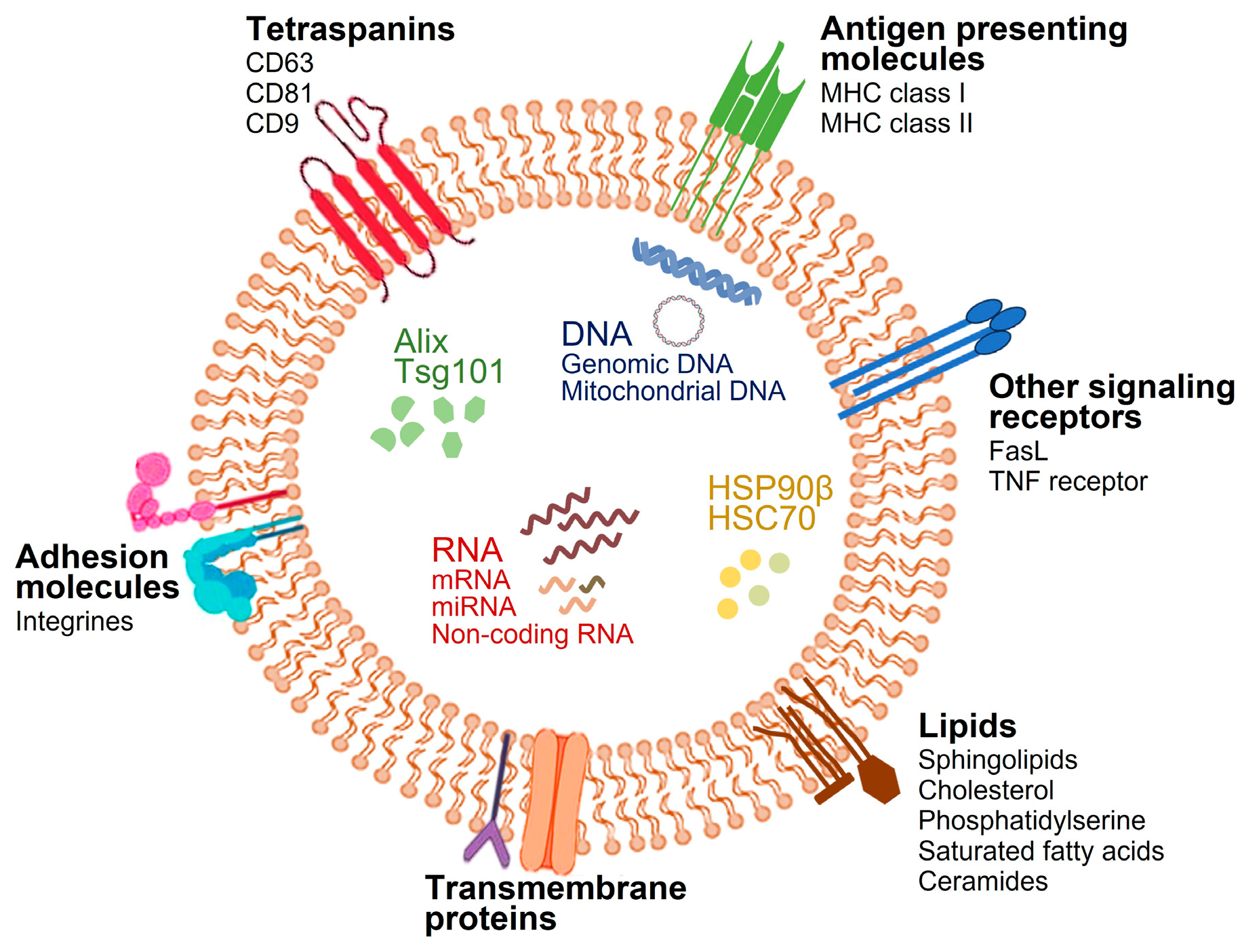
2.1. Biogenesis and Composition of Exosomes
2.2. Exosomal miRNAs and Their Role in Physiological and Pathophysiological Conditions
2.3. Exosomal miRNA as Biomarkers in Obesity and Type 2 Diabetes
2.4. Therapeutic Challenges of Exosomal miRNAs in Obesity and Type 2 Diabetes
2.5. Circulating miRNAs as Biomarkers and Therapeutic Targets of Insulin Resistance—Challenges and Limitations
2.6. Future Perspectives for miRNA-Based Therapies and Biomarker Development
3. Obesity-Derived Exosomal miRNAs: Bridging the Gap Between Insulin Resistance and Type 2 Diabetes
3.1. miRNAs as New Messengers in Intercellular Communication
3.2. The Role of Circulating miRNA in Obesity, Insulin Resistance and Type 2 Diabetes
3.3. The Involvement of Adipose Tissue-Derived Exosomal miRNAs in Adipose Tissue Inflammation During Obesity
3.4. The Involvement of Adipose Tissue-Derived Exosomal miRNAs in the Induction of Insulin Resistance and Type 2 Diabetes
3.5. The Effect of Adipose Tissue-Derived Exosomal miRNAs on the Homeostasis of Pancreatic β-Cells
| miRNA | Target Molecules | Signaling Pathways | Role in Obesity, Insulin Resistance and T2D | Reference |
|---|---|---|---|---|
| miR-27a | PPAR-γ | PPAR | Increased insulin resistance and impairment of insulin-dependent glucose uptake in skeletal muscle | [106] [107] |
| miR-29a | PPAR-δ | PPAR | Promotes insulin resistance in adipocytes, myocytes, and hepatocytes | [108] |
| miR-222 | IRS1, GLUT4 | Insulin signaling | Induces insulin resistance in skeletal muscle and hepatocytes. Reduces glucose uptake in adipocytes | [109] [110] |
| miR-144 | IRS1 | Insulin signaling | Impairs insulin signaling A potential therapeutic target in T2D | [88] [89] |
| miR-378a | C/EBPα, PGC-1β | PGC-1, adipogenesis | Enhances lipid storage and reduces thermogenesis | [113] |
| miR-103/107 | Caveolin-1 | Insulin signaling | Impair insulin sensitivity and glucose homeostasis. Promote adipocyte expansion | [114] |
| miR-34a | KLF4 | - | Inhibits M2 macrophage activation | [102] |
| miR-130b | PPAR-γ | PPAR | Inhibits M2 macrophage activation | [103] |
| miR-1224 | MSI2 | Wnt/β-catenin | Inhibits M2 macrophage activation | [101] |
| miR-155 | STAT1, STAT6, PPAR-γ/GLUT4 | JAK/STAT, PPAR | Regulates inflammation in obesity. Modulates insulin signaling and glucose tolerance | [51] [58] [105] [100] |
4. Conclusions and Future Directions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Phelps, N.H.; Singleton, R.K.; Zhou, B.; Heap, R.A.; Mishra, A.; Bennett, J.E.; Paciorek, C.J.; Lhoste, V.P.; Carrillo-Larco, R.M.; Stevens, G.A.; et al. Worldwide Trends in Underweight and Obesity from 1990 to 2022: A Pooled Analysis of 3663 Population-Representative Studies with 222 Million Children, Adolescents, and Adults. Lancet 2024, 403, 1027–1050. [Google Scholar] [CrossRef] [PubMed]
- Loos, R.J.F.; Yeo, G.S.H. The Genetics of Obesity: From Discovery to Biology. Nat. Rev. Genet. 2022, 23, 120–133. [Google Scholar] [CrossRef]
- Adams, K.F.; Schatzkin, A.; Harris, T.B.; Kipnis, V.; Mouw, T.; Ballard-Barbash, R.; Hollenbeck, A.; Leitzmann, M.F. Overweight, Obesity, and Mortality in a Large Prospective Cohort of Persons 50 to 71 Years Old. N. Engl. J. Med. 2006, 355, 763–778. [Google Scholar] [CrossRef]
- Bray, G.A. Obesity: A 100 Year Perspective. Int. J. Obes. 2024. [Google Scholar] [CrossRef]
- Deehan, E.C.; Mocanu, V.; Madsen, K.L. Effects of Dietary Fibre on Metabolic Health and Obesity. Nat. Rev. Gastroenterol. Hepatol. 2024, 21, 301–318. [Google Scholar] [CrossRef]
- Hu, T.; Liu, C.-H.; Lei, M.; Zeng, Q.; Li, L.; Tang, H.; Zhang, N. Metabolic Regulation of the Immune System in Health and Diseases: Mechanisms and Interventions. Signal Transduct. Target. Ther. 2024, 9, 268. [Google Scholar] [CrossRef]
- Pescador, N.; Pérez-Barba, M.; Ibarra, J.M.; Corbatón, A.; Martínez-Larrad, M.T.; Serrano-Ríos, M. Serum Circulating microRNA Profiling for Identification of Potential Type 2 Diabetes and Obesity Biomarkers. PLoS ONE 2013, 8, e77251. [Google Scholar] [CrossRef]
- Fabbri, M. MicroRNAs and miRceptors: A New Mechanism of Action for Intercellular Communication. Philos. Trans. R. Soc. B 2018, 373, 20160486. [Google Scholar] [CrossRef]
- Jung, S.H.; Park, H.S.; Kim, K.-S.; Choi, W.H.; Ahn, C.W.; Kim, B.T.; Kim, S.M.; Lee, S.Y.; Ahn, S.M.; Kim, Y.K.; et al. Effect of Weight Loss on Some Serum Cytokines in Human Obesity: Increase in IL-10 after Weight Loss. J. Nutr. Biochem. 2008, 19, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Hagberg, C.E.; Spalding, K.L. White Adipocyte Dysfunction and Obesity-Associated Pathologies in Humans. Nat. Rev. Mol. Cell Biol. 2024, 25, 270–289. [Google Scholar] [CrossRef]
- Wensveen, F.M.; Jelenčić, V.; Valentić, S.; Šestan, M.; Wensveen, T.T.; Theurich, S.; Glasner, A.; Mendrila, D.; Štimac, D.; Wunderlich, F.T.; et al. NK Cells Link Obesity-Induced Adipose Stress to Inflammation and Insulin Resistance. Nat. Immunol. 2015, 16, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.M.; Edwards, J.P. Exploring the Full Spectrum of Macrophage Activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.; Martinez, F.O. Alternative Activation of Macrophages: Mechanism and Functions. Immunity 2010, 32, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Hotamisligil, G.S.; Arner, P.; Caro, J.F.; Atkinson, R.L.; Spiegelman, B.M. Increased Adipose Tissue Expression of Tumor Necrosis Factor-Alpha in Human Obesity and Insulin Resistance. J. Clin. Investig. 1995, 95, 2409–2415. [Google Scholar] [CrossRef]
- Fink, L.N.; Costford, S.R.; Lee, Y.S.; Jensen, T.E.; Bilan, P.J.; Oberbach, A.; Blüher, M.; Olefsky, J.M.; Sams, A.; Klip, A. Pro-Inflammatory Macrophages Increase in Skeletal Muscle of High fat-Fed Mice and Correlate with Metabolic Risk Markers in Humans. Obesity 2014, 22, 747–757. [Google Scholar] [CrossRef]
- Lumeng, C.N.; Bodzin, J.L.; Saltiel, A.R. Obesity Induces a Phenotypic Switch in Adipose Tissue Macrophage Polarization. J. Clin. Investig. 2007, 117, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Morinaga, H.; Mayoral, R.; Heinrichsdorff, J.; Osborn, O.; Franck, N.; Hah, N.; Walenta, E.; Bandyopadhyay, G.; Pessentheiner, A.R.; Chi, T.J.; et al. Characterization of Distinct Subpopulations of Hepatic Macrophages in HFD/Obese Mice. Diabetes 2015, 64, 1120–1130. [Google Scholar] [CrossRef]
- Ivey, K.N.; Srivastava, D. microRNAs as Developmental Regulators. Cold Spring Harb. Perspect. Biol. 2015, 7, a008144. [Google Scholar] [CrossRef]
- McKernan, K.; Varghese, M.; Patel, R.; Singer, K. Role of TLR4 in the Induction of Inflammatory Changes in Adipocytes and Macrophages. Adipocyte 2020, 9, 212–222. [Google Scholar] [CrossRef]
- Mukherjee, S.; Skrede, S.; Haugstøyl, M.; López, M.; Fernø, J. Peripheral and Central Macrophages in Obesity. Front. Endocrinol. 2023, 14, 1232171. [Google Scholar] [CrossRef]
- Margolis, L.; Sadovsky, Y. The Biology of Extracellular Vesicles: The Known Unknowns. PLoS Biol. 2019, 17, e3000363. [Google Scholar] [CrossRef] [PubMed]
- Onyango, E.M.; Onyango, B.M. The Rise of Noncommunicable Diseases in Kenya: An Examination of the Time Trends and Contribution of the Changes in Diet and Physical Inactivity. J. Epidemiol. Glob. Health 2018, 8, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kadiki, O.A.; Reddy, M.R.S.; Marzouk, A.A. Incidence of Insulin-Dependent Diabetes (IDDM) and Non-Insulin-Dependent Diabetes (NIDDM) (0–34 Years at Onset) in Benghazi, Libya. Diabetes Res. Clin. Pract. 1996, 32, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Glass, C.K.; Olefsky, J.M. Inflammation and Lipid Signaling in the Etiology of Insulin Resistance. Cell Metab. 2012, 15, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Hotamisligil, G.S.; Shargill, N.S.; Spiegelman, B.M. Adipose Expression of Tumor Necrosis Factor-α: Direct Role in Obesity-Linked Insulin Resistance. Science 1993, 259, 87–91. [Google Scholar] [CrossRef]
- Hotamisligil, G.S.; Peraldi, P.; Budavari, A.; Ellis, R.; White, M.F.; Spiegelman, B.M. IRS-1-Mediated Inhibition of Insulin Receptor Tyrosine Kinase Activity in TNF-α- and Obesity-Induced Insulin Resistance. Science 1996, 271, 665–670. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Xie, Q.; Pan, X.; Zhang, R.; Zhang, X.; Peng, G.; Zhang, Y.; Shen, S.; Tong, N. Type 2 Diabetes Mellitus in Adults: Pathogenesis, Prevention and Therapy. Signal Transduct. Target. Ther. 2024, 9, 262. [Google Scholar] [CrossRef] [PubMed]
- Merrill, A.H. De Novo Sphingolipid Biosynthesis: A Necessary, but Dangerous, Pathway. J. Biol. Chem. 2002, 277, 25843–25846. [Google Scholar] [CrossRef] [PubMed]
- Matoba, K.; Takeda, Y.; Nagai, Y.; Kawanami, D.; Utsunomiya, K.; Nishimura, R. Unraveling the Role of Inflammation in the Pathogenesis of Diabetic Kidney Disease. Int. J. Mol. Sci. 2019, 20, 3393. [Google Scholar] [CrossRef]
- Sheu, M.L.; Ho, F.M.; Yang, R.S.; Chao, K.F.; Lin, W.W.; Lin-Shiau, S.Y.; Liu, S.-H. High Glucose Induces Human Endothelial Cell Apoptosis Through a Phosphoinositide 3-Kinase–Regulated Cyclooxygenase-2 Pathway. Arter. Thromb. Vasc. Biol. 2005, 25, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Zhao, H.; Song, H. Shared Signaling Pathways and Targeted Therapy by Natural Bioactive Compounds for Obesity and Type 2 Diabetes. Crit. Rev. Food Sci. Nutr. 2024, 64, 5039–5056. [Google Scholar] [CrossRef] [PubMed]
- Yung, J.H.M.; Giacca, A. Role of C-Jun N-Terminal Kinase (JNK) in Obesity and Type 2 Diabetes. Cells 2020, 9, 706. [Google Scholar] [CrossRef]
- Solinas, G.; Becattini, B. JNK at the Crossroad of Obesity, Insulin Resistance, and Cell Stress Response. Mol. Metab. 2017, 6, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-F.; Luh, F.; Ho, Y.-S.; Yen, Y. Exosomes: A Review of Biologic Function, Diagnostic and Targeted Therapy Applications, and Clinical Trials. J. Biomed. Sci. 2024, 31, 67. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, Biologic Function and Clinical Potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Larios, J.; Mercier, V.; Roux, A.; Gruenberg, J. ALIX- and ESCRT-III–Dependent Sorting of Tetraspanins to Exosomes. J. Cell Biol. 2020, 219, e201904113. [Google Scholar] [CrossRef] [PubMed]
- Murphy, D.E.; De Jong, O.G.; Brouwer, M.; Wood, M.J.; Lavieu, G.; Schiffelers, R.M.; Vader, P. Extracellular Vesicle-Based Therapeutics: Natural versus Engineered Targeting and Trafficking. Exp. Mol. Med. 2019, 51, 32. [Google Scholar] [CrossRef] [PubMed]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide Triggers Budding of Exosome Vesicles into Multivesicular Endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef] [PubMed]
- Skotland, T.; Sagini, K.; Sandvig, K.; Llorente, A. An Emerging Focus on Lipids in Extracellular Vesicles. Adv. Drug Deliv. Rev. 2020, 159, 308–321. [Google Scholar] [CrossRef]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA Delivery by Extracellular Vesicles in Mammalian Cells and Its Applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef]
- Mori, M.A.; Ludwig, R.G.; Garcia-Martin, R.; Brandão, B.B.; Kahn, C.R. Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab. 2019, 30, 656–673. [Google Scholar] [CrossRef] [PubMed]
- Shang, R.; Lee, S.; Senavirathne, G.; Lai, E.C. microRNAs in Action: Biogenesis, Function and Regulation. Nat. Rev. Genet. 2023, 24, 816–833. [Google Scholar] [CrossRef] [PubMed]
- Landrier, J.-F.; Derghal, A.; Mounien, L. MicroRNAs in Obesity and Related Metabolic Disorders. Cells 2019, 8, 859. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.; Rheinheimer, S.; Meder, B.; Stähler, C.; Meese, E.; et al. Distribution of miRNA Expression across Human Tissues. Nucleic Acids Res. 2016, 44, 3865–3877. [Google Scholar] [CrossRef] [PubMed]
- Correia De Sousa, M.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef] [PubMed]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An Overview of microRNAs: Biology, Functions, Therapeutics, and Analysis Methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Bhome, R.; Del Vecchio, F.; Lee, G.-H.; Bullock, M.D.; Primrose, J.N.; Sayan, A.E.; Mirnezami, A.H. Exosomal microRNAs (exomiRs): Small Molecules with a Big Role in Cancer. Cancer Lett. 2018, 420, 228–235. [Google Scholar] [CrossRef]
- Van den Brande, S.; Gijbels, M.; Wynant, N.; Santos, D.; Mingels, L.; Gansemans, Y.; Van Nieuwerburgh, F.; Vanden Broeck, J. The Presence of Extracellular microRNAs in the Media of Cultured Drosophila Cells. Sci. Rep. 2018, 8, 17312. [Google Scholar] [CrossRef] [PubMed]
- Isaac, R.; Reis, F.C.G.; Ying, W.; Olefsky, J.M. Exosomes as Mediators of Intercellular Crosstalk in Metabolism. Cell Metab. 2021, 33, 1744–1762. [Google Scholar] [CrossRef]
- Castaño, C.; Kalko, S.; Novials, A.; Párrizas, M. Obesity-Associated Exosomal miRNAs Modulate Glucose and Lipid Metabolism in Mice. Proc. Natl. Acad. Sci. USA 2018, 115, 12158–12163. [Google Scholar] [CrossRef]
- Kwan, H.Y.; Chen, M.; Xu, K.; Chen, B. The Impact of Obesity on Adipocyte-Derived Extracellular Vesicles. Cell. Mol. Life Sci. 2021, 78, 7275–7288. [Google Scholar] [CrossRef]
- He, X.; Kuang, G.; Wu, Y.; Ou, C. Emerging Roles of Exosomal miRNAs in Diabetes Mellitus. Clin. Transl. Med. 2021, 11, e468. [Google Scholar] [CrossRef]
- Lange, T.; Stracke, S.; Rettig, R.; Lendeckel, U.; Kuhn, J.; Schlüter, R.; Rippe, V.; Endlich, K.; Endlich, N. Identification of miR-16 as an Endogenous Reference Gene for the Normalization of Urinary Exosomal miRNA Expression Data from CKD Patients. PLoS ONE 2017, 12, e0183435. [Google Scholar] [CrossRef]
- Ruze, R.; Liu, T.; Zou, X.; Song, J.; Chen, Y.; Xu, R.; Yin, X.; Xu, Q. Obesity and Type 2 Diabetes Mellitus: Connections in Epidemiology, Pathogenesis, and Treatments. Front. Endocrinol. 2023, 14, 1161521. [Google Scholar] [CrossRef]
- Benavides-Aguilar, J.A.; Torres-Copado, A.; Isidoro-Sánchez, J.; Pathak, S.; Duttaroy, A.K.; Banerjee, A.; Paul, S. The Regulatory Role of MicroRNAs in Obesity and Obesity-Derived Ailments. Genes 2023, 14, 2070. [Google Scholar] [CrossRef]
- Heyn, G.S.; Corrêa, L.H.; Magalhães, K.G. The Impact of Adipose Tissue–Derived miRNAs in Metabolic Syndrome, Obesity, and Cancer. Front. Endocrinol. 2020, 11, 563816. [Google Scholar] [CrossRef] [PubMed]
- Tonyan, Z.N.; Barbitoff, Y.A.; Nasykhova, Y.A.; Danilova, M.M.; Kozyulina, P.Y.; Mikhailova, A.A.; Bulgakova, O.L.; Vlasova, M.E.; Golovkin, N.V.; Glotov, A.S. Plasma microRNA Profiling in Type 2 Diabetes Mellitus: A Pilot Study. Int. J. Mol. Sci. 2023, 24, 17406. [Google Scholar] [CrossRef] [PubMed]
- Brandao, B.B.; Lino, M.; Kahn, C.R. Extracellular miRNAs as Mediators of Obesity-Associated Disease. J. Physiol. 2022, 600, 1155–1169. [Google Scholar] [CrossRef] [PubMed]
- Lino, M.; Garcia-Martin, R.; Muñoz, V.R.; Ruiz, G.P.; Nawaz, A.; Brandão, B.B.; Dreyfus, J.; Pan, H.; Kahn, C.R. Multi-Step Regulation of microRNA Expression and Secretion into Small Extracellular Vesicles by Insulin. Cell Rep. 2024, 43, 114491. [Google Scholar] [CrossRef]
- Ji, C.; Guo, X. The Clinical Potential of Circulating microRNAs in Obesity. Nat. Rev. Endocrinol. 2019, 15, 731–743. [Google Scholar] [CrossRef]
- Grasedieck, S.; Sorrentino, A.; Langer, C.; Buske, C.; Döhner, H.; Mertens, D.; Kuchenbauer, F. Circulating microRNAs in Hematological Diseases: Principles, Challenges, and Perspectives. Blood 2013, 121, 4977–4984. [Google Scholar] [CrossRef] [PubMed]
- Kupec, T.; Bleilevens, A.; Iborra, S.; Najjari, L.; Wittenborn, J.; Maurer, J.; Stickeler, E. Stability of Circulating microRNAs in Serum. PLoS ONE 2022, 17, e0268958. [Google Scholar] [CrossRef]
- Gareev, I.; Beylerli, O.; Yang, G.; Sun, J.; Pavlov, V.; Izmailov, A.; Shi, H.; Zhao, S. The Current State of MiRNAs as Biomarkers and Therapeutic Tools. Clin. Exp. Med. 2020, 20, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Momin, M.Y.; Gaddam, R.R.; Kravitz, M.; Gupta, A.; Vikram, A. The Challenges and Opportunities in the Development of MicroRNA Therapeutics: A Multidisciplinary Viewpoint. Cells 2021, 10, 3097. [Google Scholar] [CrossRef] [PubMed]
- Pagoni, M.; Cava, C.; Sideris, D.C.; Avgeris, M.; Zoumpourlis, V.; Michalopoulos, I.; Drakoulis, N. miRNA-Based Technologies in Cancer Therapy. J. Pers. Med. 2023, 13, 1586. [Google Scholar] [CrossRef] [PubMed]
- Segal, M.; Slack, F.J. Challenges Identifying Efficacious miRNA Therapeutics for Cancer. Expert Opin. Drug Discov. 2020, 15, 987–991. [Google Scholar] [CrossRef]
- Grillone, K.; Caridà, G.; Luciano, F.; Cordua, A.; Di Martino, M.T.; Tagliaferri, P.; Tassone, P. A Systematic Review of Non-Coding RNA Therapeutics in Early Clinical Trials: A New Perspective against Cancer. J. Transl. Med. 2024, 22, 731. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.S.; Kang, Y.-K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.-L.; Kim, T.-Y.; et al. Phase 1 Study of MRX34, a Liposomal miR-34a Mimic, in Patients with Advanced Solid Tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef]
- Dai, M.; Li, L.; Qin, X. Clinical Value of miRNA-122 in the Diagnosis and Prognosis of Various Types of Cancer. Oncol. Lett. 2019, 17, 3919–3929. [Google Scholar] [CrossRef]
- Carpi, S.; Daniele, S.; De Almeida, J.F.M.; Gabbia, D. Recent Advances in miRNA-Based Therapy for MASLD/MASH and MASH-Associated HCC. Int. J. Mol. Sci. 2024, 25, 12229. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S. New Molecular Biomarkers in Precise Diagnosis and Therapy of Type 2 Diabetes. Health Technol. 2020, 10, 601–608. [Google Scholar] [CrossRef]
- Abbasi, A.; Sahlqvist, A.-S.; Lotta, L.; Brosnan, J.M.; Vollenweider, P.; Giabbanelli, P.; Nunez, D.J.; Waterworth, D.; Scott, R.A.; Langenberg, C.; et al. A Systematic Review of Biomarkers and Risk of Incident Type 2 Diabetes: An Overview of Epidemiological, Prediction and Aetiological Research Literature. PLoS ONE 2016, 11, e0163721. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.; Chakraborty, A.; Sarkar, D.; Langthasa, M.; Rahman, M.; Bari, M.; Singha, R.S.; Malakar, A.K.; Chakraborty, S. Interplay between miRNAs and Human Diseases. J. Cell. Physiol. 2018, 233, 2007–2018. [Google Scholar] [CrossRef]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of Extracellular Circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Gayosso-Gómez, L.V.; Ortiz-Quintero, B. Circulating MicroRNAs in Blood and Other Body Fluids as Biomarkers for Diagnosis, Prognosis, and Therapy Response in Lung Cancer. Diagnostics 2021, 11, 421. [Google Scholar] [CrossRef]
- Wang, F.; Chen, C.; Wang, D. Circulating microRNAs in Cardiovascular Diseases: From Biomarkers to Therapeutic Targets. Front. Med. 2014, 8, 404–418. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Su, Y.; Sharma, M.; Singh, S.; Kim, S.; Peavey, J.J.; Suerken, C.K.; Lockhart, S.N.; Whitlow, C.T.; Craft, S.; et al. MicroRNA Expression in Extracellular Vesicles as a Novel Blood-based Biomarker for Alzheimer’s Disease. Alzheimer’s Dement. 2023, 19, 4952–4966. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Cui, J.; Wu, H.; Lu, Q. The Emerging Role of Circulating microRNAs as Biomarkers in Autoimmune Diseases. Autoimmunity 2014, 47, 419–429. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, H.; Zhao, M.; Chang, C.; Lu, Q. Clinical Significance of miRNAs in Autoimmunity. J. Autoimmun. 2020, 109, 102438. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; He, Y.; Mackowiak, B.; Gao, B. MicroRNAs as Regulators, Biomarkers and Therapeutic Targets in Liver Diseases. Gut 2021, 70, 784–795. [Google Scholar] [CrossRef]
- Liu, X.; Pan, Q.; Cao, H.; Xin, F.; Zhao, Z.; Yang, R.; Zeng, J.; Zhou, H.; Fan, J. Lipotoxic Hepatocyte-Derived Exosomal MicroRNA 192-5p Activates Macrophages Through Rictor/Akt/Forkhead Box Transcription Factor O1 Signaling in Nonalcoholic Fatty Liver Disease. Hepatology 2020, 72, 454–469. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Rao, L.V.M. The Role of microRNAs in Inflammation. Int. J. Mol. Sci. 2022, 23, 15479. [Google Scholar] [CrossRef]
- Conti, I.; Varano, G.; Simioni, C.; Laface, I.; Milani, D.; Rimondi, E.; Neri, L.M. miRNAs as Influencers of Cell–Cell Communication in Tumor Microenvironment. Cells 2020, 9, 220. [Google Scholar] [CrossRef]
- Sepúlveda, F.; Mayorga-Lobos, C.; Guzmán, K.; Durán-Jara, E.; Lobos-González, L. EV-miRNA-Mediated Intercellular Communication in the Breast Tumor Microenvironment. Int. J. Mol. Sci. 2023, 24, 13085. [Google Scholar] [CrossRef]
- Zatterale, F.; Longo, M.; Naderi, J.; Raciti, G.A.; Desiderio, A.; Miele, C.; Beguinot, F. Chronic Adipose Tissue Inflammation Linking Obesity to Insulin Resistance and Type 2 Diabetes. Front. Physiol. 2020, 10, 1607. [Google Scholar] [CrossRef] [PubMed]
- Chao, Y.; Gu, T.; Zhang, Z.; Wu, T.; Wang, J.; Bi, Y. The Role of miRNAs Carried by Extracellular Vesicles in Type 2 Diabetes and Its Complications. J. Diabetes 2023, 15, 838–852. [Google Scholar] [CrossRef] [PubMed]
- Karolina, D.S.; Armugam, A.; Tavintharan, S.; Wong, M.T.K.; Lim, S.C.; Sum, C.F.; Jeyaseelan, K. MicroRNA 144 Impairs Insulin Signaling by Inhibiting the Expression of Insulin Receptor Substrate 1 in Type 2 Diabetes Mellitus. PLoS ONE 2011, 6, e22839. [Google Scholar] [CrossRef]
- Karolina, D.S.; Armugam, A.; Tavintharan, S.; Wong, M.T.K.; Lim, S.C.; Sum, C.F.; Jeyaseelan, K. Correction: MicroRNA 144 Impairs Insulin Signaling by Inhibiting the Expression of Insulin Receptor Substrate 1 in Type 2 Diabetes Mellitus. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Azzimato, V.; Chen, P.; Barreby, E.; Morgantini, C.; Levi, L.; Vankova, A.; Jager, J.; Sulen, A.; Diotallevi, M.; Shen, J.X.; et al. Hepatic miR-144 Drives Fumarase Activity Preventing NRF2 Activation During Obesity. Gastroenterology 2021, 161, 1982–1997.e11. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Hong, J.; Cao, Y.; Shi, J.; Gu, W.; Ning, G.; Zhang, Y.; Wang, W. Elevated Circulating microRNA-122 Is Associated with Obesity and Insulin Resistance in Young Adults. Eur. J. Endocrinol. 2015, 172, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Hess, A.L.; Larsen, L.H.; Udesen, P.B.; Sanz, Y.; Larsen, T.M.; Dalgaard, L.T. Levels of Circulating miR-122 Are Associated with Weight Loss and Metabolic Syndrome. Obesity 2020, 28, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Willeit, P.; Skroblin, P.; Moschen, A.R.; Yin, X.; Kaudewitz, D.; Zampetaki, A.; Barwari, T.; Whitehead, M.; Ramírez, C.M.; Goedeke, L.; et al. Circulating MicroRNA-122 Is Associated With the Risk of New-Onset Metabolic Syndrome and Type 2 Diabetes. Diabetes 2017, 66, 347–357. [Google Scholar] [CrossRef]
- Párrizas, M.; Brugnara, L.; Esteban, Y.; González-Franquesa, A.; Canivell, S.; Murillo, S.; Gordillo-Bastidas, E.; Cussó, R.; Cadefau, J.A.; García-Roves, P.M.; et al. Circulating miR-192 and miR-193b Are Markers of Prediabetes and Are Modulated by an Exercise Intervention. J. Clin. Endocrinol. Metab. 2015, 100, E407–E415. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Zhao, M.; Li, Z.; Nie, H.; He, J.; Chen, Z.; Yuan, J.; Guo, H.; Zhang, X.; Yang, H.; et al. Plasma miR-193b-3p Is Elevated in Type 2 Diabetes and Could Impair Glucose Metabolism. Front. Endocrinol. 2022, 13, 814347. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, A.; Zollinger, L.; Saely, C.H.; Muendlein, A.; Evangelakos, I.; Nasias, D.; Charizopoulou, N.; Schofield, J.D.; Othman, A.; Soran, H.; et al. Circulating microRNAs -192 and -194 Are Associated with the Presence and Incidence of Diabetes Mellitus. Sci. Rep. 2018, 8, 14274. [Google Scholar] [CrossRef]
- Latouche, C.; Natoli, A.; Reddy-Luthmoodoo, M.; Heywood, S.E.; Armitage, J.A.; Kingwell, B.A. MicroRNA-194 Modulates Glucose Metabolism and Its Skeletal Muscle Expression Is Reduced in Diabetes. PLoS ONE 2016, 11, e0155108. [Google Scholar] [CrossRef]
- Jones, A.; Danielson, K.M.; Benton, M.C.; Ziegler, O.; Shah, R.; Stubbs, R.S.; Das, S.; Macartney-Coxson, D. miRNA Signatures of Insulin Resistance in Obesity. Obesity 2017, 25, 1734–1744. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ren, Y.; Chang, K.; Wu, W.; Griffiths, H.R.; Lu, S.; Gao, D. Adipose Tissue Macrophages as Potential Targets for Obesity and Metabolic Diseases. Front. Immunol. 2023, 14, 1153915. [Google Scholar] [CrossRef]
- Tryggestad, J.B.; Teague, A.M.; Sparling, D.P.; Jiang, S.; Chernausek, S.D. Macrophage-Derived microRNA-155 Increases in Obesity and Influences Adipocyte Metabolism by Targeting Peroxisome Proliferator-Activated Receptor Gamma. Obesity 2019, 27, 1856–1864. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Yao, X.; Teng, Y.; Zhao, T.; Lin, L.; Li, Y.; Shang, H.; Jin, Y.; Jin, Q. Adipocytes-Derived Exosomal microRNA-1224 Inhibits M2 Macrophage Polarization in Obesity-Induced Adipose Tissue Inflammation via MSI2-Mediated Wnt/β-Catenin Axis. Mol. Nutr. Food Res. 2022, 66, 2100889. [Google Scholar] [CrossRef]
- Pan, Y.; Hui, X.; Hoo, R.L.C.; Ye, D.; Chan, C.Y.C.; Feng, T.; Wang, Y.; Lam, K.S.L.; Xu, A. Adipocyte-Secreted Exosomal microRNA-34a Inhibits M2 Macrophage Polarization to Promote Obesity-Induced Adipose Inflammation. J. Clin. Investig. 2019, 129, 834–849. [Google Scholar] [CrossRef]
- Zhang, M.; Zhou, Z.; Wang, J.; Li, S. MiR-130b Promotes Obesity Associated Adipose Tissue Inflammation and Insulin Resistance in Diabetes Mice through Alleviating M2 Macrophage Polarization via Repression of PPAR-γ. Immunol. Lett. 2016, 180, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.-M.; Lin, X.; Xu, F.; Shan, S.-K.; Guo, B.; Li, F.-X.-Z.; Zheng, M.-H.; Wang, Y.; Xu, Q.-S.; Yuan, L.-Q. Exosomes and Obesity-Related Insulin Resistance. Front. Cell Dev. Biol. 2021, 9, 651996. [Google Scholar] [CrossRef] [PubMed]
- Ying, W.; Riopel, M.; Bandyopadhyay, G.; Dong, Y.; Birmingham, A.; Seo, J.B.; Ofrecio, J.M.; Wollam, J.; Hernandez-Carretero, A.; Fu, W.; et al. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell 2017, 171, 372–384.e12. [Google Scholar] [CrossRef] [PubMed]
- Payet, T.; Gabinaud, E.; Landrier, J.; Mounien, L. Role of micro-RNAs Associated with Adipose-derived Extracellular Vesicles in Metabolic Disorders. Obes. Rev. 2024, 25, e13755. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Du, H.; Wei, S.; Feng, L.; Li, J.; Yao, F.; Zhang, M.; Hatch, G.M.; Chen, L. Adipocyte-Derived Exosomal MiR-27a Induces Insulin Resistance in Skeletal Muscle Through Repression of PPARγ. Theranostics 2018, 8, 2171–2188. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Sun, Y.-C.; Cheng, P.; Shao, H.-G. Adipose Tissue Macrophage-Derived Exosomal miR-29a Regulates Obesity-Associated Insulin Resistance. Biochem. Biophys. Res. Commun. 2019, 515, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Song, H.; Shuo, L.; Wang, L.; Xie, P.; Li, W.; Liu, J.; Tong, Y.; Zhang, C.-Y.; Jiang, X.; et al. Gonadal White Adipose Tissue-Derived Exosomal MiR-222 Promotes Obesity-Associated Insulin Resistance. Aging 2020, 12, 22719–22743. [Google Scholar] [CrossRef]
- Shi, Z.; Zhao, C.; Guo, X.; Ding, H.; Cui, Y.; Shen, R.; Liu, J. Differential Expression of MicroRNAs in Omental Adipose Tissue From Gestational Diabetes Mellitus Subjects Reveals miR-222 as a Regulator of ERα Expression in Estrogen-Induced Insulin Resistance. Endocrinology 2014, 155, 1982–1990. [Google Scholar] [CrossRef]
- Dang, S.-Y.; Leng, Y.; Wang, Z.-X.; Xiao, X.; Zhang, X.; Wen, T.; Gong, H.-Z.; Hong, A.; Ma, Y. Exosomal Transfer of Obesity Adipose Tissue for Decreased miR-141-3p Mediate Insulin Resistance of Hepatocytes. Int. J. Biol. Sci. 2019, 15, 351–368. [Google Scholar] [CrossRef] [PubMed]
- Gesmundo, I.; Pardini, B.; Gargantini, E.; Gamba, G.; Birolo, G.; Fanciulli, A.; Banfi, D.; Congiusta, N.; Favaro, E.; Deregibus, M.C.; et al. Adipocyte-Derived Extracellular Vesicles Regulate Survival and Function of Pancreatic β Cells. JCI Insight 2021, 6, e141962. [Google Scholar] [CrossRef] [PubMed]
- Machado, I.F.; Teodoro, J.S.; Palmeira, C.M.; Rolo, A.P. miR-378a: A New Emerging microRNA in Metabolism. Cell. Mol. Life Sci. 2020, 77, 1947–1958. [Google Scholar] [CrossRef] [PubMed]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 Regulate Insulin Sensitivity. Nature 2011, 474, 649–653. [Google Scholar] [CrossRef]
- Palihaderu, P.; Mendis, B.; Premarathne, J.; Dias, W.; Yeap, S.K.; Ho, W.Y.; Dissanayake, A.; Rajapakse, I.; Karunanayake, P.; Senarath, U.; et al. Therapeutic Potential of miRNAs for Type 2 Diabetes Mellitus: An Overview. Genet. Epigenet. 2022, 15, 25168657221130041. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vukelić, I.; Šuša, B.; Klobučar, S.; Buljević, S.; Liberati Pršo, A.-M.; Belančić, A.; Rahelić, D.; Detel, D. Exosome-Derived microRNAs: Bridging the Gap Between Obesity and Type 2 Diabetes in Diagnosis and Treatment. Diabetology 2024, 5, 706-724. https://doi.org/10.3390/diabetology5070052
Vukelić I, Šuša B, Klobučar S, Buljević S, Liberati Pršo A-M, Belančić A, Rahelić D, Detel D. Exosome-Derived microRNAs: Bridging the Gap Between Obesity and Type 2 Diabetes in Diagnosis and Treatment. Diabetology. 2024; 5(7):706-724. https://doi.org/10.3390/diabetology5070052
Chicago/Turabian StyleVukelić, Iva, Branislav Šuša, Sanja Klobučar, Sunčica Buljević, Ana-Marija Liberati Pršo, Andrej Belančić, Dario Rahelić, and Dijana Detel. 2024. "Exosome-Derived microRNAs: Bridging the Gap Between Obesity and Type 2 Diabetes in Diagnosis and Treatment" Diabetology 5, no. 7: 706-724. https://doi.org/10.3390/diabetology5070052
APA StyleVukelić, I., Šuša, B., Klobučar, S., Buljević, S., Liberati Pršo, A.-M., Belančić, A., Rahelić, D., & Detel, D. (2024). Exosome-Derived microRNAs: Bridging the Gap Between Obesity and Type 2 Diabetes in Diagnosis and Treatment. Diabetology, 5(7), 706-724. https://doi.org/10.3390/diabetology5070052








