Abstract
Methanotrophs are bacteria that can consume methane as their sole carbon and energy source to produce a wide variety of high-value products such as lipids, biopolymers, ectoine, and single cell proteins (SCPs). Collected samples from various sources were subjected to DNA extraction followed by 16S rRNA analysis to determine the identity and relative abundance of their microbial population. Several taxa of methanotrophs were detected in the samples including Type I (Methylobacter), Type X (Methylocaldum), Type II (Methylocystis, Methylosinus, and Beijerinckia), and Type III (Verrucomicrobium). This paper expounds the effects of environmental/cultivation conditions on the growth and population of different types of methanotrophs. The results could be used to systematically identify source(s) of natural consortia that can be enriched and developed to produce specific target product(s) under a given cultivation conditions/limitations.
1. Introduction
In recent years, the increase in greenhouse gas (GHG) emissions, causing global warming, has been a pressing issue due to its evident harmful environmental effects. Methane (CH4) is considered as the second most prominent GHG produced next to carbon dioxide (CO2) and has a global warming potential of 27–30 times higher than CO2 over 100 years []. Methane is the key component of natural gas, which is typically used for power, fuel, and heat. The advancement in shale gas production has resulted in the instability of natural gas prices in the past decade, e.g., $8.86 per million British thermal unit (MMBtu) in 2008, $2.05/MMBtu in 2020, and $7.88/MMBtu in September 2022 []. As a result, large volumes of CH4 are vented and flared into the atmosphere mainly due to unprofitability, operational safety, and costly connection to the pipeline []. In 2021 alone, the US flared about 8764 million cubic meters (MCM) of natural gas with ~23 million metric ton of carbon dioxide equivalent (MMT CO2e) emission and with an equivalent economic value of over 1 billion dollars [], causing significant negative environmental impacts and lost revenues. The growing concerns toward climate change mitigation led to the continuous quest for economically viable technologies to reduce these GHG emissions. Hence, there is an opportunity to develop processes to economically convert CH4 to high-value products. One such process is the utilization of CH4 as substrate for microbial bioconversion instead of using expensive sugar-based feedstocks [,,].
Methanotrophs are gaining interests because of their ability to utilize CH4 as their sole carbon and energy source []. They play a vital role in carbon cycling as they can convert CH4 into a wide variety of valuable bioproducts such as lipids, biopolymers, ectoine, and single cell proteins (SCPs) [,,,]. These lipids can be used to produce renewable diesel/green energy or as feedstock for oleochemical manufacturing. Biopolymers such as polyhydroxyalkanoates (PHAs) are biodegradable, non-toxic, and thermoplastic molecules which can be applied in various energy and environmental applications as well as a potential replacement of conventional plastics [,,]. On the other hand, ectoine is broadly employed in cosmetics industry, dermatology, and it is also an effective stabilizer for nucleic acids, enzymes, and DNA-protein complexes applied in pharmaceutical industries [,,,], while SCPs can be used as an alternative protein source that has the advantage of being independent of agricultural products (e.g., soybean) as a staring material [,].
Methanotrophs are gram-negative members of Proteobacteria that are ubiquitous in nature commonly found in soil, natural gas fields, wetlands, sewage sludges, and waste treatment facilities [,,,]. They are classified into taxonomic groups based on their 16S rRNA gene sequence, their cell morphology, ultra-structure, phylogeny, and metabolic pathways [,,] as shown in Figure 1.
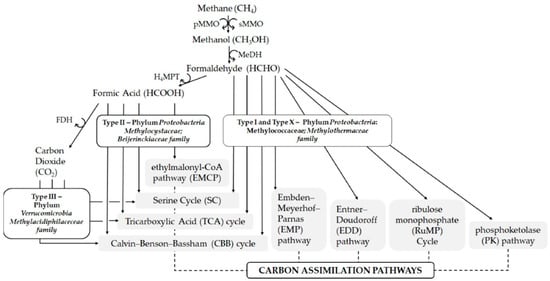
Figure 1.
Different types of methanotrophs and their metabolic pathways: pMMO—particulate monooxygenase, sMMO—soluble monooxygenase, MeDH—methanol dehydrogenase, H4MPT—tetrahydromethanopterin, FDH—formate dehydrogenase. Modified with permission from Kalyuzhnaya et al. [].
Distinct types of methanotrophs react differently to changing environmental conditions. The knowledge on the behavior of methanotrophs under different conditions is critical for choosing the suitable type of methanotrophs for culture enrichment and cultivation optimization tailored for producing a certain bio-product. In this work, natural microbial consortia present in samples collected from various sources (particularly low and high O2 levels) were tested for the presence of methanotrophs. The results could be used to identify possible sources of seed-stocks that contain certain type(s) of methanotrophs for further studies.
2. Materials and Methods
Samples were collected from various sites including sediments from three open drainage ditches and sludges from different wastewater treatment plants (WWTPs) located in Lafayette, LA, USA—South WWTP (activated sludge and aerobic digester), East WWTP (activated sludge) and Ambassador Caffery WWTP (activated sludge). The samples were immediately subjected to DNA extraction using DNeasy® Powersoil® Pro Kit (Qiagen, Germany) and the extracts were subjected to full length 16S rRNA gene diversity analysis using bTEFAP® technology (Mr. DNA Molecular Research LP, Shallowater, TX, USA). Sequencing was performed on a MiSeq following the manufacturer’s protocols and sequence data were processed using ribosomal and functional gene analysis pipeline. The final zero-radius operational taxonomic units (zOTUs) were classified using BLASTn against a curated database derived from the National Center for Biotechnology Information (NCBI).
3. Results and Discussion
In all the samples, low but detectable levels of methanotrophs were identified including Type I (Methylobacter), Type X (Methylocaldum), Type II (Methylocystis, Methylosinus, and Beijerinckia), and Type III (Verrucomicrobium) (see Table 1).

Table 1.
Methanotroph composition from various sources.
Intensive studies involving methanotrophs has revealed that changing parameters such as CH4 and O2 concentrations, nitrogen sources, copper content, pH, and temperature promote the growth and enhance the population of particular types of methanotrophs. In general, Type I methanotrophs prefer low CH4 and high O2 concentrations while Type II methanotrophs favor high CH4 and low O2 concentrations [,]. This is evident from the results in Table 1, showing that Type I methanotrophs were not detected in samples collected from drainage sediments (DS1, DS2, and DS3). These samples were collected in low O2 environments (i.e., under <1 foot of stagnant muddy water), and thus, favored Type II methanotrophs. In contrast, samples collected from WWTPs (EWAS, SWAS, SWDS, and AWAS) contain Types I, X and II. These samples were collected from aerobic treatment units (i.e., high O2 environments) that favor Type I. Nevertheless, localized low O2 regions within these treatment units might have allowed the proliferations of Type II methanotrophs as well.
Among the parameters that affect methanotrophs growth, only the CH4 and O2 levels do not require chemical analyses of the growth environment. In particular, the level of O2 can be easily speculated as illustrated above. In this work, only the level of O2 was used as parameter for choosing the source of consortia. Nevertheless, whenever chemical assays are feasible, the following can be used in deciding the source of seed consortium. In terms of nitrogen, Type I methanotrophs preferred an environment with high nitrogen content or lower carbon to nitrogen (C/N) ratio while Type II methanotrophs are more common in N-limited (or high C/N ratio) conditions []. Type II methanotrophs and some strains of Methylobacter (Type I) can fix atmospheric N2 because they possess the nitrogenase enzyme. Moreover, studies revealed that methanotrophs grow better on inorganic nitrogen sources (nitrate or ammonia) than atmospheric N2 [,]. Copper content, on the other hand, greatly influences the growth of methanotrophs that have the particulate monooxygenase (pMMO) since copper regulates the expression of this enzyme [,].
Methanotrophs are not known to produce neutral lipids (e.g., triglycerides, waxes). Membrane lipids in the form of phospholipids are the only class of lipids typically found in these microbes. As such, the amount of phospholipids that can be obtained from methanotrophs is directly proportional to the biomass produced during cultivation. However, the type of phospholipids is dependent on the type of methanotrophs. For example, phosphatidyl dimethyl ethanolamine and phosphatidyl methyl ethanolamine are found in Type I (Methylobacter), Type II (Methylocystis and Methylosinus), and Type X (Methylocaldum) methanotrophs []. Therefore, if these types of phospholipids are the target products, the consortium in EWAS is the most suitable seed for cultivation. The most studied species for ectoine production is Methylomicrobium alcaliphilum 20Z (Type I), but ectoine can also be synthesized by Methylosinus sporium (Type II) and Methylobacter marinus 7C (Type I) []. Any of the samples collected can be used as seed for ectoine production because Methylosinus sporium was detected in all of them. However, the most suitable might be DS1 as it contains the highest concentration of this species (Table 1). Favorable characteristics of methanotrophs that can produce SCP should have a rapid growth rate, easy to cultivate, and with high protein production capacity []. Methylocystis sp. (Type II) is one of the methanotrophs species that had been used for SCP production [] at broad pH and temperature ranges. Although any of the samples can be used as seed consortium for SCP production, DS1 is the best choice if abundance is required. Otherwise, if abundance and species diversity is sought, SWDS should be chosen.
Poly(3-hydroxybutyrate) or PHB, which is a member of the PHA family, is another potential high-value product from methanotrophs. The PHBs are accumulated in all Type II methanotrophs as a survival mechanism under nutrients starvation [,]. The results (Table 1) suggest that any of the samples could be used as seed for PHB production. The final choice comes down to whether abundance or diversity or both is required by the cultivation. For PHB or for any of the target products, the ultimate choice of which seed to use will also depend on the cultivation conditions. Generally, the growth conditions that lead to PHB accumulation include: (i) low N level (ammonia or nitrate), (ii) copper deficiency, and (iii) fed-batch cultivation []. Additionally, AlSayed et al. [] reported that most PHB accumulation studies were conducted at temperature from 20 to 40 °C and pH of 6–7. Some reports suggest that increasing the medium acidity also increased PHB accumulation in Type II methanotrophs []. The PHB accumulation in Methylocystis sp. GB25 DSM 7674 was successfully enhanced under N-limited condition during fed-batch cultivation []. In any case, the cultivation temperature and pH should be considered, noting that some species are more tolerant to drastic conditions than others (Table 2) and might necessitate species diversity over abundance. Additionally, the composition of feed gas should also be considered. As indicated in Figure 1, Types II and III methanotrophs can simultaneously consume CO2 and CH4 and should preferably be used for cultivation involving biogases (a mixture mainly composed of CH4 and CO2).

Table 2.
Temperature and pH growth conditions of different types of methanotrophs.
4. Conclusions
In this work, samples from several locations, including sediments from three open drainage ditches and sludges from different WWTPs, were collected to identify and quantitate different types of methanotrophs. Based on the 16S rRNA analysis, the samples from each location were composed of diverse types of methanotrophs including Type I (Methylobacter), Type X (Methylocaldum), Type II (Methylocystis, Methylosinus, and Beijerinckia), and Type III (Verrucomicrobium). Although different parameters could affect growth and proliferation of methanotrophs, this work focused mainly on O2 levels. As anticipated, samples collected from locations with low O2 levels (i.e., drainage ditches) contained non-detectable levels of Type I methanotrophs. The results of this work emphasize the importance of environmental conditions on the choice of a natural source of methanotrophic consortium. It should be noted, however, that other parameters might still need to be considered, along with target product(s) and cultivation conditions, to identify the most suitable natural consortium source for further studies.
Author Contributions
Conceptualization, E.D.R., M.E.Z., R.A.H., A.C., D.L.B.F. and L.S.H.D.; methodology, E.D.R., W.E.H. and L.S.H.D.; software, E.D.R. and L.S.H.D.; validation, E.D.R. and L.S.H.D.; formal analysis, E.D.R. and L.S.H.D.; investigation, L.S.H.D. and R.S.B.; writing—original draft preparation, L.S.H.D.; writing—review and editing, E.D.R., R.A.H., A.C., D.L.B.F., M.E.Z. and L.S.H.D.; supervision, E.D.R., R.A.H., A.C., D.L.B.F. and M.E.Z.; project administration, E.D.R.; funding acquisition, E.D.R., M.E.Z., R.A.H., A.C. and D.L.B.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by NASA EPSCoR (Grant No. 80NSSC18M0062) and Louisiana Board of Regents (ITRS) (Grant No. LEQSF(2019-22)-RD-B-06).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
This research was conducted with the support of the staff and students of the Energy Institute of Louisiana (EIL) at the University of Louisiana at Lafayette. The authors also acknowledge the financial supports from NASA EPSCoR and Louisiana Board of Regents (ITRS).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- EPA. Understanding Global Warming Potentials. Available online: https://www.epa.gov/ghgemissions/understanding-global-warming-potentials#:~:text=Methane%20(CH4)%20is%20estimated,less%20time%20than%20CO2 (accessed on 5 October 2022).
- EIA. Natural gas: Henry Hub Natural Gas Spot Price. Available online: https://www.eia.gov/dnav/ng/hist/rngwhhdm.htm (accessed on 7 August 2022).
- DOE. Natural Gas Flaring and Venting: State and Federal Regulatory Overview, Trends, and Impacts; U.S. Department of Energy: 2019. Available online: https://www.energy.gov/sites/prod/files/2019/08/f65/Natural%20Gas%20Flaring%20and%20Venting%20Report.pdf (accessed on 25 October 2022).
- The World Bank. Global Gas Flaring Reduction Partnership (GGFR). Available online: https://www.worldbank.org/en/programs/gasflaringreduction/global-flaring-data (accessed on 25 October 2022).
- Fei, Q.; Guarnieri, M.T.; Tao, L.; Laurens, L.M.; Dowe, N.; Pienkos, P.T. Bioconversion of natural gas to liquid fuel: Opportunities and challenges. Biotechnol. Adv. 2014, 32, 596–614. [Google Scholar] [CrossRef]
- AlSayed, A.; Fergala, A.; Eldyasti, A. Enhancement of the cultivation process conditions of mixed culture methanotrophic Proteobacteria phylum enriched from waste activated sludge as the first step for value added recovery process. J. Biosci. Bioeng. 2019, 127, 602–608. [Google Scholar] [CrossRef]
- Semrau, J.D.; Di Spirito, A.A.; Yoon, S. Methanotrophs and copper. FEMS Microbiol. Rev. 2010, 34, 496–531. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.M.; Su, Y.; Zhang, H.T.; Chen, M.; He, R. Responses of methanotrophic activity, community and EPS production to CH4 and O2 concentrations in waste biocover soils. Waste Manag. 2015, 42, 118–127. [Google Scholar] [CrossRef]
- Ge, X.; Yang, L.; Sheets, J.P.; Yu, Z.; Li, Y. Biological conversion of methane to liquid fuels: Status and opportunities. Biotechnol. Adv. 2014, 32, 1460–1475. [Google Scholar] [CrossRef] [PubMed]
- AlSayed, A.; Fergala, A.; Khattab, S.; Eldyasti, A. Kinetics of type I methanotrophs mixed culture enriched from waste activated sludge. Biochem. Eng. J. 2018, 132, 60–67. [Google Scholar] [CrossRef]
- Kasprzycka, A.; Lalak-Kańczugowska, J.; Walkiewicz, A.; Bulak, P.; Proc, K.; Stępień, Ł. Biocatalytic conversion of methane – selected aspects. Curr. Opin. Chem. Eng. 2019, 26, 28–32. [Google Scholar] [CrossRef]
- Jawaharraj, K.; Shrestha, N.; Chilkoor, G.; Dhiman, S.S.; Islam, J.; Gadhamshetty, V. Valorization of methane from environmental engineering applications: A critical review. Water Res. 2020, 187, 116400. [Google Scholar] [CrossRef] [PubMed]
- Pastor, J.M.; Salvador, M.; Argandoña, M.; Bernal, V.; Reina-Bueno, M.; Csonka, L.N.; Iborra, J.L.; Vargas, C.; Nieto, J.J.; Cánovas, M. Ectoines in cell stress protection: Uses and biotechnological production. Biotechnol. Adv. 2010, 28, 782–801. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, K.K.; Goswami, G.; Das, D. Biotransformation of Methane and Carbon Dioxide Into High-Value Products by Methanotrophs: Current State of Art and Future Prospects. Front. Microbiol. 2021, 12, 636486. [Google Scholar] [CrossRef] [PubMed]
- Cantera, S.; Lebrero, R.; Rodríguez, E.; García-Encina, P.A.; Muñoz, R. Continuous abatement of methane coupled with ectoine production by Methylomicrobium alcaliphilum 20Z in stirred tank reactors: A step further towards greenhouse gas biorefineries. J. Clean. Prod. 2017, 152, 134–141. [Google Scholar] [CrossRef]
- Guerrero-Cruz, S.; Vaksmaa, A.; Horn, M.A.; Niemann, H.; Pijuan, M.; Ho, A. Methanotrophs: Discoveries, Environmental Relevance, and a Perspective on Current and Future Applications. Front. Microbiol. 2021, 12, 678057. [Google Scholar] [CrossRef]
- Hanson, R.S.; Hanson, T.E. Methanotrophic bacteria. Microbiol. Rev. 1996, 60, 439–471. Available online: https://www.ncbi.nlm.nih.gov/pubmed/8801441 (accessed on 25 October 2022). [CrossRef] [PubMed]
- Karthikeyan, O.P.; Chidambarampadmavathy, K.; Nadarajan, S.; Heimann, K. Influence of nutrients on oxidation of low level methane by mixed methanotrophic consortia. Env. Sci. Pollut. Res. Int. 2016, 23, 4346–4357. [Google Scholar] [CrossRef] [PubMed]
- Kalyuzhnaya, M.G.; Puri, A.W.; Lidstrom, M.E. Metabolic engineering in methanotrophic bacteria. Metab. Eng. 2015, 29, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Amaral, J.A.; Knowles, R. Growth of methanotrophs in methane and oxygen counter gradients. FEMS Microbiol. Lett. 1995, 126, 215–220. [Google Scholar] [CrossRef]
- AlSayed, A.; Fergala, A.; Eldyasti, A. Sustainable biogas mitigation and value-added resources recovery using methanotrophs intergrated into wastewater treatment plants. Rev. Environ. Sci. Bio/Technol. 2018, 17, 351–393. [Google Scholar] [CrossRef]
- Bowman, J. The Methanotrophs—The Families Methylococcaceae and Methylocystaceae. In The Prokaryotes; Springer: New York, NY, USA, 2006; pp. 266–289. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).