In Silico Analysis of Toehold-Aptamer Sequences Targeting the SARS-CoV-2 Nucleocapsid Protein Gene for Biosensor Development †
Abstract
:1. Introduction
2. Computational Methods
2.1. Structure Retrieval
2.2. Multiple Sequence Alignment Analysis
2.3. Aptamers’ 2D-Structure Prediction
2.4. Aptamers’ 3D Modeling
2.5. Molecular Docking
3. Results and Discussion
3.1. Multiple Sequence Alignment Results
3.2. Aptamers’ 2D Structure and Free Energy
3.3. Aptamers’ 3D Modeling
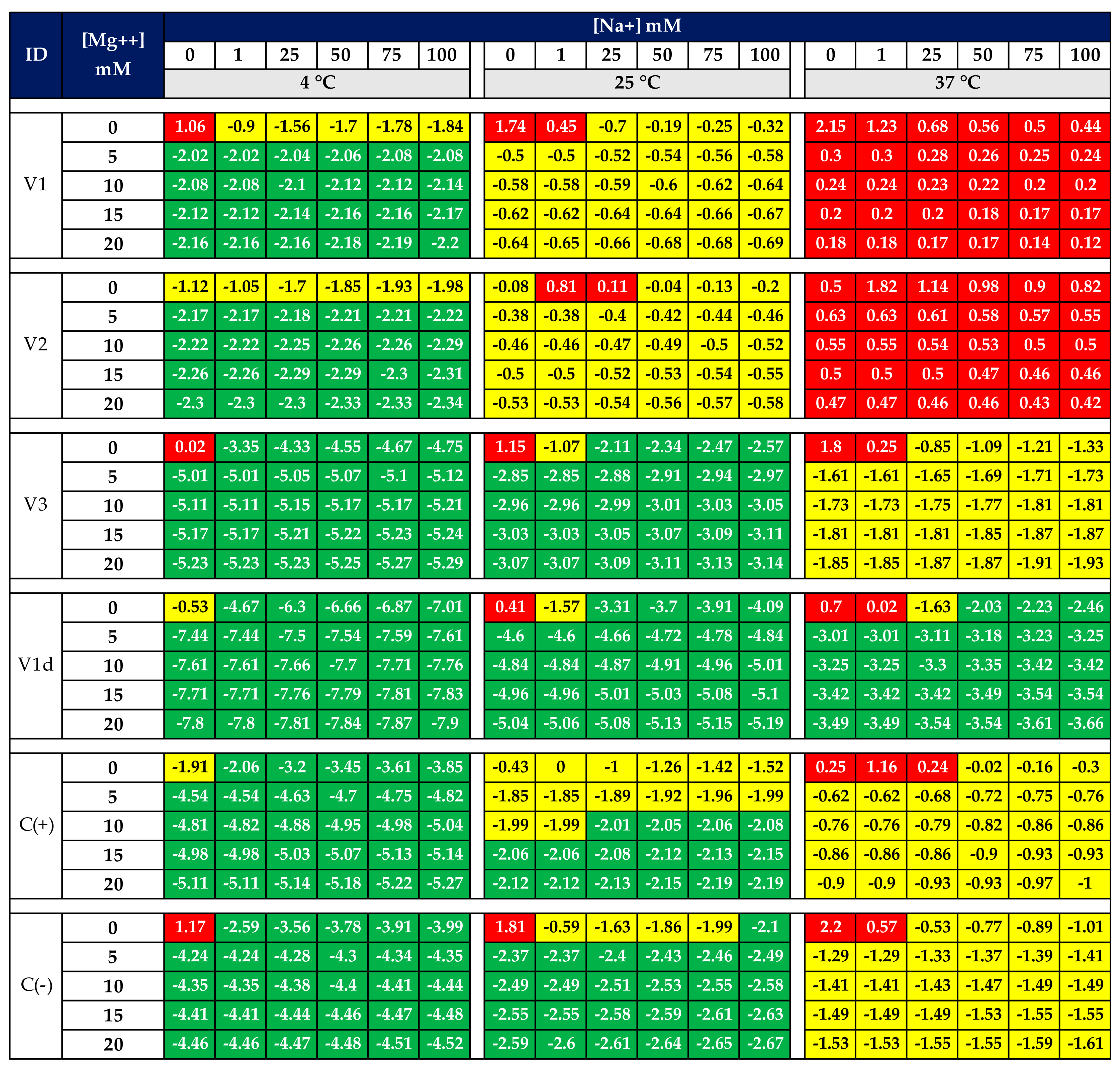
3.4. Molecular Docking
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| ID | Trigger | Length | Location |
|---|---|---|---|
| v1 | 5′- TCTTGGTTCACCGCTCTCACTCAA -3′ | 24 | 28,424–28,447 |
| v2 | 5′- CGCTCTCACTCAACATGGCAAGG -3′ | 23 | 28,435–28,457 |
| v3 | 5′- CTACGCAGAAGGGAGCAGAGGCGG -3′ | 24 | 28,786–28,809 |
| C(+) | 3′- GCAAATCTAGGCTTGCTGTTTGGG -5′ | 24 | |
| C(−) | 3′- TGCAGAAAAAAACCATGGGTTGGG -5′ | 24 |
References
- Wandtke, T.; Wędrowska, E.; Szczur, M.; Przybylski, G.; Libura, M.; Kopiński, P. Aptamers—Diagnostic and Therapeutic Solution in SARS-CoV-2. Int. J. Mol. Sci. 2022, 23, 1412. [Google Scholar] [CrossRef] [PubMed]
- Santosh, T.S.; Parmar, R.; Anand, H.; Srikanth, K.; Saritha, M. A review of salivary diagnostics and its potential implication in detection of COVID-19. Cureus 2020, 12, e7708. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Chen, Q.; Li, J.; Liu, Z. Diagnostic techniques for COVID-19: A mini-review. J. Virol. Methods 2022, 301, 114437. [Google Scholar] [CrossRef] [PubMed]
- García-López, R.; Laresgoiti-Servitje, E.; Lemus-Martin, R.; Sanchez-Flores, A.; Sanders-Velez, C. The New SARS-CoV-2 Variants and Their Epidemiological Impact in Mexico. Mbio 2022, 13, e01060-21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xiang, R.; Huo, S.; Zhou, Y.; Jiang, S.; Wang, Q.; Yu, F. Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduct. Target. Ther. 2021, 6, 233. [Google Scholar] [CrossRef] [PubMed]
- Gorgels, K.M.; Dingemans, J.; van der Veer, B.M.; Hackert, V.; Hensels, A.Y.; den Heijer, C.D.; van Alphen, L.B.; Savelkoul, P.H.M.; Hoebe, C.J. Linked nosocomial COVID-19 outbreak in three facilities for people with intellectual and developmental disabilities due to SARS-CoV-2 variant B. 1.1. 519 with spike mutation T478K in the Netherlands. BMC Infect. Dis. 2022, 22, 139. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Maldonado, A.P.; Vázquez-Pérez, J.A.; Cedro-Tanda, A.; Taboada, B.; Boukadida, C.; Wong-Arámbula, C.; Nuñez-García, T.E.; Cruz-Ortiz, N.; Barrera-Badillo, G.; Hernández-Rivas, L.; et al. Emergence and spread of the potential variant of interest (VOI) B. 1.1. 519 of SARS-CoV-2 predominantly present in Mexico. Arch. Virol. 2021, 166, 3173–3177. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.; Navale, G.R.; Dharne, M.S. Biosensors: Frontiers in rapid detection of COVID-19. 3 Biotech 2020, 10, 385. [Google Scholar] [CrossRef] [PubMed]
- Soler, M.; Estevez, M.C.; Cardenosa-Rubio, M.; Astua, A.; Lechuga, L.M. How nanophotonic label-free biosensors can contribute to rapid and massive diagnostics of respiratory virus infections: COVID-19 case. ACS Sens. 2020, 5, 2663–2678. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Stavins, R.; Kindratenko, V.; Baek, J.; Wang, L.; White, K.; Kumar, J.; Valera, E.; King, W.P.; Bashir, R. Microfluidic point-of-care device for detection of early strains and B. 1.1. 7 variant of SARS-CoV-2 virus. Lab Chip 2022, 22, 1297–1309. [Google Scholar] [CrossRef] [PubMed]
- Ozer, T.; Geiss, B.J.; Henry, C.S. Review Chemical and Biological Sensors for Viral Detection. J. Electrochem. Soc. 2020, 167, 37523. [Google Scholar] [CrossRef] [PubMed]
- Tothill, I.E.; Turner, A.P.F. Biosensors. 2003. Available online: https://www.sciencedirect.com/science/article/pii/B012227055X013742 (accessed on 12 January 2023).
- El-Sherif, D.M.; Abouzid, M.; Gaballah, M.S.; Ahmed, A.A.; Adeel, M.; Sheta, S.M. New approach in SARS-CoV2 surveillance using biosensor technology: A review. Environ. Sci. Pollut. Res. 2022, 29, 1677–1695. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Wang, T.; Zhang, Z.; Sheng, S.; Yu, W.; Xie, G. A novel colorimetric biosensor for detecting target DNA and human alpha thrombin based on associative toehold activation concatemer induced catalyzed hairpin assembly amplification. Sens. Actuators Chem. 2017, 239, 447–454. [Google Scholar] [CrossRef]
- Zhang, L.; Fang, X.; Liu, X.; Ou, H.; Zhang, H.; Wang, J.; Li, Q.; Cheng, H.; Zhang, W.; Luo, Z. Discovery of sandwich type COVID-19 nucleocapsid protein DNA aptamers. Chem. Commun. 2020, 56, 10235–10238. [Google Scholar] [CrossRef] [PubMed]
- Kothandan, R.; Uthayasooriyan, P.; Vairamani, S. Search for RNA aptamers against non-structural protein of SARS-CoV-2: Design using molecular dynamics approach. Beni-Suef Univ. J. Basic Appl. Sci. 2021, 10, 64. [Google Scholar] [CrossRef] [PubMed]
- Poolsup, S.; Zaripov, E.; Hüttmann, N.; Minic, Z.; Artyushenko, P.V.; Shchugoreva, I.A.; Tomilin, F.N.; Kichkailo, A.S.; Berezovski, M.V. Discovery of DNA aptamers targeting SARS-CoV-2 nucleocapsid protein and protein-binding epitopes for label-free COVID-19 diagnostics. Mol. Ther. Nucleic Acids 2023, 31, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wu, Q.; Chen, J.; Ni, X.; Dai, J. A DNA aptamer based method for detection of SARS-CoV-2 nucleocapsid protein. Virol. Sin. 2020, 35, 351–354. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.; Feng, J.; Zhang, J.; Hu, K.; Wang, D.; Zha, L.; Hu, X.; Li, R. Aptamer/antibody sandwich method for digital detection of SARS-CoV2 nucleocapsid protein. Talanta 2022, 236, 122847. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sczepanski, J.T. Targeting a conserved structural element from the SARS-CoV-2 genome using l-DNA aptamers. RSC Chem. Biol. 2022, 3, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Lin, C.; Hu, Y.; Zan, H.; Xu, X.; Sun, C.; Zou, H.; Li, Y. Sensitive fluorescence biosensor for SARS-CoV-2 nucleocapsid protein detection in cold-chain food products based on DNA circuit and g-CNQDs@ Zn-MOF. LWT 2022, 169, 114032. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esquivel-Ortiz, K.M.; Antonio-Pérez, A.; Torres-Huerta, A.L. In Silico Analysis of Toehold-Aptamer Sequences Targeting the SARS-CoV-2 Nucleocapsid Protein Gene for Biosensor Development. Eng. Proc. 2023, 35, 21. https://doi.org/10.3390/IECB2023-14718
Esquivel-Ortiz KM, Antonio-Pérez A, Torres-Huerta AL. In Silico Analysis of Toehold-Aptamer Sequences Targeting the SARS-CoV-2 Nucleocapsid Protein Gene for Biosensor Development. Engineering Proceedings. 2023; 35(1):21. https://doi.org/10.3390/IECB2023-14718
Chicago/Turabian StyleEsquivel-Ortiz, Karla M., Aurora Antonio-Pérez, and Ana L. Torres-Huerta. 2023. "In Silico Analysis of Toehold-Aptamer Sequences Targeting the SARS-CoV-2 Nucleocapsid Protein Gene for Biosensor Development" Engineering Proceedings 35, no. 1: 21. https://doi.org/10.3390/IECB2023-14718
APA StyleEsquivel-Ortiz, K. M., Antonio-Pérez, A., & Torres-Huerta, A. L. (2023). In Silico Analysis of Toehold-Aptamer Sequences Targeting the SARS-CoV-2 Nucleocapsid Protein Gene for Biosensor Development. Engineering Proceedings, 35(1), 21. https://doi.org/10.3390/IECB2023-14718







