Data-Driven Drug Repurposing in Diabetes Mellitus through an Enhanced Knowledge Graph †
Abstract
:1. Introduction
2. Materials and Methods
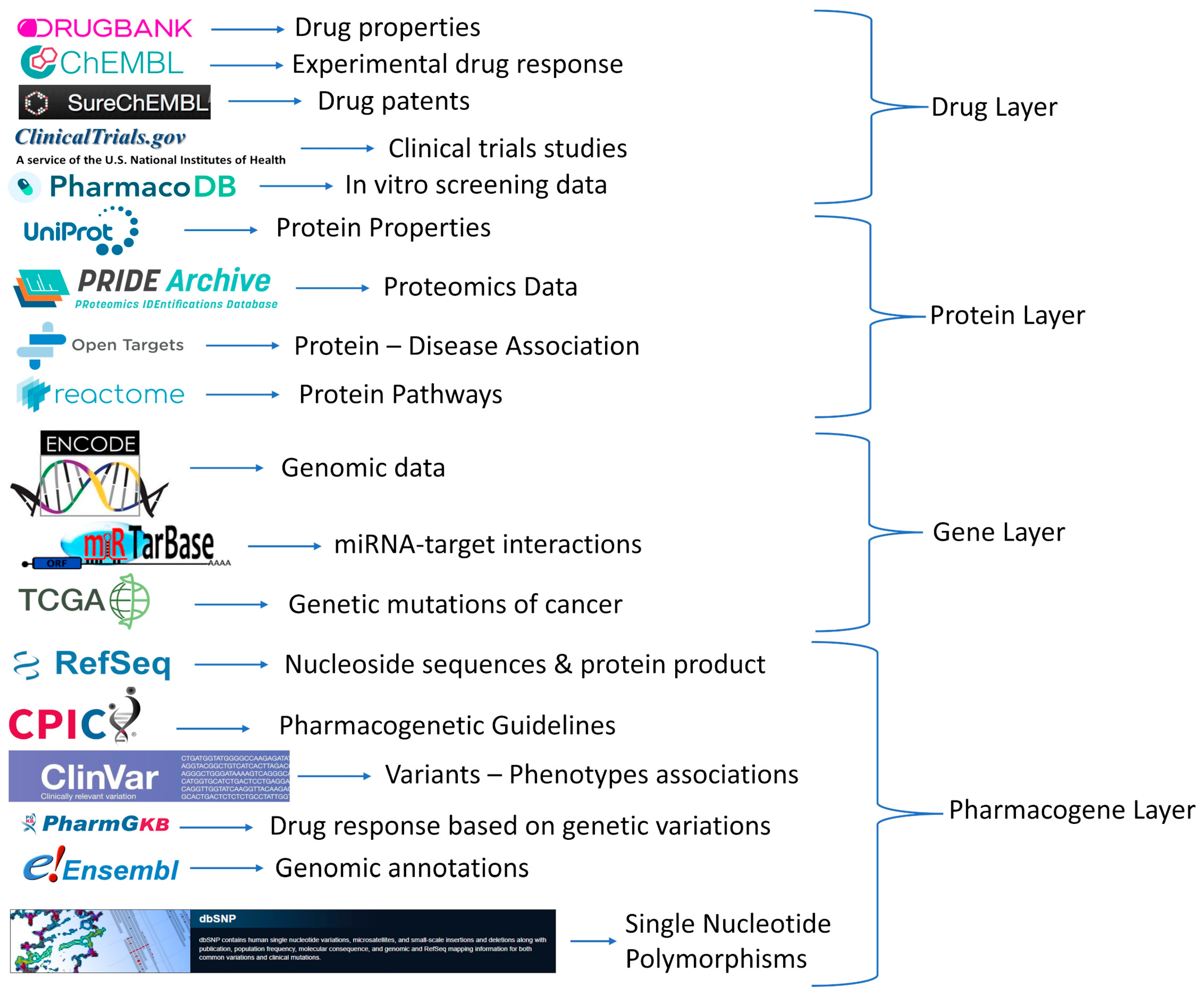
2.1. Databases and Repositories
2.2. Data Gathering
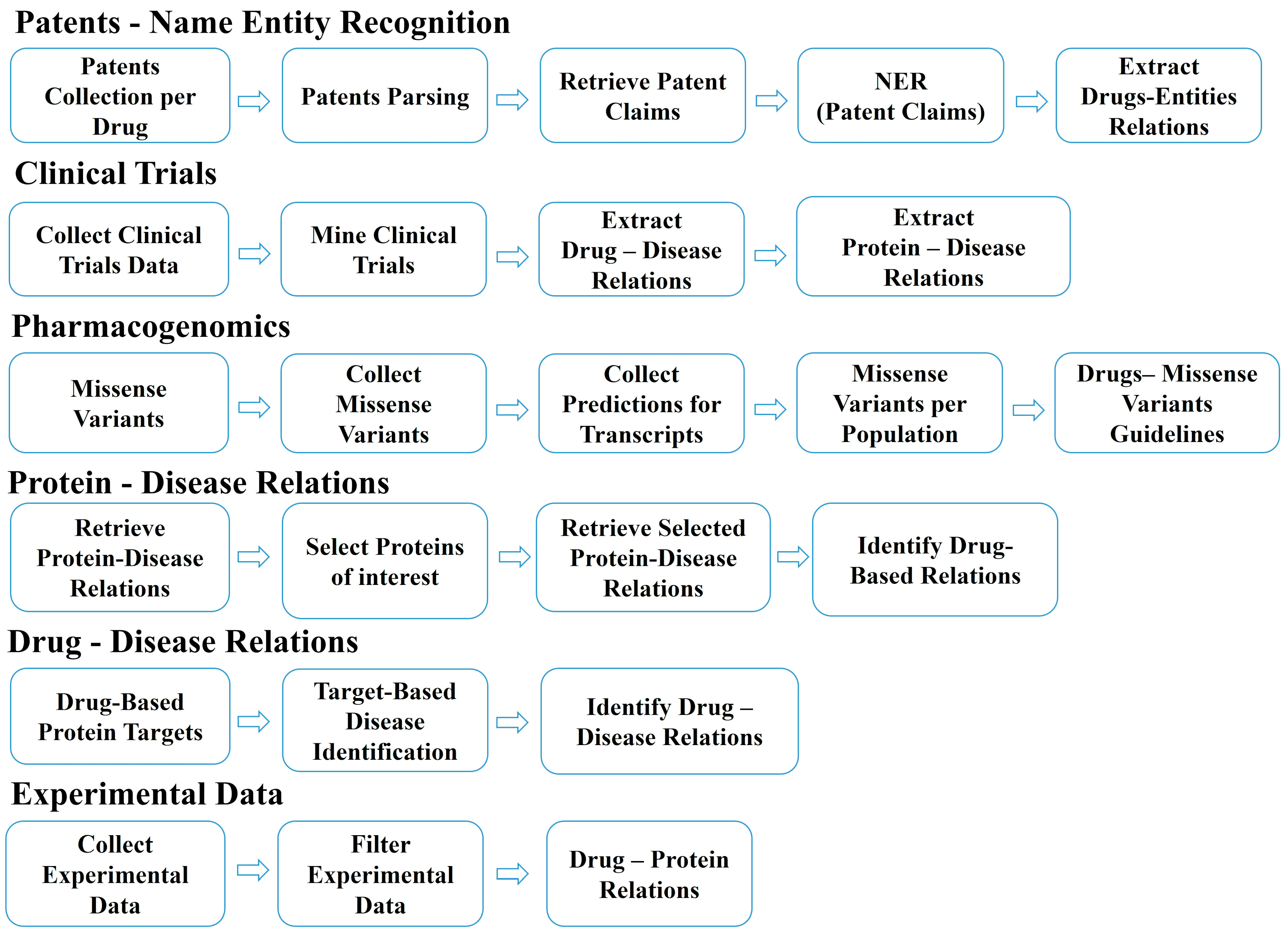
2.3. Information Extraction per Data Type
2.4. Molecular Docking
2.5. Data Integration in an Enhanced Knowledge Graph
2.6. Graph-Based Machine Learning
3. Results
3.1. Link Prediction through Machine Learning
- Random Forest: 500 trees (ntrees) with 65 features sampled during splitting at each node (mtry).
- Support Vector Machines: radial kernel basis function as kernel, sigma equal to 0.0043, and the cost of constraints violation (C) set to 1.
- k–Nearest Neighbors: 9 neighbors (k).
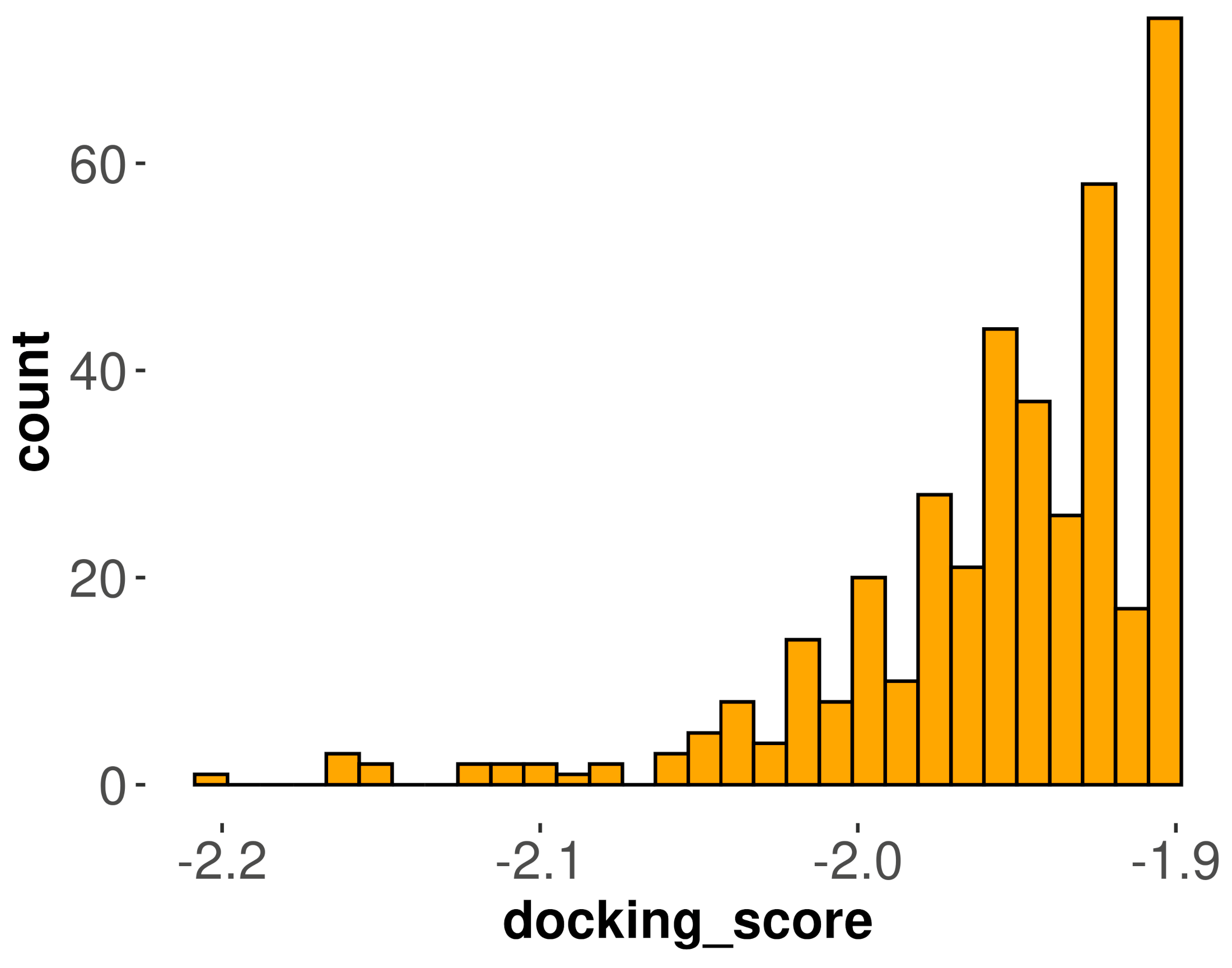
3.2. Molecular Docking Analysis for DPP-4 Inhibitors
3.3. Identifying Drug Repurposing Candidates
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- International Diabetes Federation. Five questions on the IDF Diabetes Atlas. Diabetes Res. Clin. Pract. 2013, 102, 147–148. [Google Scholar] [CrossRef] [PubMed]
- Banday, M.Z.; Sameer, A.S.; Nissar, S. Pathophysiology of Diabetes: An Overview. Avicenna J. Med. 2020, 10, 174–188. [Google Scholar] [CrossRef] [PubMed]
- Kahaly, G.J.; Hansen, M.P. Type 1 Diabetes Associated Autoimmunity. Autoimmun. Rev. 2016, 15, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Muoio, D.M.; Newgard, C.B. Mechanisms of Disease: Molecular and Metabolic Mechanisms of Insulin Resistance and β-Cell Failure in Type 2 Diabetes. Nat. Rev. Mol. Cell Biol. 2008, 9, 193–205. [Google Scholar] [CrossRef]
- Dahlén, A.D.; Dashi, G.; Maslov, I.; Attwood, M.M.; Jonsson, J.; Trukhan, V.; Schiöth, H.B. Trends in Antidiabetic Drug Discovery: FDA Approved Drugs, New Drugs in Clinical Trials and Global Sales. Front. Pharmacol. 2022, 12, 807548. [Google Scholar] [CrossRef]
- Chaudhury, A.; Duvoor, C.; Reddy Dendi, V.S.; Kraleti, S.; Chada, A.; Ravilla, R.; Marco, A.; Shekhawat, N.S.; Montales, M.T.; Kuriakose, K.; et al. Clinical Review of Antidiabetic Drugs: Implications for Type 2 Diabetes Mellitus Management. Front. Endocrinol. 2017, 8, 6. [Google Scholar] [CrossRef]
- Zhu, S.; Bai, Q.; Li, L.; Xu, T. Drug Repositioning in Drug Discovery of T2DM and Repositioning Potential of Antidiabetic Agents. Comput. Struct. Biotechnol. J. 2022, 20, 2839–2847. [Google Scholar] [CrossRef]
- Jarada, T.N.; Rokne, J.G.; Alhajj, R. A Review of Computational Drug Repositioning: Strategies, Approaches, Opportunities, Challenges, and Directions. J. Cheminformatics 2020, 12, 1–23. [Google Scholar] [CrossRef]
- Zeng, X.; Tu, X.; Liu, Y.; Fu, X.; Su, Y. Toward Better Drug Discovery with Knowledge Graph. Curr. Opin. Struct. Biol. 2022, 72, 114–126. [Google Scholar] [CrossRef]
- Ghorbanali, Z.; Zare-Mirakabad, F.; Akbari, M.; Salehi, N.; Masoudi-Nejad, A. DrugRep-KG: Toward Learning a Unified Latent Space for Drug Repurposing Using Knowledge Graphs. J. Chem. Inf. Model. 2023, 63, 2532–2545. [Google Scholar] [CrossRef]
- Zhu, Y.; Che, C.; Jin, B.; Zhang, N.; Su, C.; Wang, F. Knowledge-Driven Drug Repurposing Using a Comprehensive Drug Knowledge Graph. Health Informatics J. 2020, 26, 2737–2750. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Martin, M.J.; O’Donovan, C.; Magrane, M.; Alpi, E.; Antunes, R.; Bely, B.; Bingley, M.; Bonilla, C.; Britto, R.; et al. UniProt: The Universal Protein Knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar] [CrossRef]
- Papadatos, G.; Davies, M.; Dedman, N.; Chambers, J.; Gaulton, A.; Siddle, J.; Koks, R.; Irvine, S.A.; Pettersson, J.; Goncharoff, N.; et al. SureChEMBL: A Large-Scale, Chemically Annotated Patent Document Database. Nucleic Acids Res. 2016, 44, D1220–D1228. [Google Scholar] [CrossRef] [PubMed]
- Zarin, D.A.; Tse, T.; Williams, R.J.; Califf, R.M.; Ide, N.C. The ClinicalTrials.Gov Results Database—Update and Key Issues. N. Engl. J. Med. 2011, 364, 852–860. [Google Scholar] [CrossRef] [PubMed]
- Van Den Boom, D.; Wjst, M.; Everts, R.E. PharmGKB: The Pharmacogenomics Knowledge Base Caroline. Methods Mol. Biol. 2013, 1015, 71–85. [Google Scholar] [CrossRef]
- Türei, D.; Korcsmáros, T.; Saez-Rodriguez, J. OmniPath: Guidelines and Gateway for Literature-Curated Signaling Pathway Resources. Nat. Methods 2016, 13, 966–967. [Google Scholar] [CrossRef]
- Gillespie, M.; Jassal, B.; Stephan, R.; Milacic, M.; Rothfels, K.; Senff-Ribeiro, A.; Griss, J.; Sevilla, C.; Matthews, L.; Gong, C.; et al. The Reactome Pathway Knowledgebase 2022. Nucleic Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef]
- Howe, K.L.; Achuthan, P.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Ridwan Amode, M.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2021. Nucleic Acids Res. 2021, 49, D884–D891. [Google Scholar] [CrossRef]
- Relling, M.V.; Klein, T.E. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther. 2011, 89, 464–467. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving Access to Variant Interpretations and Supporting Evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef] [PubMed]
- Sherry, S.T.; Ward, M.; Sirotkin, K. DbSNP—Database for Single Nucleotide Polymorphisms and Other Classes of Minor Genetic Variation. Genome Res. 1999, 9, 677–679. [Google Scholar] [CrossRef] [PubMed]
- Mendez, D.; Gaulton, A.; Bento, A.P.; Chambers, J.; De Veij, M.; Félix, E.; Magariños, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. ChEMBL: Towards Direct Deposition of Bioassay Data. Nucleic Acids Res. 2019, 47, D930–D940. [Google Scholar] [CrossRef]
- Huang, H.Y.; Lin, Y.C.D.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. MiRTarBase Update 2022: An Informative Resource for Experimentally Validated MiRNA-Target Interactions. Nucleic Acids Res. 2022, 50, D222–D230. [Google Scholar] [CrossRef]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Sander, C.; Stuart, J.M.; Chang, K.; Creighton, C.J.; et al. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Feizi, N.; Nair, S.K.; Smirnov, P.; Beri, G.; Eeles, C.; Esfahani, P.N.; Nakano, M.; Tkachuk, D.; Mammoliti, A.; Gorobets, E.; et al. PharmacoDB 2.0: Improving Scalability and Transparency of in Vitro Pharmacogenomics Analysis. Nucleic Acids Res. 2022, 50, D1348–D1357. [Google Scholar] [CrossRef]
- Ochoa, D.; Hercules, A.; Carmona, M.; Suveges, D.; Gonzalez-Uriarte, A.; Malangone, C.; Miranda, A.; Fumis, L.; Carvalho-Silva, D.; Spitzer, M.; et al. Open Targets Platform: Supporting Systematic Drug-Target Identification and Prioritisation. Nucleic Acids Res. 2021, 49, D1302–D1310. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Ouzounis, S.; Panagiotopoulos, V.; Bafiti, V.; Zoumpoulakis, P.; Cavouras, D.; Kalatzis, I.; Matsoukas, M.-T.; Katsila, T. Molecular Representations Predicts CYP450 Inhibition: Toward Precision in Drug Repurposing. OMICS A J. Integr. Biol. 2023, 27, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, T.M.; Bauer, M.R.; Boeckler, F.M. Applying DEKOIS 2.0 in Structure-Based Virtual Screening to Probe the Impact of Preparation Procedures and Score Normalization. J. Cheminformatics 2015, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Sultan, S.F.; Tian, Y.; Chen, M.; Skiena, S. Fast and Accurate Network Embeddings via Very Sparse Random Projection. In Proceedings of the 28th ACM International Conference on Information and Knowledge Management, Beijing, China, 3–7 November 2019; pp. 399–408. [Google Scholar] [CrossRef]

| Metric | 10-Fold Cross-Validation | External Test Set | ||||
|---|---|---|---|---|---|---|
| RF | SVM | KNN | RF | SVM | KNN | |
| Accuracy | 95.23 | 96.62 | 95.89 | 95.12 | 96.68 | 95.89 |
| Precision | 97.32 | 98.91 | 97.76 | 97.17 | 98.94 | 97.71 |
| Recall | 85.75 | 86.26 | 87.42 | 85.86 | 86.47 | 87.68 |
| MCC | 0.83 | 0.88 | 0.86 | 0.83 | 0.88 | 0.86 |
| AUC | 0.96 | 0.97 | 0.97 | 0.96 | 0.97 | 0.97 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ouzounis, S.; Kanterakis, A.; Panagiotopoulos, V.; Cavouras, D.; Zoumpoulakis, P.; Matsoukas, M.-T.; Katsila, T.; Kalatzis, I. Data-Driven Drug Repurposing in Diabetes Mellitus through an Enhanced Knowledge Graph. Eng. Proc. 2023, 50, 9. https://doi.org/10.3390/engproc2023050009
Ouzounis S, Kanterakis A, Panagiotopoulos V, Cavouras D, Zoumpoulakis P, Matsoukas M-T, Katsila T, Kalatzis I. Data-Driven Drug Repurposing in Diabetes Mellitus through an Enhanced Knowledge Graph. Engineering Proceedings. 2023; 50(1):9. https://doi.org/10.3390/engproc2023050009
Chicago/Turabian StyleOuzounis, Sotiris, Alexandros Kanterakis, Vasilis Panagiotopoulos, Dionisis Cavouras, Panagiotis Zoumpoulakis, Minos-Timotheos Matsoukas, Theodora Katsila, and Ioannis Kalatzis. 2023. "Data-Driven Drug Repurposing in Diabetes Mellitus through an Enhanced Knowledge Graph" Engineering Proceedings 50, no. 1: 9. https://doi.org/10.3390/engproc2023050009
APA StyleOuzounis, S., Kanterakis, A., Panagiotopoulos, V., Cavouras, D., Zoumpoulakis, P., Matsoukas, M.-T., Katsila, T., & Kalatzis, I. (2023). Data-Driven Drug Repurposing in Diabetes Mellitus through an Enhanced Knowledge Graph. Engineering Proceedings, 50(1), 9. https://doi.org/10.3390/engproc2023050009









