Grading of Melanoma Tissues by Raman MicroSpectroscopy †
Abstract
:1. Introduction
2. Materials and Methods
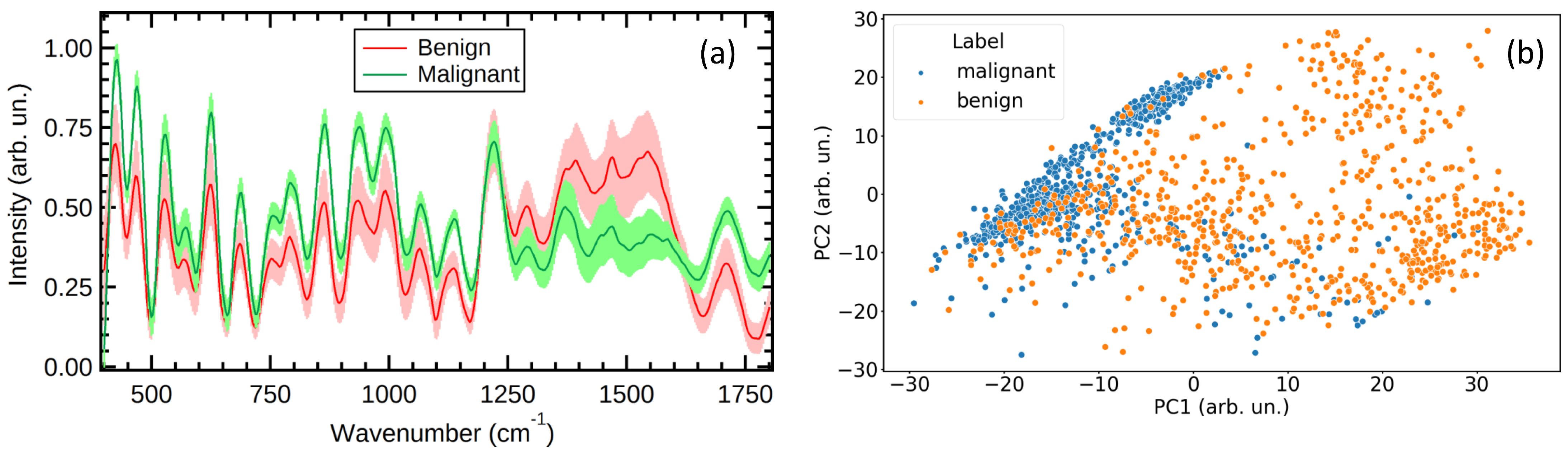
3. Results
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Larson, A.R.; Konat, E.; Alani, R.M. Melanoma biomarkers: Current status and vision for the future. Nat. Clin. Pract. Oncol. 2009, 6, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Araújo, D.C.; Veloso, A.A.; de Oliveira Filho, R.S.; Giraud, M.N.; Raniero, L.J.; Ferreira, L.M.; Bitar, R.A. Finding reduced Raman spectroscopy fingerprint of skin samples for melanoma diagnosis through machine learning. Artif. Intell. Med. 2021, 120, 102161. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, T.G.; Lee, S.H.; Kim, W.; Bang, A.; Moon, S.W.; Song, J.; Shin, J.H.; Yu, J.S.; Choi, S. Label-free surface-enhanced Raman spectroscopy biosensor for on-site breast cancer detection using human tears. ACS Appl. Mater. Interfaces 2020, 12, 7897–7904. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, R.; Daniel, A.; Lyng, F.M. Raman Spectroscopy for Early Detection of Cervical Cancer, a Global Women’s Health Issue—A Review. Molecules 2023, 28, 2502. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Li, Y.; Huang, F.; Dong, J.; Li, F.; Yang, X.; Zhu, S.; Yang, M. Identification and analysis of serum samples by surface-enhanced Raman spectroscopy combined with characteristic ratio method and PCA for gastric cancer detection. J. Innov. Opt. Health Sci. 2019, 12, 1950003. [Google Scholar] [CrossRef]
- Fox, S.A.; Shanblatt, A.A.; Beckman, H.; Strasswimmer, J.; Terentis, A.C. Raman spectroscopy differentiates squamous cell carcinoma (SCC) from normal skin following treatment with a high-powered CO2 laser. Lasers Surg. Med. 2014, 46, 757–772. [Google Scholar] [CrossRef] [PubMed]
- Baria, E.; Cicchi, R.; Malentacchi, F.; Mancini, I.; Pinzani, P.; Pazzagli, M.; Pavone, F.S. Supervised learning methods for the recognition of melanoma cell lines through the analysis of their Raman spectra. J. Biophotonics 2021, 14, 202000365. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Lui, H.; McLean, D.I.; Zeng, H. Automated autofluorescence background subtraction algorithm for biomedical Raman spectroscopy. Appl. Spectrosc. 2007, 61, 1225–1232. [Google Scholar] [CrossRef] [PubMed]
- Gautam, R.; Vanga, S.; Ariese, F.; Umapathy, S. Review of multidimensional data processing approaches for Raman and infrared spectroscopy. EPJ Tech. Instrum. 2015, 2, 1–38. [Google Scholar] [CrossRef]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef] [PubMed]
- Fawagreh, K.; Gaber, M.M.; Elyan, E. Random forests: From early developments to recent advancements. Syst. Sci. Control. Eng. Open Access J. 2014, 2, 602–609. [Google Scholar] [CrossRef]
- Stone, M. Cross-validation: A review. Stat. J. Theor. Appl. Stat. 1978, 9, 127–139. [Google Scholar]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Lima, A.M.F.; Daniel, C.R.; Navarro, R.S.; Bodanese, B.; Pasqualucci, C.A.; Pacheco, M.T.T.; Zângaro, R.A.; Silveira, L., Jr. Discrimination of non-melanoma skin cancer and keratosis from normal skin tissue in vivo and ex vivo by Raman spectroscopy. Vib. Spectrosc. 2019, 100, 131–141. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lazzini, G.; D’Acunto, M. Grading of Melanoma Tissues by Raman MicroSpectroscopy. Eng. Proc. 2023, 51, 10. https://doi.org/10.3390/engproc2023051010
Lazzini G, D’Acunto M. Grading of Melanoma Tissues by Raman MicroSpectroscopy. Engineering Proceedings. 2023; 51(1):10. https://doi.org/10.3390/engproc2023051010
Chicago/Turabian StyleLazzini, Gianmarco, and Mario D’Acunto. 2023. "Grading of Melanoma Tissues by Raman MicroSpectroscopy" Engineering Proceedings 51, no. 1: 10. https://doi.org/10.3390/engproc2023051010





