Abstract
One of the most widely used imaging techniques in medicine is magnetic resonance imaging (MRI). It is a tool that doctors use to comprehend human anatomy and carry out more accurate analyses. In the study of brain anatomy, image processing super resolution technology has become important to overcome physical restrictions due to image deterioration caused by hardware constraints, lengthier scanning periods, and artefacts. Super resolution is an approach to raise an image’s resolution while improving the image’s quality from a low-resolution (LR) image to a higher-resolution (HR) image. The study provides an overview of deep learning techniques for creating super-resolution (SR) MRI brain images. A widely used deep learning (DL) technique, accessible brain MRI dataset, and quantity evaluation matrices have been presented, mostly used for image super resolution. Factors affecting hardware constraints and artifacts, including magnetic field homogeneity, gradient nonlinearity, radiofrequency (RF) coil sensitivity, signal-to-noise ratio (SNR), and gradient coil performance, have been taken into account. This research focuses mostly on brain MRI images as a contribution to the medical industry for super resolution.
1. Introduction
Super resolution image processing techniques, such as MRI, are crucial in image processing and computer vision. They recover high-resolution images from low-resolution images, providing insights into specific diseases and aiding doctors in diagnosing brain illnesses [1,2]. Interpolation methods, including reconstruction, learning, and interpolation, are used to reconstruct high-resolution images from low-resolution data [3,4]. The paper’s primary objectives are:
- (a)
- Examine the deep learning framework used for super resolution, specifically for MRI brain images, to overcome physical constraints and artefacts that reduce image quality.
- (b)
- Analyze the brain MRI dataset commonly used for the super resolution approach to deliver exact diagnosis.
- (c)
- Image quality assessment (IQA) for evaluating an image’s visual quality is covered for several imaging modalities.
2. Materials and Methods/Methodology
The study uses deep convolutional layers in SRCNN architecture to learn intricate features between low-resolution and high-resolution picture patches [5]. The network learns in hierarchical formats and directly maps low-resolution patches to high-resolution patches without intermediary stages [6,7]. The study minimizes mean squared error and employs techniques like wavelet-based MRI denoising, reconstruction, noise filtering, noise estimation, and restoration.
2.1. Supervised or Discriminative Learning
To perform specific data-processing tasks, the entire model is trained using input-output samples [8].
2.1.1. Convolutional Neural Network for Super-Resolution for MRI Brain Image
A CNN is an artificial neural network with few connections between the layers and an emphasis on maintaining spatial correlations in the data. With the input structured in a grid and transferred across layers that maintain these linkages, each layer action in a CNN operates on a small portion of the preceding layer [9,10].
2.1.2. Densely Connected Architecture for Super Solution of the MRI Dataset
The application of deep learning, CNN, has greatly improved the resolution of MRI datasets. As can be seen, it has used (DCSRN) 3D Densely Connected Super-Resolution Networks [11]. When techniques like bicubic interpolation, nearest neighbor up-sampling, and three other deep learning models— 3D-FSRCNN, DCSRN, and 2D-FSRCNN—were compared, it was shown that the DCSRN-trained model was the fastest deep learning model [12].
2.1.3. Residual Learning for Super-Resolution of the MRI Dataset
The dilated convolutional neural network model uses skip-connection, which mixes local texture information with global abstraction information, to achieve better super-resolution performance [13]. The gradient-guided residual network in [14,15] is therefore based on two intuitions. The recovery of higher-frequency information in a high-resolution image is facilitated by the CNN-based SR approach and gradient features of the LR image [16].
2.2. Unsupervised or Generative Learning
A deep learning method called unsupervised super-resolution enhances image resolution without the need for annotated data. An unsupervised super-resolution method known as “Generative Adversarial Networks” (GANs) consists of two networks: a generator network and a discriminator network [17].
2.2.1. Generative Adversarial Networks (GAN) for Super-Resolution of MRI Brain Image
Generative modeling is a machine learning task that uses unsupervised learning to identify patterns in input data. It has been compared to modern models like Pix2Pix, DECNN, LA-GANs, and MedGAN, demonstrating improved performance in brain MRIs [18].
2.2.2. Reduce Scan Time Using GAN Methods
Potential applications exist for a deep-learning SR approach that interpolates medical images using a 3D perceptually tuned GAN network [19]. Deep learning architecture is used for de-aliasing and conditional generative adversarial network-based rapid CS-MRI. Generator network architecture (GAN) was used for skip connections and the GAN training technique that is constantly enhanced [20].
2.3. Hybrid Architecture
Deep learning hybrid models combine generative and discriminative models to tackle practical issues [21]. Three types include combining multiple models, fused models, and non-deep learning classifiers. Brain MRI images can be generated using deep learning models like CNNs, MobileNetV2, ResNet152V2, and GAN-based augmentation methods. A mixed convolutions algorithm is proposed for this purpose [22,23,24,25].
3. Results and Discussions
When digital photos are acquired, processed, compressed, stored, transmitted, or reproduced, a number of distortions might occur that could degrade the visual quality. Image quality assessment (IQA) is used to evaluate an image’s authenticity to its intended or original images. IQA is broken into two types: (i) subjective evaluation, which is evaluated by humans, and (ii) objective assessment, which is calculated using mathematical methods.
3.1. Qualitative and Quantitative Analysis
3.1.1. Peak Signal-to-Noise Ratio (PSNR)
PSNR is the most often used quality measures for loss reconstruction. For the purpose of calculating the PSNR for image super-resolution, the MSE between images and the maximum pixel value is employed.
3.1.2. Structure Similarity Index (SSIM)
A flawless reference image and a test image are compared side by side for brightness, contrast, and structural attributes via SSIM. A few elements were then added together to determine the total similarity.
3.1.3. Optimizers
ADAM (Adaptive Moment Estimation): ADAM adjusts the learning rates based on the first and second moments of the gradients for each parameter. ADAM uses bias correction in the early training phases to correct the initial bias and guarantee stronger convergence. Regardless of the quantity of arguments, ADAM keeps a certain number of state variables. As a result, it functions effectively with huge models and uses less RAM.
SGD (Stochastic Gradient Descent): SGD is suited for large datasets due to its stochastic nature, which speeds up processing. SGD is more effective than batch gradient descent because it changes the model parameters after handling each mini-batch. Faster convergence is possible thanks to frequent parameter updates, particularly in noisy or non-conv situations.
RMSProp: The learning rate for each parameter in the model is altered by RMSProp according to the historical average of squared gradients. The learning rate is dynamically altered by computing a running average of the magnitudes of earlier gradients. Some of the difficulties involved in manually choosing an acceptable learning rate are lessened by this adaptive learning rate.
3.1.4. Loss Function
The following loss functions are used during experiments. These are Mean Square Error (MSE), Mean Absolute Error (MAE), and Perceptual Loss.
3.2. Performance of ADAM Optimizer and Use of Loss Functions Using Super Resolution Convolution Neural Network (SRCNN)
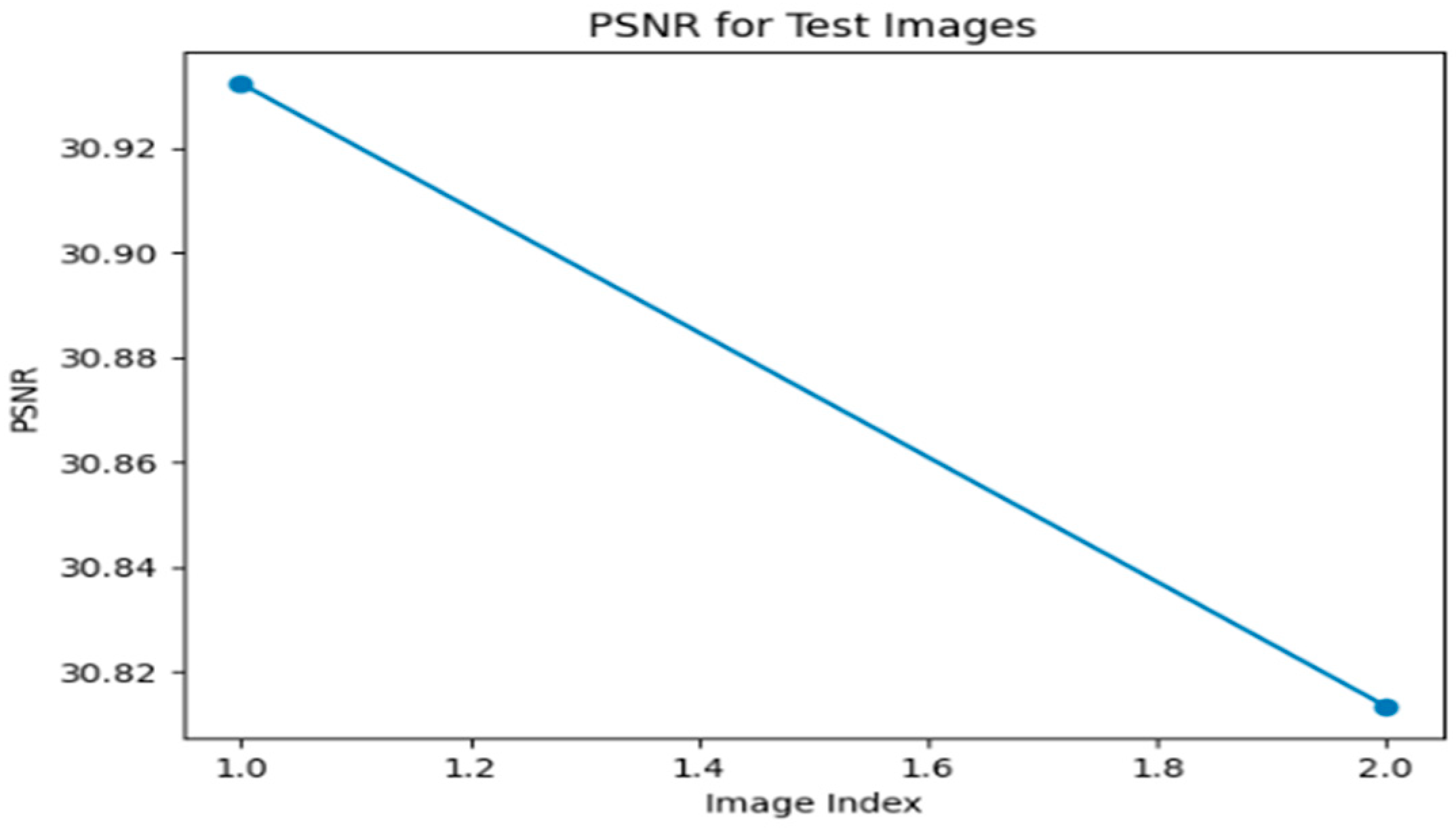
Figure 1, Figure 2, Figure 3, Figure 4, Figure 5 and Figure 6 show the performance of ADAM optimizer and MSE, MAE, and perceptual loss functions which are used for error computation using the Super Resolution Convolution Neural Network (SRCNN).

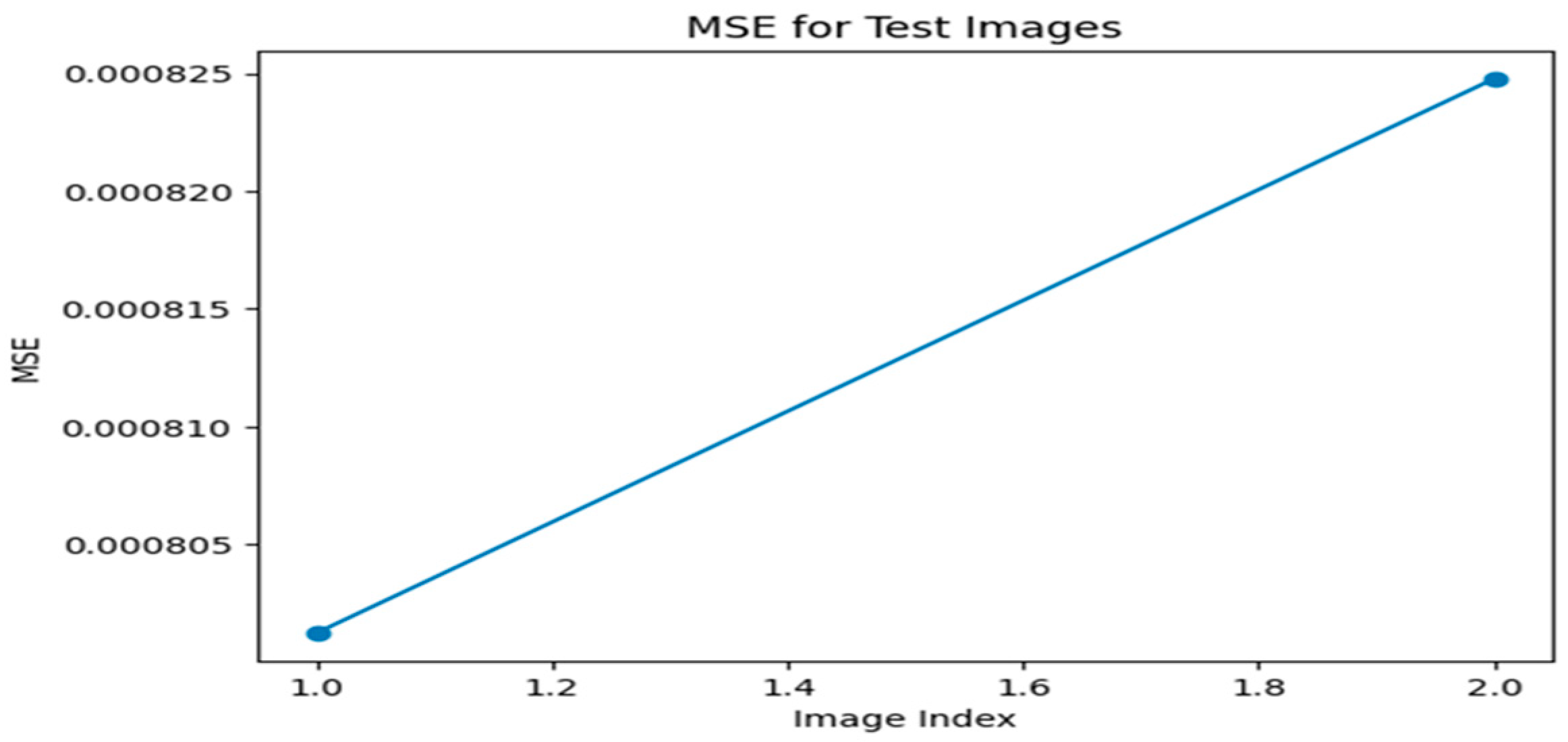
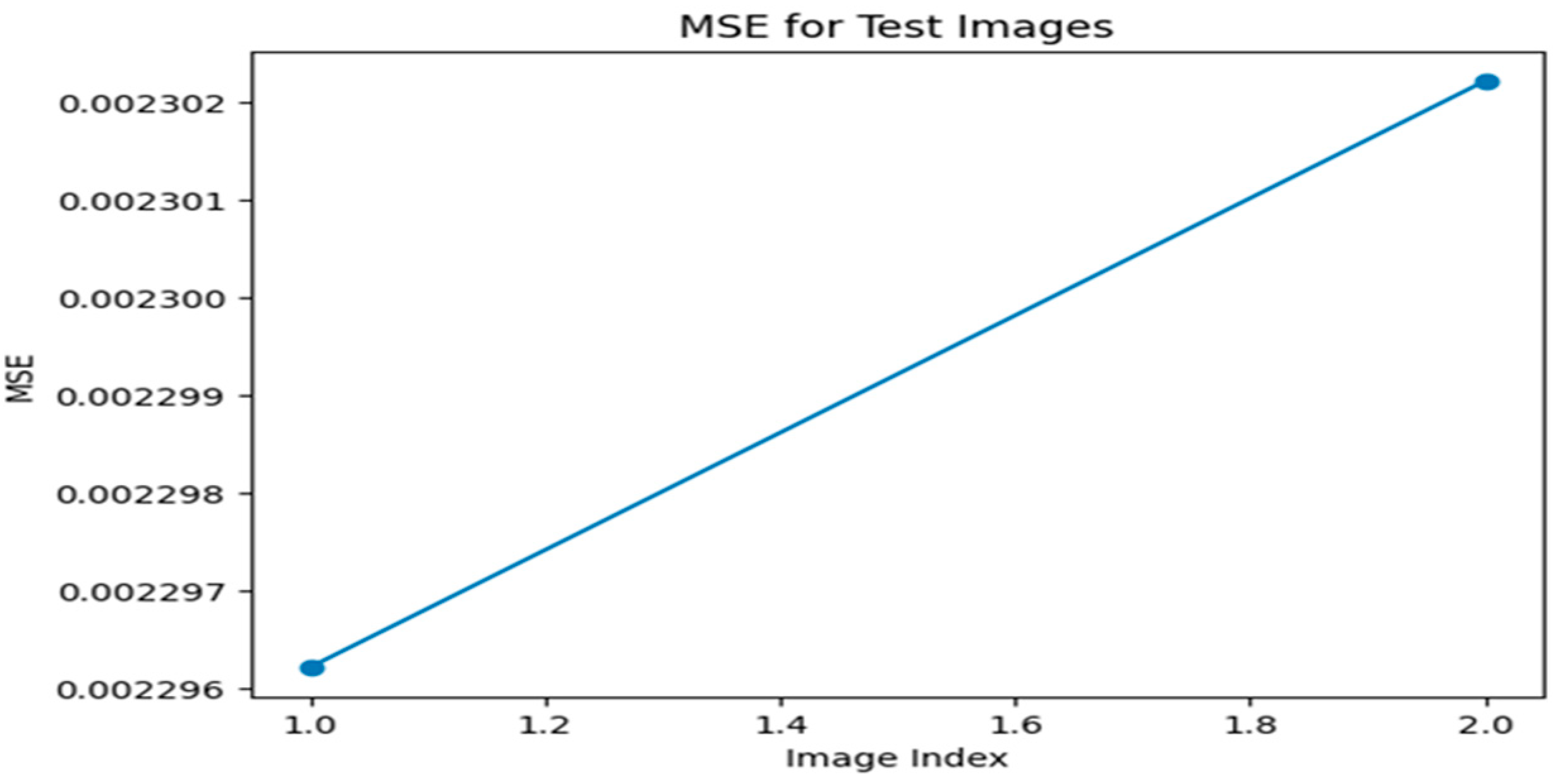
Figure 1.
MSE graph for Optimizer—ADAM, Loss function—MSE.

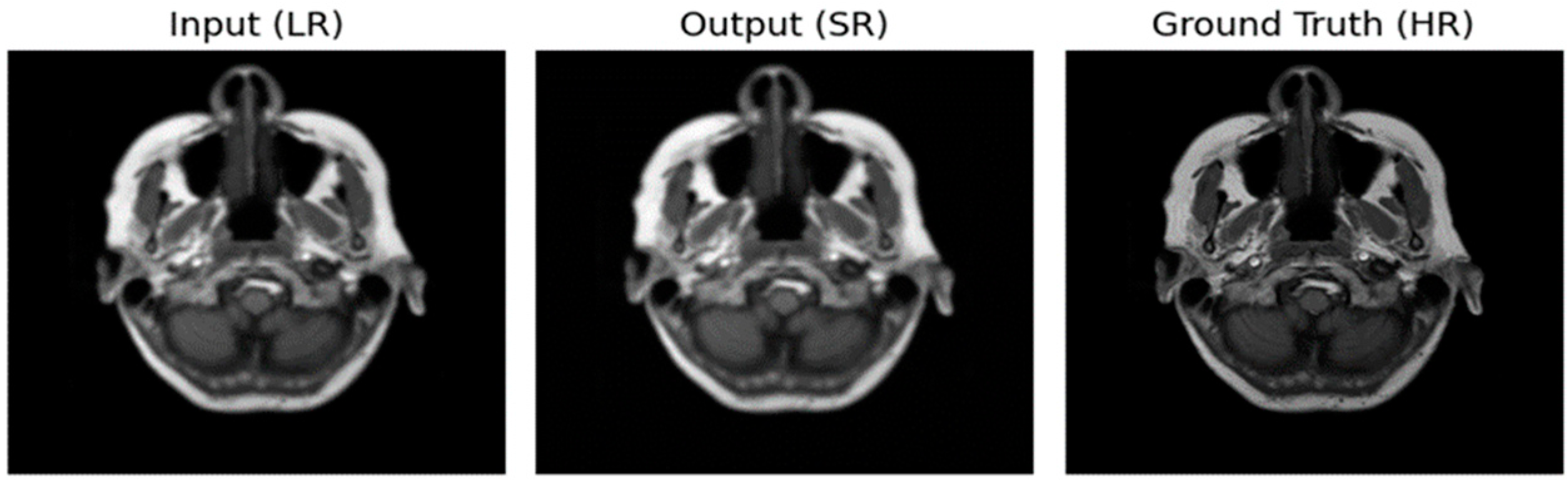
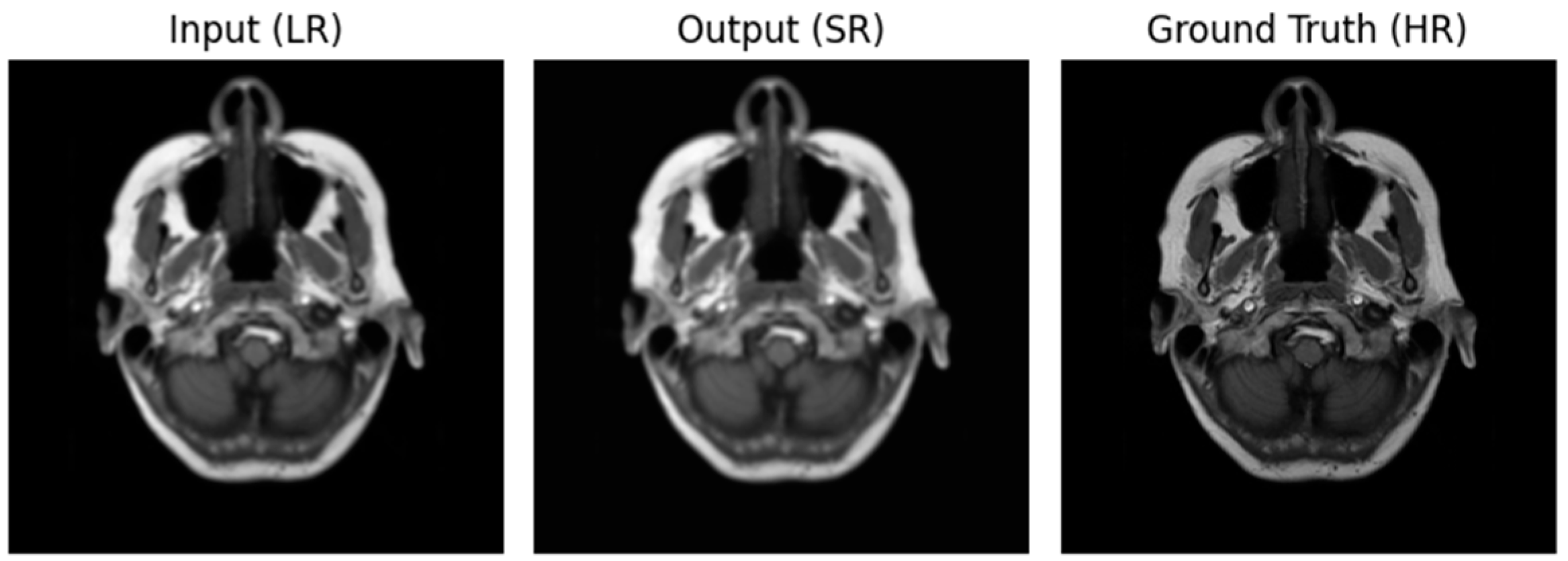
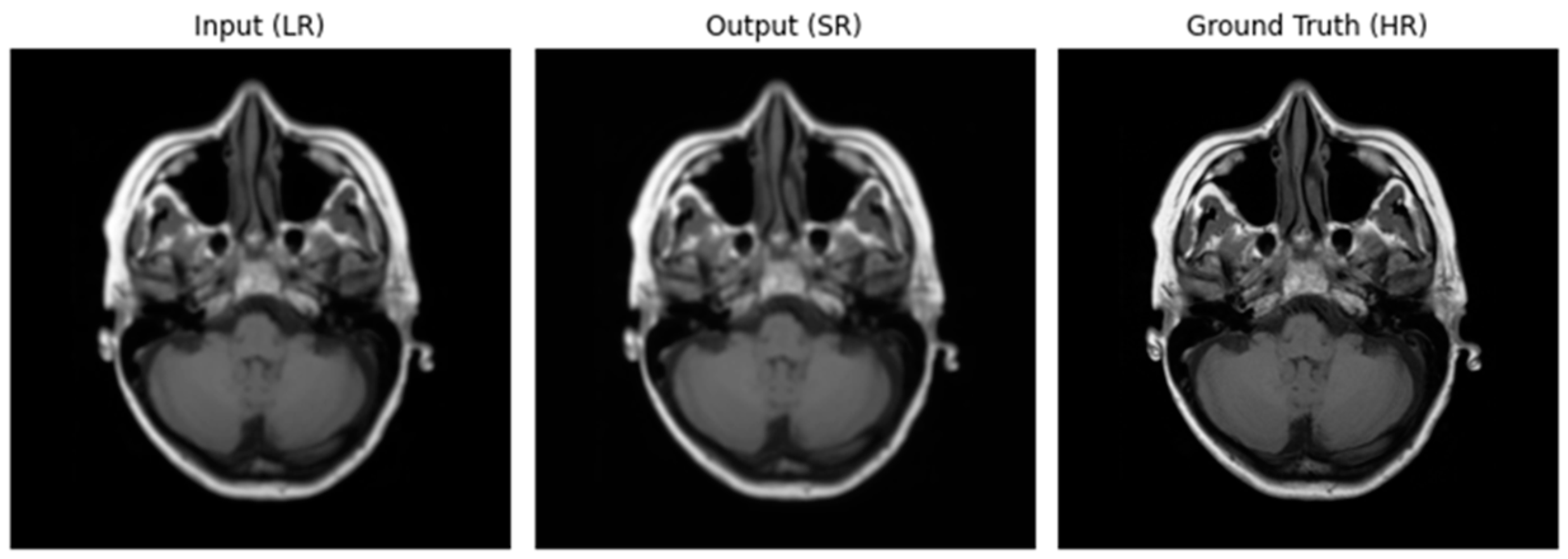
Figure 2.
Image using Optimizer—ADAM, Loss function—MSE.

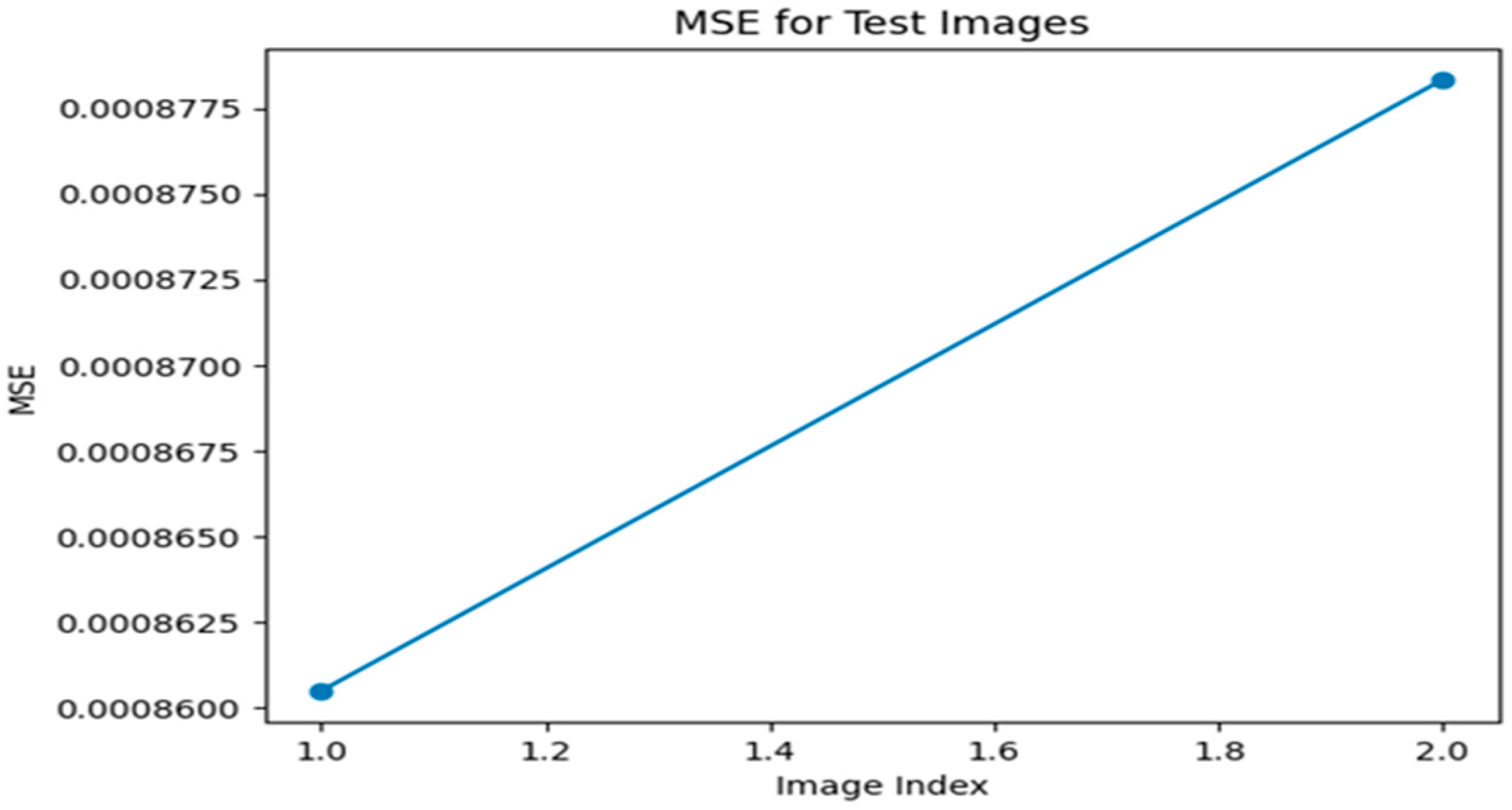
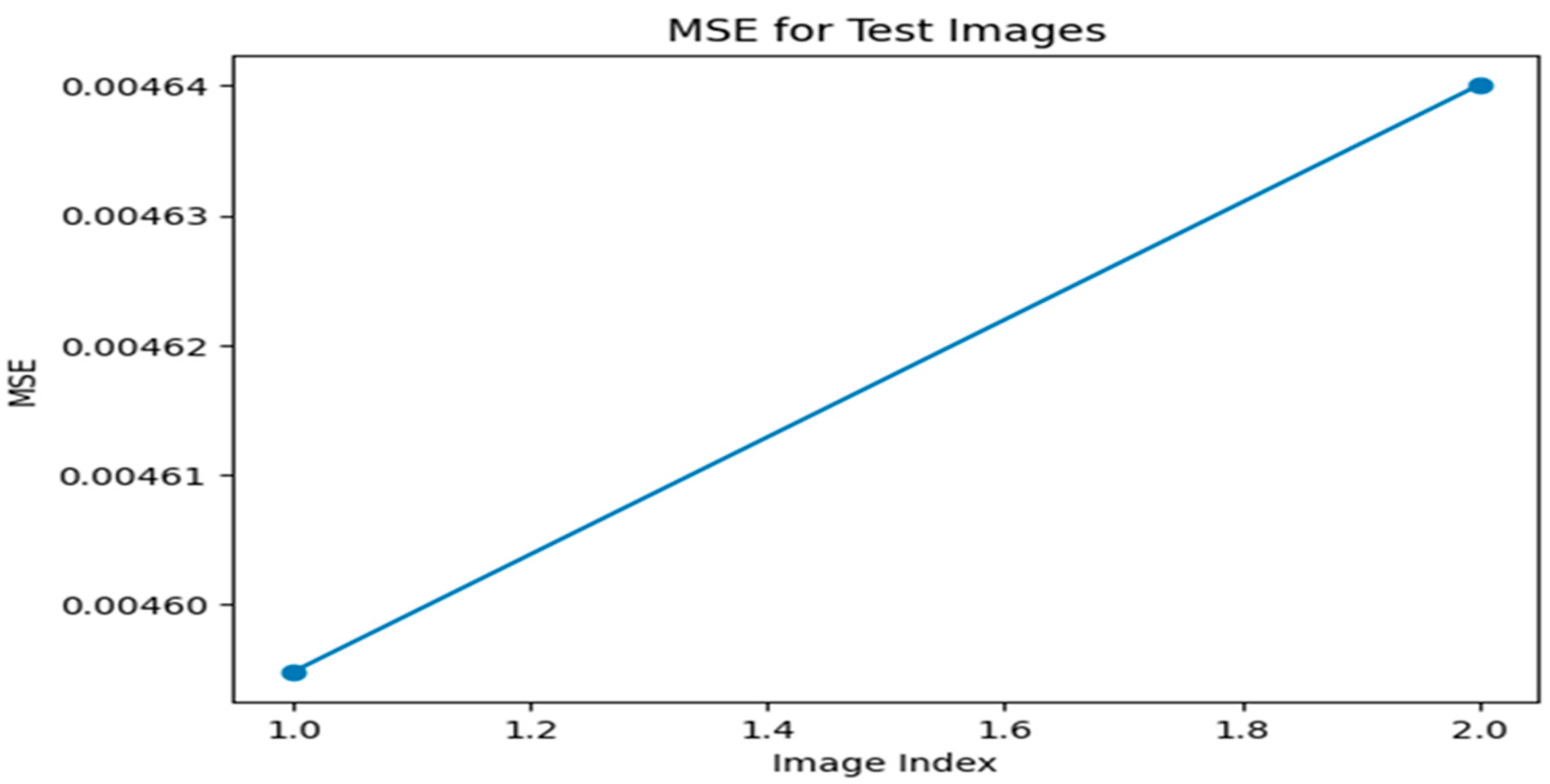
Figure 3.
MSE graph for Optimizer—ADAM, loss function—MAE.

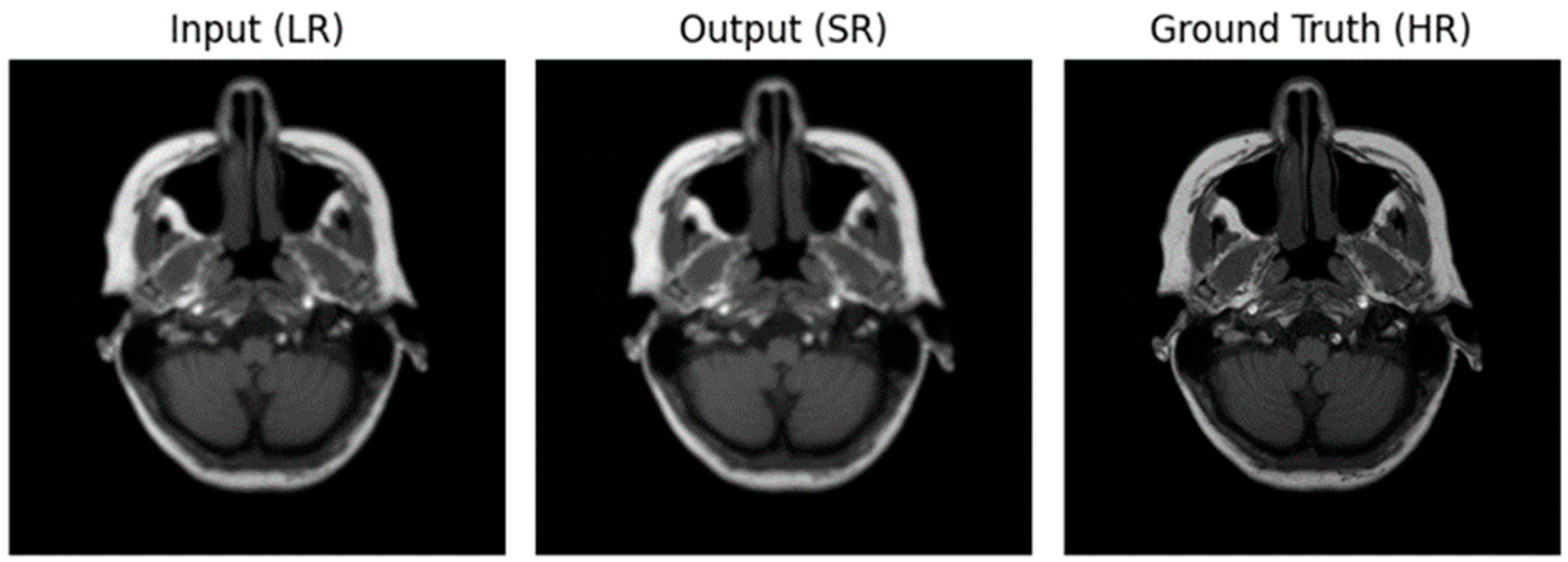
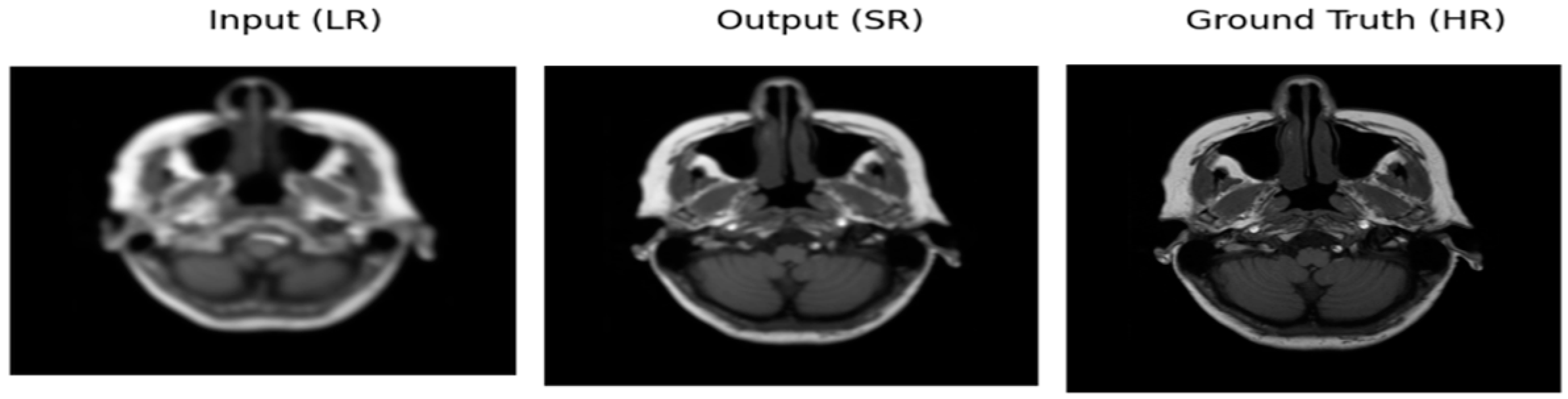
Figure 4.
Image for Optimizer—ADAM, loss function—MAE.

Figure 5.
MSE graph for Optimizer—Adam, loss function—Perceptual.

Figure 6.
Image using the Optimizer as Adam and loss function as Perceptual.
3.3. Performance of ADAM Optimizer and Use of Loss Functions Using Super Resolution Residual Network (SR ResNet)
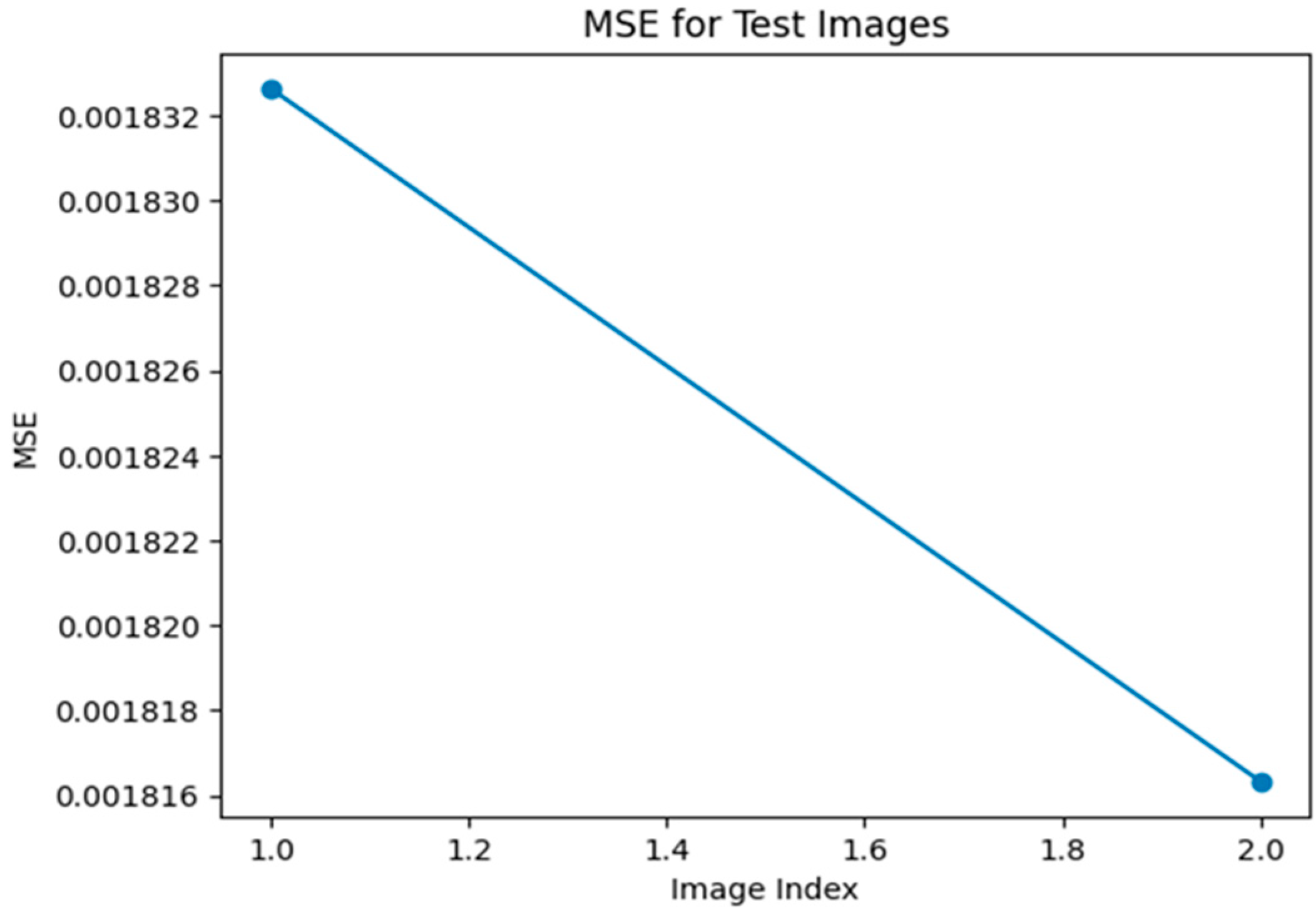
In Figure 7, Figure 8, Figure 9, Figure 10, Figure 11 and Figure 12, we show the performance of the ADAM optimizer and MSE, MAE, and perceptual loss functions, which are used for error computation using a Super Resolution Residual Network (SR ResNet).

Figure 7.
MSE graph for Optimizer—Adam, loss—MSE.

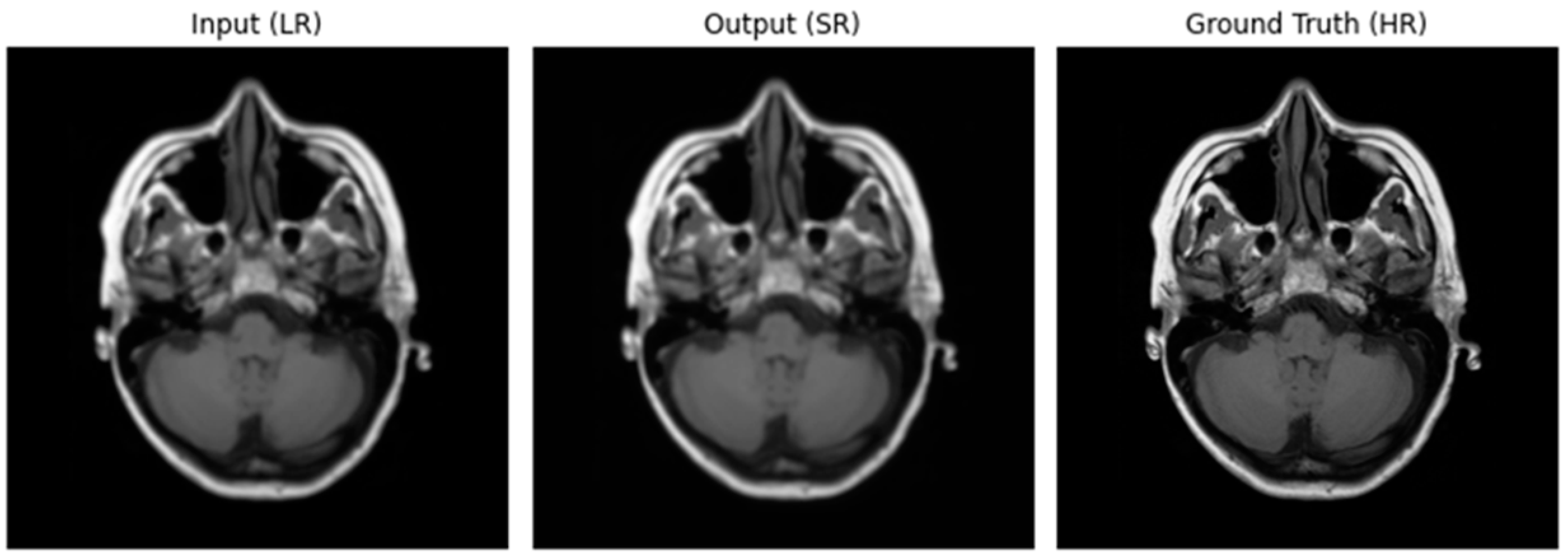
Figure 8.
Image after using Adam Optimizer and MSE loss function.

Figure 9.
MSE graph for Optimizer—Adam, loss—MAE.

Figure 10.
Image using Optimizer as ADAM and loss function as MAE.
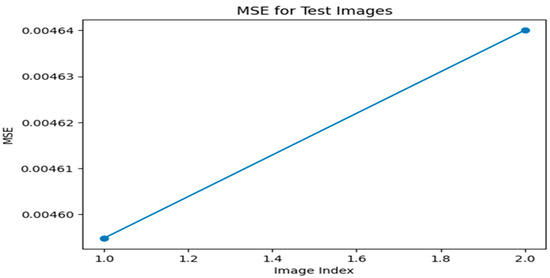
Figure 11.
MSE graph for Optimizer—Adam, loss—Perceptual Loss.

Figure 12.
Image using Optimizer as ADAM and loss function as Perceptual Loss.
3.4. Comparative Analysis Table of SRCNN and SR ResNet with ADAM Optimizer and Loss Functions
In Table 1 and Table 2, the comparative analysis of SRCNN and SR ResNet with the ADAM optimizer and loss functions are shown below:

Table 1.
SRCNN (ADAM optimizer and loss functions).

Table 2.
SR ResNet (ADAM optimizer and loss functions).
4. Discussion
The study uses the ADAM optimizer to improve MRI image quality using SRCNN and SR ResNet. SR ResNet achieves higher PSNR and SSIM values, while SRCNN produces super-resolved images with higher accuracy and less noise. The study focuses on improving precision, clarity, and dependability in medical imaging, and addressing limitations like dataset representativeness and generalization concerns. Reliable evaluation methods and clinical validity are crucial for real-world application.
5. Conclusions
In our study, the optimizer used for super-resolution of input images was consistent for SRCNN and SR ResNet. The motive behind using the ADAM optimizer for both networks was for drawing a relative comparison on a selected set of loss functions. In future research, other optimizers, such as SGD, RMSProp, etc., may be used for the comparison of super-resolution of low-resolution images.
Author Contributions
Conceptualization, M.R. and B.U.; methodology, M.R., J.R. and B.U.; software, J.R.; validation, B.U.; formal analysis, J.R.; investigation, J.R.; resources, M.R.; data curation, B.U.; writing—original draft preparation, J.R.; writing—review and editing, M.R. and K.S.; visualization, K.S.; supervision, K.S.; project administration, M.R. and K.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Endowment fund/TMA Pai Research Grant, Grant number: 6100/SMIT/R&D/Project/13/2022.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zhang, Z.; Wang, X.; Jung, C. DCSR: Dilated convolutions for single image super-resolution. IEEE Trans. Image Process. 2018, 28, 1625–1635. [Google Scholar] [CrossRef] [PubMed]
- Gayathri, P.; Dhavileswarapu, A.; Ibrahim, S.; Paul, R.; Gupta, R. Exploring the Potential of VGG-16 Architecture for Accurate Brain Tumor Detection Using Deep Learning. J. Comput. Mech. Manag. 2023, 2, 23056. [Google Scholar] [CrossRef]
- Zhang, Y.; Shi, F.; Cheng, J.; Wang, L.; Yap, P.-T.; Shen, D. Longitudinally Guided Super-Resolution of Neonatal Brain Magnetic Resonance Images. IEEE Trans. Cybern. 2019, 49, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Thurnhofer-Hemsi, K.; López-Rubio, E.; Roé-Vellvé, N.; Domínguez, E.; Molina-Cabello, M.A. Super-resolution of 3D Magnetic Resonance Images by Random Shifting and Convolutional Neural Networks. In Proceedings of the 2018 International Joint Conference on Neural Networks (IJCNN), Rio de Janeiro, Brazil, 8–13 July 2018; pp. 1–8. [Google Scholar] [CrossRef]
- Sindel, A.; Breininger, K.; Käßer, J.; Hess, A.; Maier, A.; Köhler, T. Learning from a Handful Volumes: MRI Resolution Enhancement with Volumetric Super-Resolution Forests. In Proceedings of the 2018 25th IEEE International Conference on Image Processing (ICIP), Athens, Greece, 7–10 October 2018; pp. 1453–1457. [Google Scholar] [CrossRef]
- Irina, S.; Vilaplana, V. Brain MRI super-resolution using 3D generative adversarial networks. arXiv 2018, arXiv:1812.11440. [Google Scholar]
- Chen, Y.; Xie, Y.; Zhou, Z.; Shi, F.; Christodoulou, A.G.; Li, D. Brain MRI super resolution using 3D deep densely connected neural networks. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018. [Google Scholar]
- Du, J.; Wang, L.; Gholipour, A.; He, Z.; Jia, Y. Accelerated Super-resolution MR Image Reconstruction via a 3D Densely Connected Deep Convolutional Neural Network. In Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Madrid, Spain, 3–6 December 2018; pp. 349–355. [Google Scholar] [CrossRef]
- Mzoughi, H.; Njeh, I.; Slima, M.B.; Hamida, A.B. Histogram equalization-based techniques for contrast enhancement of MRI brain Glioma tumor images: Comparative study. In Proceedings of the 2018 4th International Conference on Advanced Technologies for Signal and Image Processing (ATSIP), Sousse, Tunisia, 21–24 March 2018; pp. 1–6. [Google Scholar] [CrossRef]
- Chen, Y.; Shi, F.; Christodoulou, A.G.; Xie, Y.; Zhou, Z.; Li, D. Efficient and accurate MRI super-resolution using a generative adversarial network and 3D multi-level densely connected network. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, 16–20 September 2018; Proceedings, Part I. Springer International Publishing: Cham, Switzerland, 2018. [Google Scholar]
- Zheng, H.; Zeng, K.; Guo, D.; Ying, J.; Yang, Y.; Peng, X.; Huang, F.; Chen, Z.; Qu, X. Multi-Contrast Brain MRI Image Super-Resolution with Gradient-Guided Edge Enhancement. IEEE Access 2018, 6, 57856–57867. [Google Scholar] [CrossRef]
- Li, Y.; Song, B.; Guo, J.; Du, X.; Guizani, M. Super-Resolution of Brain MRI Images Using Overcomplete Dictionaries and Nonlocal Similarity. IEEE Access 2019, 7, 25897–25907. [Google Scholar] [CrossRef]
- Li, Z.; Yu, J.; Wang, Y.; Zhou, H.; Yang, H.; Qiao, Z. DeepVolume: Brain Structure and Spatial Connection-Aware Network for Brain MRI Super-Resolution. IEEE Trans. Cybern. 2021, 51, 3441–3454. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; He, Y. Gradient-Guided Convolutional Neural Network for MRI Image Super-Resolution. Appl. Sci. 2019, 9, 4874. [Google Scholar] [CrossRef]
- Liang, Z.; Zhang, J. Mouse brain MR super-resolution using a deep learning network trained with optical imaging data. Front. Radiol. 2023, 3, 1155866. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Zhang, X.; Tian, Y.; Wang, W.; Xue, J.-H. Deep learning for single image super-resolution: A brief review. IEEE Trans. Multimed. 2019, 21, 3106–3121. [Google Scholar] [CrossRef]
- Liu, C.; Sun, X.; Chen, C.; Rosin, P.L.; Yan, Y.; Jin, L.; Peng, X. Multi-Scale Residual Hierarchical Dense Networks for Single Image Super-Resolution. IEEE Access 2019, 7, 60572–60583. [Google Scholar] [CrossRef]
- Shende, P.; Pawar, M.; Kakde, S. A Brief Review on: MRI Images Reconstruction using GAN. In Proceedings of the 2019 International Conference on Communication and Signal Processing (ICCSP), Chennai, India, 4–6 April 2019; pp. 139–142. [Google Scholar] [CrossRef]
- Zhu, J.; Yang, G.; Lio, P. How can we make GAN perform better in single medical image super-resolution? A lesion focused multi-scale approach. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019. [Google Scholar]
- Zhao, X.; Zhang, Y.; Qin, Y.; Wang, Q.; Zhang, T.; Li, T. Single MR image super-resolution via channel splitting and serial fusion network. Knowl. -Based Syst. 2022, 246, 108669. [Google Scholar] [CrossRef]
- Du, J.; Wang, L.; Liu, Y.; Zhou, Z.; He, Z.; Jia, Y. Brain MRI Super-Resolution Using 3D Dilated Convolutional Encoder–Decoder Network. IEEE Access 2020, 8, 18938–18950. [Google Scholar] [CrossRef]
- Mane, V.; Jadhav, S.; Lal, P. Image super-resolution for MRI images using 3D faster super-resolution convolutional neural network architecture. In Proceedings of the International Conference on Automation, Computing and Communication 2020 (ICACC 2020), Navi Mumbai, India, 27–28 June 2020. [Google Scholar]
- Tan, C.; Zhu, J.; Lio, P. Arbitrary scale super-resolution for brain MRI images. In Proceedings of the Artificial Intelligence Applications and Innovations: 16th IFIP WG 12.5 International Conference, AIAI 2020, Neos Marmaras, Greece, 5–7 June 2020; Proceedings, Part I. Springer International Publishing: Cham, Switzerland, 2020. [Google Scholar]
- Wang, L.; Du, J.; Zhu, H.; He, Z.; Jia, Y. Brain MR Image Super-resolution using 3D Feature Attention Network. In Proceedings of the 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Seoul, Republic of Korea, 16–19 December 2020; pp. 1151–1155. [Google Scholar]
- Zhang, H.; Shinomiya, Y.; Yoshida, S. 3D MRI Reconstruction Based on 2D Generative Adversarial Network Super-Resolution. Sensors 2021, 21, 2978. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).