Abstract
A brain tumor diagnosis is a complex and difficult task that requires accurate and efficient data analysis. In past years, deep learning has emerged as a promising tool for improving the accuracy of mental health diagnoses. This research article presents a review of various in-depth studies and models for mental health diagnosis and examines the performance of convolutional neural networks (CNNs), VGG16, and other deep learning models on multistate data in the brain. The results show that deep learning models can provide high accuracy and efficiency in brain tumor detection beyond imaging techniques to also discuss the clinical applications of these models, including assisting radiologists in brain diagnosis and improving patient outcomes. Overall, this work raises awareness of deep learning’s application in medicine and offers insights into the future of brain tumor research.
1. Introduction
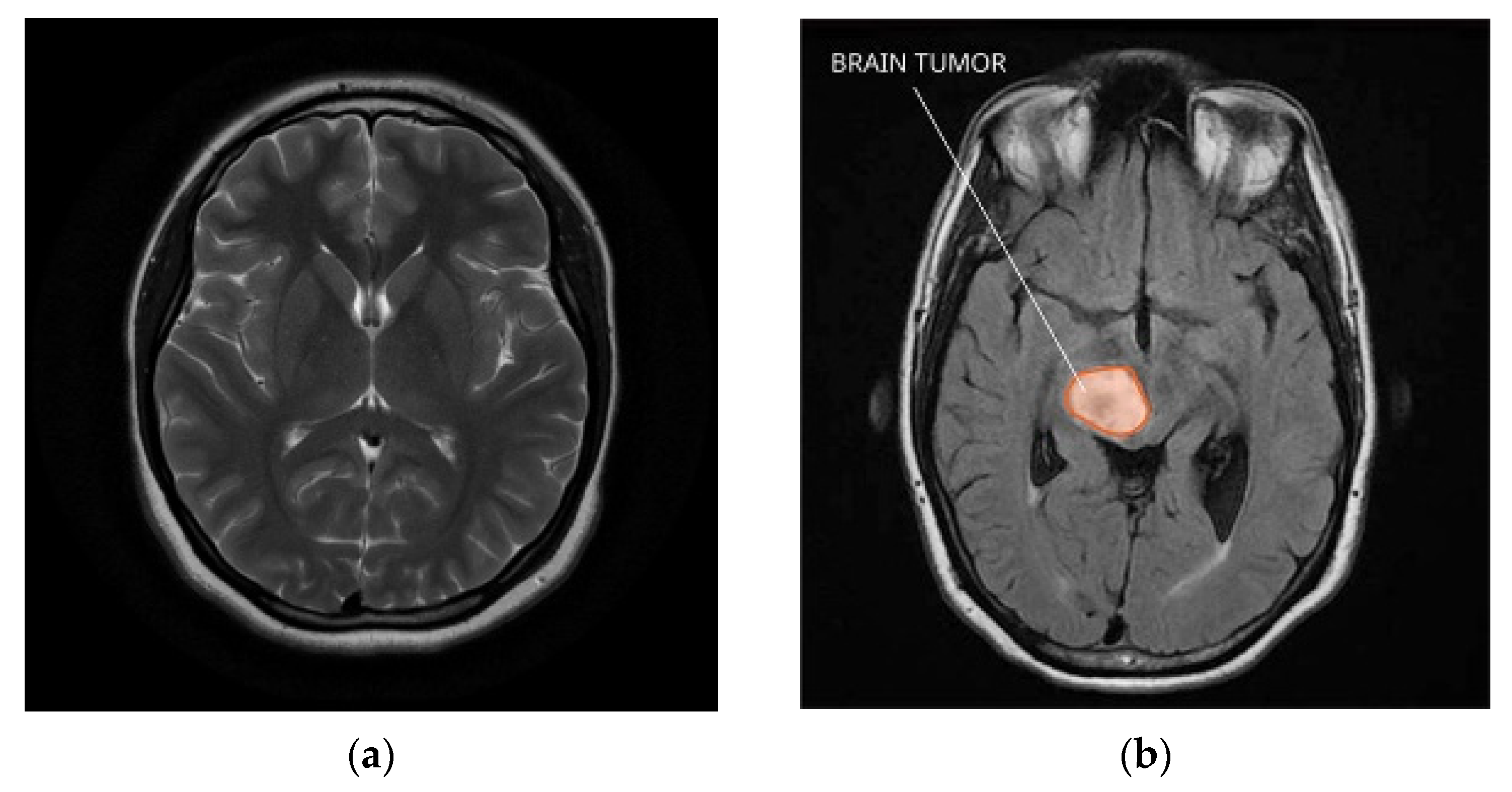
Brain tumors are among the most challenging conditions to identify and manage. They are hard to find with traditional imaging methods because of their differences in size, shape, and position. On the other hand, current developments in deep learning methods show promise for raising the precision and effectiveness of brain tumor diagnosis [1]. The technique of deep learning, a kind of machine learning, uses neural networks to model and solve complex problems. With the help of characteristics and patterns that may be utilized to create predictions or classifications, these networks are made to learn from massive datasets [2]. In clinical research, deep learning has demonstrated significant promise for increasing diagnosis accuracy and decreasing analysis time. This paper examines the use of deep learning for brain diagnostics in this model, discussing several techniques and models created for this goal, such as VGG16 and convolutional neural networks (CNNs) [3,4]. Additionally, it examines how well these models work with various kinds of neurons and contrasts the outcomes with those of more traditional models. The purpose of this work is to show how deep learning can improve the precision and efficacy of mental health diagnosis [5]. This method intends to break new ground in research and treatment options while also increasing public awareness of the use of AI in medicine. Creating a normal brain and tumor brain MRI dataset for medical research and diagnostic purposes involves several critical steps. Here is a general overview of how you can work on such a dataset for (a) a normal brain and (b) a brain tumor which is depicted in the Figure 1.
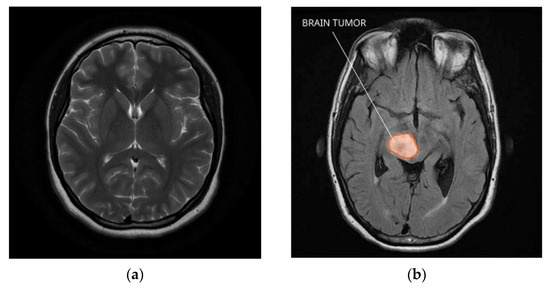
Figure 1.
Normal brain and tumor brain MRI dataset. (a) Normal brain; (b) brain tumor.
2. Literature Survey
In recent years, many in-depth studies have been conducted on the diagnosis of mental illnesses. These techniques fall into three categories: convolutional neural networks (CNNs), VGG16, and hybrid networks. A CNN is a widely used neural network for image classification. A CNN uses layers to learn the features of the image and layers to shrink the rest of the map. CNNs are widely used in brain diagnostics to report a 3D neural network design for MRI image-based brain delineation [6]. Their design is consistent with results from the Brin TS state-of-the-art database, which reported a CNN-based method for brain segmentation in MRI images [2,7]. The method they propose provides higher accuracy than traditional methods, such as fuzzy C-means clustering. The hybrid network is a combination of the CNN and RNN [8]. It has been proposed that these networks use CNNs and RNNs for brain diagnostics. Using a 3D convolutional neural network (CNN), MRI characteristics may be extracted, and scans have 92.7% accuracy [9,10]. A neural network (FCN) takes advantage of the state of the BraTS dataset for brain segmentation [11,12,13,14]. A study using transfection methods used the pre-trained Inception V3 network and adjusted it to data from MRI scans, achieving 96.05% accuracy [6,12]. Another method for the two-step deep learning-based method for brain diagnosis is MRI scans [13,15]. The author used an FCN to classify tumor sites and a CNN to classify tumors [16]. The author achieved 87.38% accuracy and 91% accuracy in tumor segmentation, and 32% was used for tumor classification using an SVM classifier for brain diagnosis with features extracted from MRI scans with an accuracy of 88.3% [1,6,17]. The classification of glioma grade using RF was performed for up to 80.2% of the actual value [18,19]. However, these algorithms require manual processing, which can take a long time and cannot capture complex images. Genetic Algorithms (GAs) can be used to search for key sites for the selection of compounds. A GA gives better results than PCA but there are some minor problems, like not finding the global optimum and solving different problems to find the solution [19,20]. In this case, PSO is applied and used for selection. A Metaheuristic, Bionic Search Technique is the best PSO [7]. The authors classified oral cancer using PSO, Bayesian LDA, and colony optimization (ACO) [21]. They displayed the variations in fuzzy C-means classification accuracy between the GA and PSO for brain tumor detection.
3. Proposed System
- Image Processing: There are many imaging techniques that use the brain, including Magnetic Resonance Imaging (MRI), Computed Tomography (CT), and Positron Emission Tomography (PET). Each model has its own strengths and limitations, and the choice of model will depend on the specific issue and capability. MRI is the most widely used for brain imaging. It has a high resolution and similar texture, making it ideal for determining the location and size of a tumor. MRI can also provide information about the blood supply to the tumor and the presence of enemas (swelling). MRI is noninvasive and contains no ionizing radiation, making it safe and versatile. However, MRI can be expensive and time-consuming, and patients may feel claustrophobic or uncomfortable during the scan. CT is another method that can be used to image brain tumors. It uses X-rays to create detailed images of the brain that can be used to determine the location and size of tumors. CT is faster, cheaper, and sometimes more effective than MRI. However, CT contains ionizing radiation, which can be dangerous to patients, and provides less contrast between soft tissues than MRI. PET is a technique that can be used to identify areas of the brain with increased metabolic activity that may indicate the presence of cancer. PET scans are often used with CT or MRI scans to give more details regarding the tumor’s dimensions and position. PET scans involve injections of antibodies that can be harmful to patients and are more expensive and more common than MRI or CT. For in-depth investigations, MRI is often preferred because of its high resolution and tissue homogeneity. MRI images can be used to create 3D volumes of the brain. They can be applied to deep learning model instructional design and evaluation. CT and PET images can also be used for in-depth investigations but may require additional pre-processing steps to improve image quality and reduce noise. In general, the choice of modality depends on the specific questions and available resources, but MRI is generally considered the gold standard for neuroimaging.
- Pre-Processing: The methods used to obtain digital images ready for examination or additional processing are referred to as image pre-processing. Enhancing the clarity or quality of the image and making it simpler to retrieve valuable information from it are the two main objectives of image pre-processing.
- Feature Extraction and Selection: Selecting a portion of the most significant characteristics from a dataset’s larger collection of features is known as feature selection. This is usually accomplished by selecting the characteristics that are most informative for the model after assessing each feature’s correlation or significance with respect to the target variable. Contrarily, feature reduction entails condensing the initial collection of features into a new set that is smaller and yet preserves the majority of the original features’ content.
- Image Classification: In computer vision, classifying images is a frequent activity that entails labeling a picture according to its visual information. Image classification aims to create a model that can correctly recognize the objects or scenes that are seen in an image and give the image the appropriate label or labels.
- VGG16: With regard to picture classification, object detection, and segmentation, among other computer operations, this deep learning system has demonstrated state-of-the-art performance. Thirteen convolutional layers and three complete layers make up the sixteen layers of the VGG16 architecture. There are fixed-size 3 × 3 filters on convolution layers and fixed-size 2 × 2 filters on pooling layers. Each convolutional layer has twice as many filters as the previous layer, with the first layer having 64 filters. The output layer features a SoftMax function for categorization, and each layer includes 4096 units. The VGG16 architecture’s simplicity and consistency are among its key characteristics. Using fixed filters and filters in each layer, this model may be readily modified for various needs. Smaller-sized filters can also aid in maintaining performance.
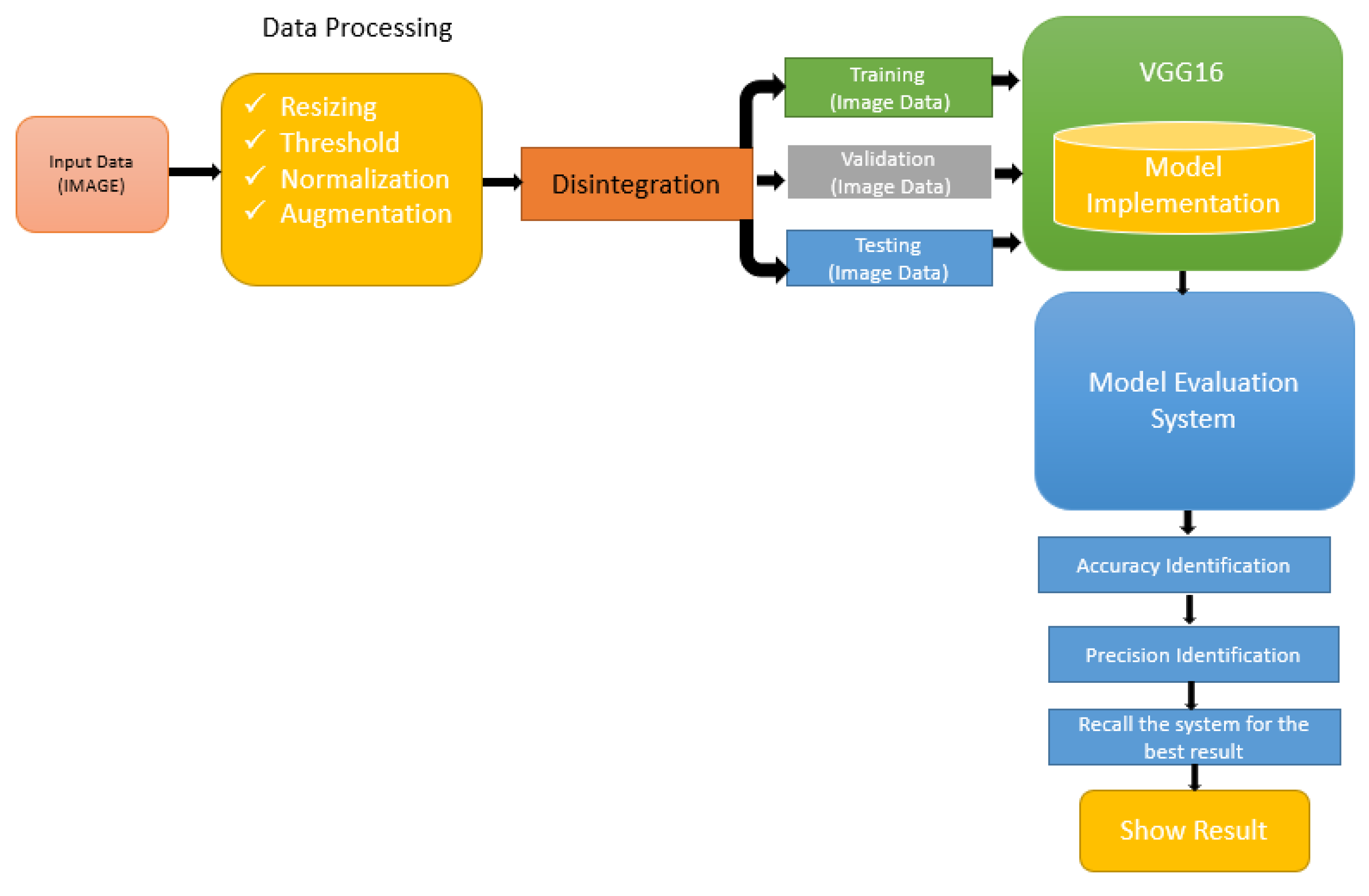
Figure 2 represents the proposed framework for the VGG166 method.
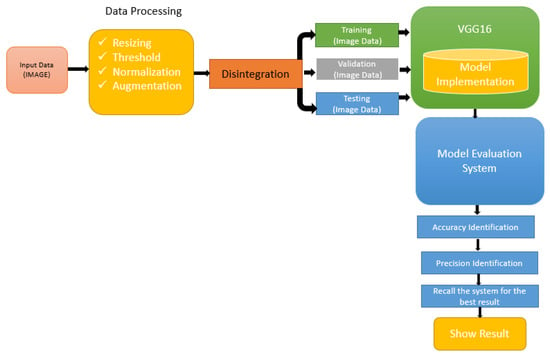
Figure 2.
Medical image processing system for the model evaluation system.
4. Result
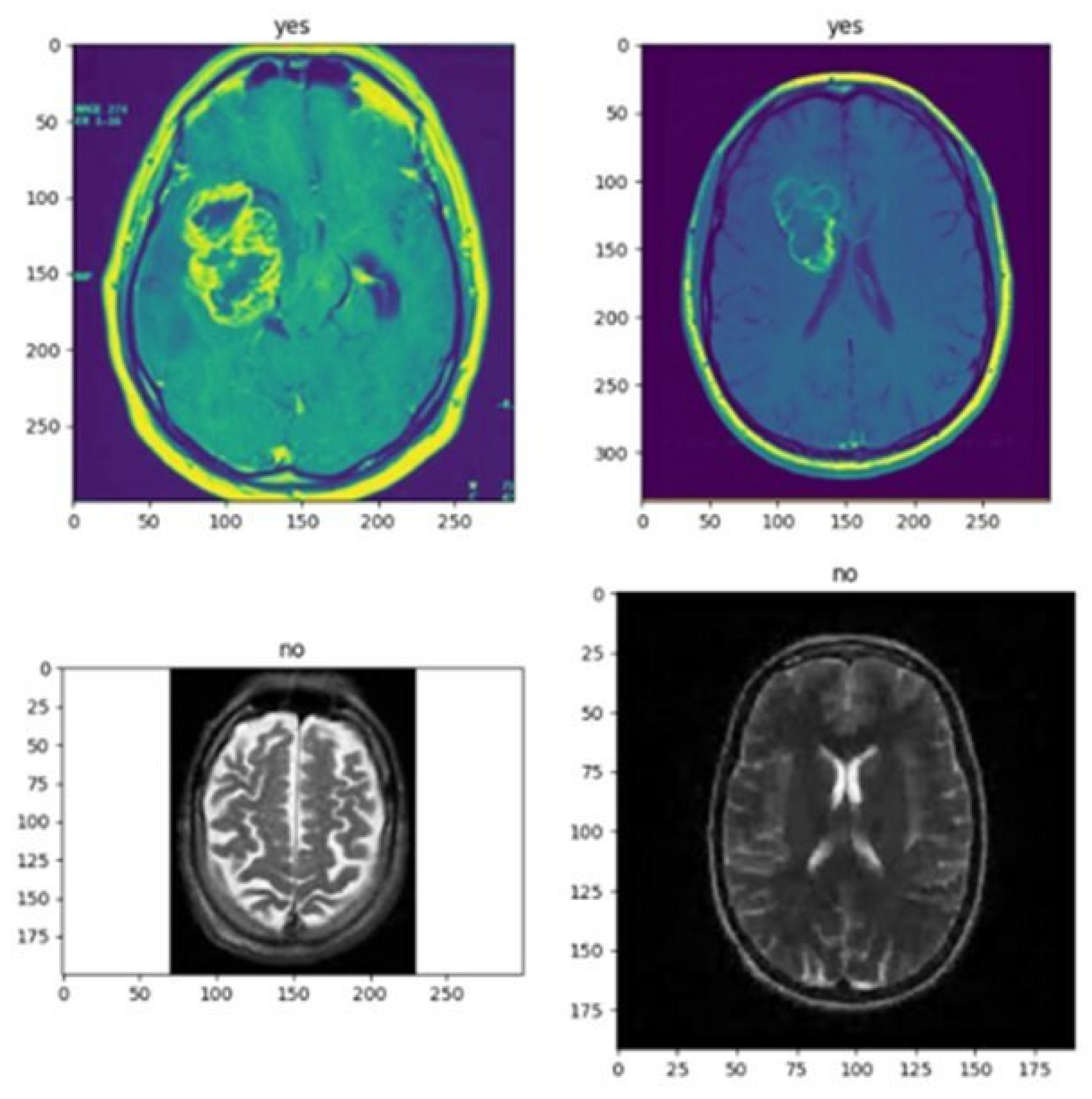
I obtained the following search results after using the correct code. I see that VGG-16 can detect brain tumors easily, whereas CNN cannot detect brain tumors accurately, so clinical-level results are unreliable. Figure 3 represents the effective result using VGG166 approaches.
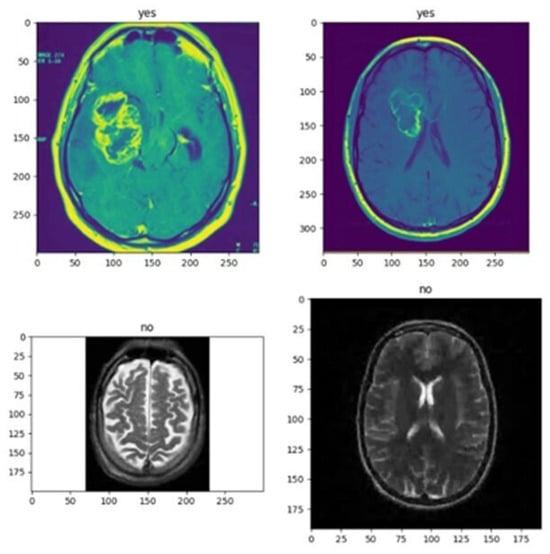
Figure 3.
This figure shows the effective result using the VGG16 method.
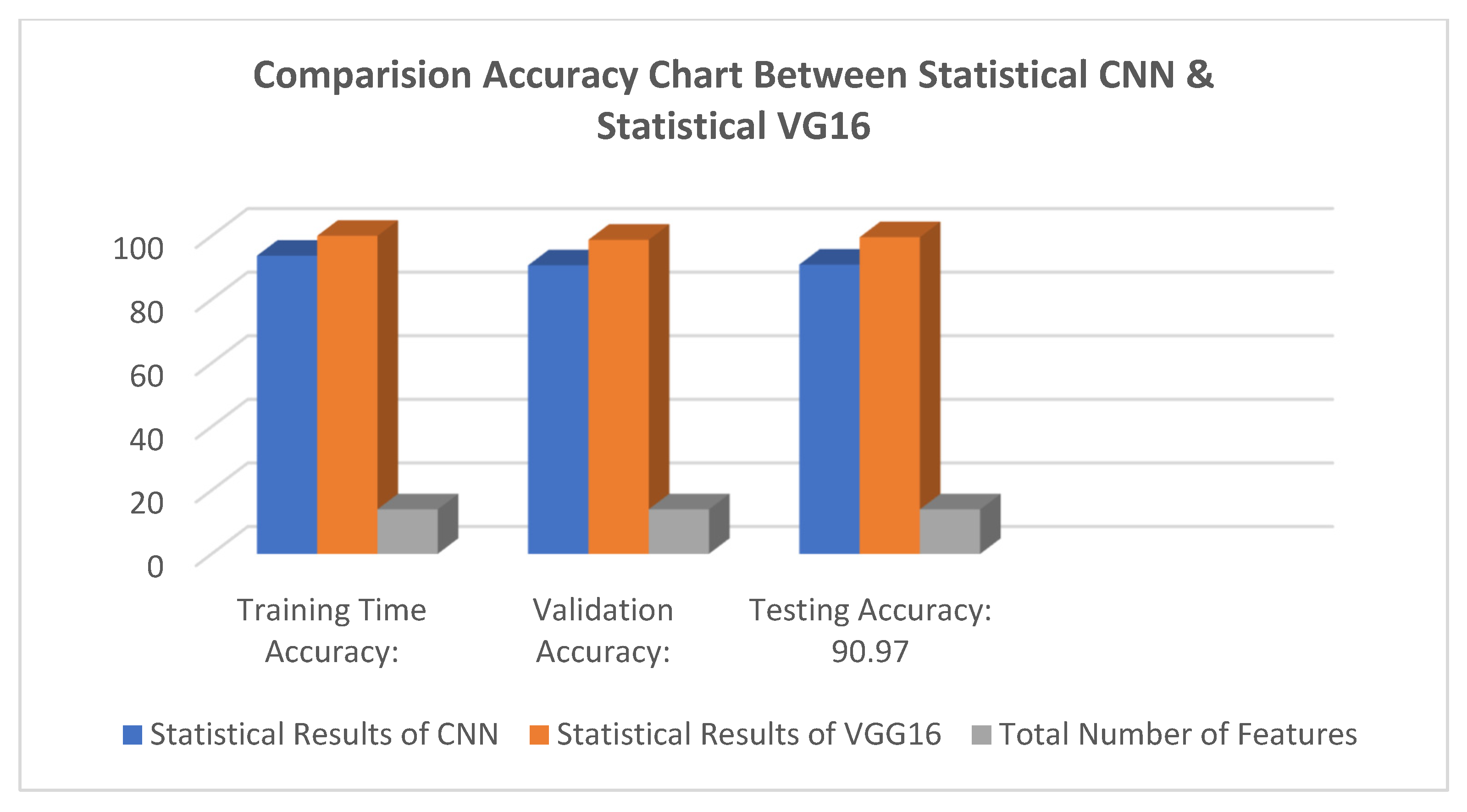
Below are the results of the CNN and VGG16 methods I used to make the decision. This includes the loss and accuracy of the model, which can be very helpful in deciding which model to choose for different problems. The statistical result shows the accuracy of the model and Figure 4 and Figure 5 Show the accuracy result between the CNN feature and value points and the VGG16 feature and value points where Table 1 shows the statistical result accuracy summary of the CNN.
- A.
- Statistical Results of CNN
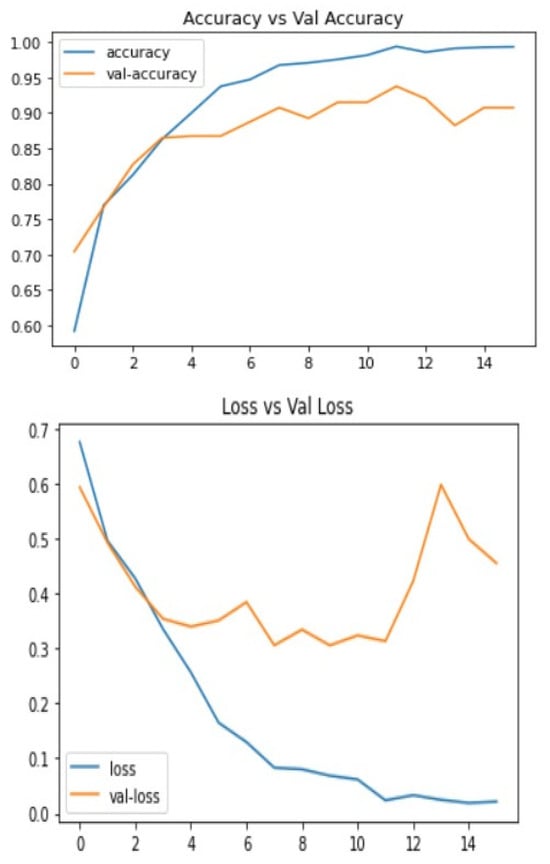 Figure 4. Statistical result of CNN features and value points.Figure 4. Statistical result of CNN features and value points.
Figure 4. Statistical result of CNN features and value points.Figure 4. Statistical result of CNN features and value points.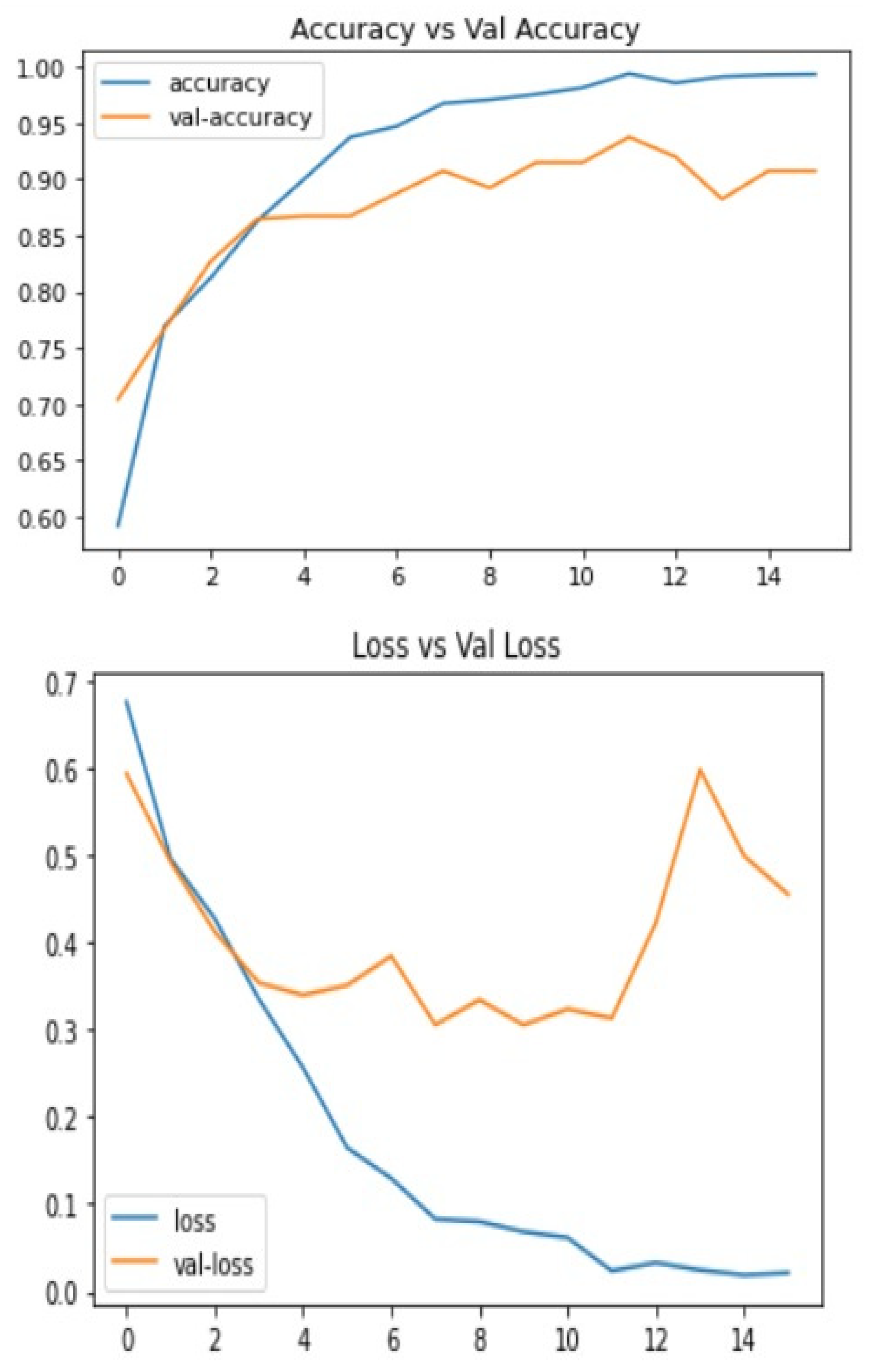
 Table 1. Statistical result accuracy summary of the CNN.Table 1. Statistical result accuracy summary of the CNN.
Table 1. Statistical result accuracy summary of the CNN.Table 1. Statistical result accuracy summary of the CNN.Accuracy Identification Feature Rates Total Number of Features 14 Training Time Accuracy 93.73 Validation Accuracy 90.73 Testing Accuracy 90.97
- B.
- Statistical Results of VGG16
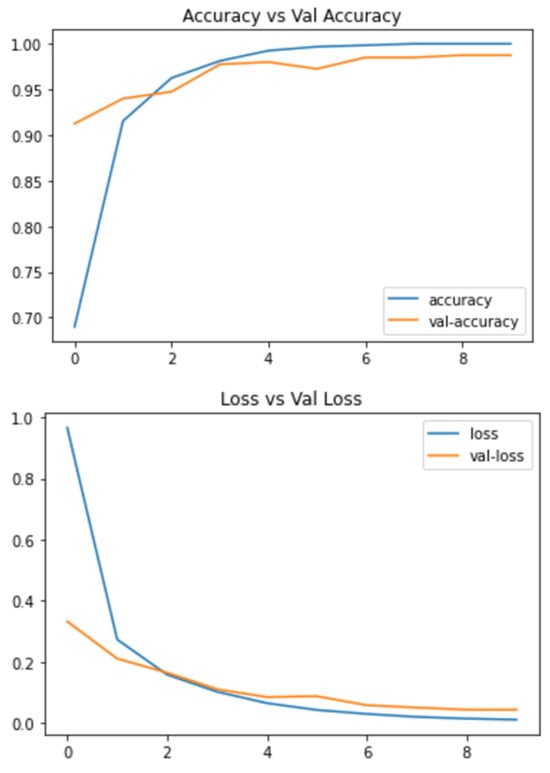 Figure 5. Statistical result of VGG16 features and value points.Figure 5. Statistical result of VGG16 features and value points.
Figure 5. Statistical result of VGG16 features and value points.Figure 5. Statistical result of VGG16 features and value points.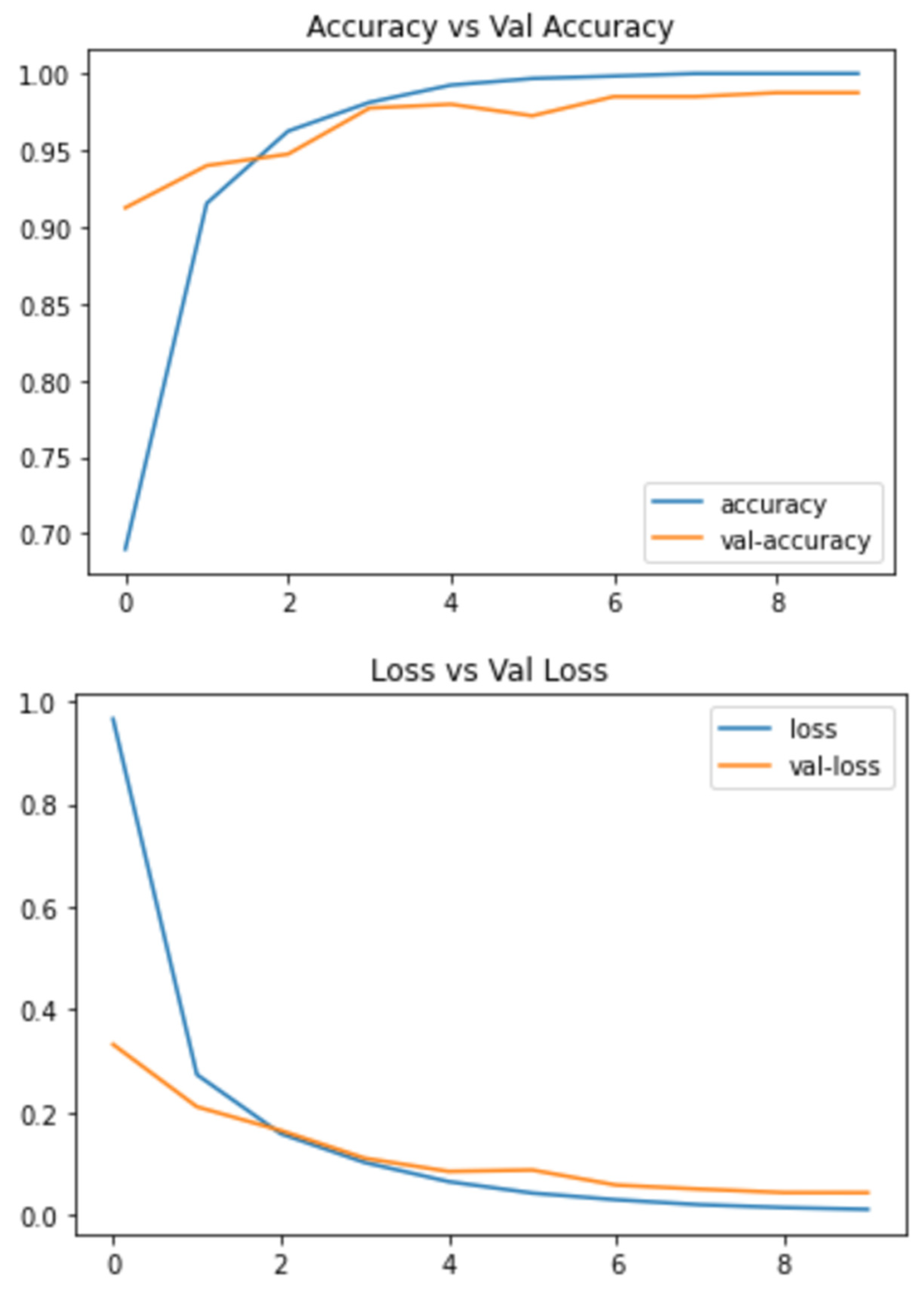
Figure 5 shows the results feature of VGG16 and Figure 6 represents the comparison result between the CNN and VGG16 approaches. Table 2 lists the number of features correctness of the training period, the test accuracy, and the accuracy of the validation verification, along with certain feature part assumptions for this study.
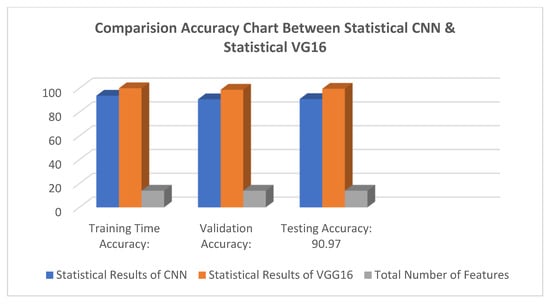
Figure 6.
Comparison result between CNN and VGG16 methods.

Table 2.
Statistical result accuracy summary of VGG16.
5. Conclusions and Future Work
In conclusion, the use of CNNs and VGG16 for brain cancer diagnosis shows great promise in improving the accuracy and speed of diagnosis. While there are still some challenges and limitations to overcome, the potential benefits are huge. As machine learning continues to evolve, expect to see more models developed that can detect and diagnose brain tumors and ultimately lead to better outcomes for patients. For future work, I would like to discuss the following: using deep learning, improving model accuracy, using image segmentation, and identifying tumors.
Author Contributions
Conceptualization, S.R. and S.D.; methodology, S.R.; software, S.R. and S.D.; validation, S.R. and S.D.; formal analysis, S.D.; investigation, S.R. and S.D.; writing—original draft preparation; writing—review and editing, S.R.; visualization, S.R. and S.D.; supervision, S.D.; project administration, S.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained with the authors and as and when required will be provided.
Acknowledgments
The Department of Information Technology at Manipal University Jaipur is acknowledged by the authors for providing the tools required to carry out this study. I would also like to thank our supervisor, Sumit Dhariwal, for his invaluable support and guidance during the course of the project, and I would like to acknowledge the contribution of doctors’ results available on Kaggle datasets and their expertise in identifying the various images of brain tumor diseases. Lastly, I want to express my gratitude to the families for their unwavering support and encouragement.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kamnitsas, K.; Ledig, C.; Newcombe VF, J.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Rueckert, D.; Glocker, B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2016, 36, 61–78. [Google Scholar] [CrossRef] [PubMed]
- Gayathri, P.; Dhavileswarapu, A.; Ibrahim, S.; Paul, R.; Gupta, R. Exploring the Potential of VGG-16 Architecture for Accurate Brain Tumor Detection Using Deep Learning. J. Comput. Mech. Manag 2023, 2, 23056. [Google Scholar] [CrossRef]
- Li, H.; Chen, H.; Qi, X.; Dou, Q.; Fu, C.W.; Heng, P.A. H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans. Med. Imaging 2018, 37, 2663–2674. [Google Scholar] [CrossRef] [PubMed]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C. Brain tumor segmentation with deep neural networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Han, X. MR-based synthetic CT generation using a deep convolutional neural network method. Med. Phys. 2017, 44, 1408–1419. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ding, C.; Li, L.; Zhu, X.; Wang, J.; Tang, Y. 3D CNN-based automatic diagnosis of brain tumor using multi-modality MR images. Comput. Biol. Med. 2019, 109, 218–226. [Google Scholar]
- Liu, F.; Zhou, Z.; Jang, H.; Samsonov, A.; Zhao, G.; Kijowski, R. Deep learning approach for evaluating knee MR images: Achieving high diagnostic performance for cartilage lesion detection. Radiology 2018, 289, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Y.; Yuan, Y.; Yue, X.; Han, Z. Deep learning-based segmentation and classification of brain tumor and stroke lesions in MRI. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 1747–1758. [Google Scholar]
- Kaur, H.; Gupta, R.; Kumar, A. A review of recent advances in deep learning for brain tumor detection using MRI. Artif. Intell. Med. 2019, 98, 43–54. [Google Scholar]
- Dhariwal, S.; Raghuwanshi, S.; Shrivastava, S. Content-Based Image Retrieval Using Normalization of Vector Approach to SVM. In Advances in Computer Science, Engineering & Applications, Proceedings of the Second International Conference on Computer Science, Engineering & Applications (ICCSEA 2012), Delhi, India, 25–27 May 2012; Springer: Berlin/Heidelberg, Germany, 2012; Volume 2, pp. 793–801. [Google Scholar]
- Dhariwal, S.; Palaniappan, S. Image Normalization and Weighted Classification Using an Efficient Approach for SVM Classifiers. Int. J. Image Graph. 2020, 20, 2050035. [Google Scholar] [CrossRef]
- Dhariwal, S.; Rawat, R.; Patearia, N. C-Queued Technique against SQL Injection Attack. Int. J. Adv. Res. Comput. Sci. 2011, 2, 461–464. [Google Scholar]
- Kumar, M.S.; Kumarasamy, M.; Madhavi, N.B.; Dhariwal, S.; Kumar, R.S.; Oyebode, O.J. Reinforcement Based Concrete Modelling in Commercial Buildings Using Machine Learning Simulations. Int. J. Intell. Syst. Appl. Eng. 2023, 11, 118–126. [Google Scholar]
- Dhariwal, S.; Raipuria, A. Impact of Green Communication and Technology System. In Emerging Technologies for Computing, Communication and Smart Cities, Proceedings of ETCCS 2021, Punjab, India, 21–22 August 2021; pp. 605–615; Springer Nature: Singapore, 2022. [Google Scholar]
- Nandha, G.N.; Karnan, M. Diagnose brain tumor through MRI using image processing clustering algorithms such as Fuzzy C Means along with intelligent optimization techniques. In Proceedings of the 2010 IEEE International Conference on Computational Intelligence and Computing Research (ICCIC 2010), Coimbatore, India, 28–29 December 2010. [Google Scholar]
- Mohanaiah, P.; Sathyanarayana, P.; GuruKumar, L. Image Texture Feature Extraction Using GLCM Approach. Int. J. Sci. Res. Publ. 2013, 3, 1–5. [Google Scholar]
- Jothi, G.; Hannah Inbarani, H. Hybrid tolerance rough set-firefly based supervised feature selection for mri brain tumor image classification. Appl. Soft Comput. 2016, 46, 639–651. [Google Scholar]
- James, K.; Eberhart, R.C. A discrete binary version of the particle swarm algorithm. In Proceedings of the 1997 IEEE International Conference on Systems, Man, and Cybernetics. Computational Cybernetics and Simulation, Orlando, FL, USA, 12–15 October 1997; Volume 5. [Google Scholar]
- Zhou, J.; Zhong, T.; He, X. Auxiliary Diagnosis of Breast Tumor Based on PNN Classifier Optimized by PCA and PSO Algorithm. In Proceedings of the 2017 9th International Conference on Intelligent Human-Machine Systems and Cybernetics (IHMSC), Hangzhou, China, 26–27 August 2017; Volume 2. [Google Scholar]
- Ahmad, I. Enhancing SVM performance in intrusion detection using optimal feature subset selection based on genetic principal components. Neural Comput. Appl. 2014, 24, 1671–1682. [Google Scholar] [CrossRef]
- Kumar, A.; Ansari, M.A. Performance Evaluation of De-Noised Medical Images After Removing Speckled Noise by Wavelet Transform. IJBET 2021, 36, 318–330. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).